Abstract
We have developed a sequencing method based on the RNA polymerase chain termination reaction with rhodamine dye attached to 3′-deoxynucleoside triphosphate (3′-dNTP). This method enables us to conduct a rapid isothermal sequencing reaction in <30 min, to reduce the amount of template required, and to do PCR direct sequencing without the elimination of primers and 2′-dNTP, which disturbs the Sanger sequencing reaction. An accurate and longer read length was made possible by newly designed four-color dye-3′-dNTPs and mutated RNA polymerase with an improved incorporation rate of 3′-dNTP. This method should be useful for large-scale sequencing in genome projects and clinical diagnosis.
High throughput DNA sequencing is essential technology for genome projects and clinical diagnosis (1). Recent developments for high throughput DNA sequencing include multiple capillary array sequencer, enzyme and fluorescent primer or fluorescent dideoxynucleotide (ddNTP) for sequencing reactions (2–5). The ideal sequencing reaction should be accurate and quick and easy to perform, enabling automation of a large number of reactions.
Currently, cycle sequencing chemistry, employing dye primers and dye terminators, is widely used. Dye-primer chemistry is useful for long-read sequencing due to the uniform incorporation of four types of ddNTP, resulting in an even peak height for each signal. However, this requires four independent reactions and the sequencing pattern is sometimes flawed by false stops at some sites with no incorporation of ddNTPs. On the other hand, dye-terminator chemistry was developed as a one-tube reaction without false stops, but shows various incorporation rates for the four color terminators resulting in failure of long-read sequencing. Recently, “ThermoSequenase”, a newly developed enzyme mutated to make the incorporation uniform, allowed the improvement of long read sequencing (5). However, cycle sequencing has the drawback of a long reaction time (2- to 3-hr reaction) because of its requirement of temperature cycling.
At present, two independent methods for preparing DNA templates are available. One is DNA cloning using a plasmid vector and the other is PCR. Although PCR has limitations on the size or sequence to be amplified, it is very convenient to prepare template(s) directly from plasmids in Escherichia coli and cells from tissues without cloning and library construction (6). From this aspect, PCR allows automation of template preparation because it can amplify very rapidly DNA fragment(s) from a large number of samples. However, in the direct sequencing of PCR products by using Dye-terminator chemistry, unreacted 2′-dNTP and primers must be eliminated to avoid interference with the subsequent sequencing reaction. Although efforts have been made to quickly purify the PCR product such as enzymatic degradation using exonuclease I and shrimp alkaline phosphatase, most protocols are time-consuming, laborious, and expensive (7, 8).
To overcome the above problems, we pursued a completely different approach. Based on the property of promoter-dependent RNAP, we predicted that the chain termination method using this group of enzymes would be useful. First, RNAP does not use primers and cannot incorporate 2′-dNTP, but can incorporate nucleoside triphosphate (NTP) and 3′-dNTP. This would allow direct sequencing without any purification steps. Second, it is much faster to process (240 bases/sec) than Taq polymerase (60 bases/sec), thus reducing the reaction time (9, 10). Third, the turnover of RNAP allows us to amplify the signal without temperature cycling. This reaction does not require a denaturation step to hybridize the sequencing primer to the template. Finally, a large amount of sequencing product can be transcriptionally amplified from a small amount of DNA template. Cunninghum et al. (11) reported that T7 RNAP can produce 600 molecules of in vitro transcripts from 1 molecule of DNA template.
Axelrod and Kramer (12) reported the use of T7 and SP6 RNAPs for chain termination reaction with radioisotope-internal labeling. However, their data showed a variation in peak heights due to variation in the incorporation of 3′-dNTP, in agreement with our unpublished data. Moreover, the chain termination method using the wild-type (wt) RNAP and 3′-dNTP produced many false ladders caused by nonspecific stopping of polymerization. Thus, despite the possibility of applying RNAP for the sequencing technique, there has been no further development due to variation in the incorporation of 3′-dNTP and the lack of any fluorescent substrate for the chain termination reaction.
In this paper, we describe a completely new RNAP-based sequencing method named, “transcriptional sequencing.” For accurate, long-read sequencing, we developed the four-color dye-3′-dNTPs (dye terminators), which carry a long carbon spacer (n = 4), connecting nucleotides and fluorescent (rhodamine) dyes, and mutated T7 RNAPs to improve the uniformity of the incorporation rate of 3′-dNTP. We also purified the RNase-free yeast pyrophosphatase (PPase) to inhibit pyrophospholysis that leads to degradation of specific 3′-dNTP-terminated fragments on transcriptional sequencing, resulting in improvement of peak uniformity. This method made possible a rapid isothermal sequencing reaction in <30 min, based on the high processivity of RNAP and complete prevention of false stops by the newly designed dye-3′-dNTP. PCR direct sequencing using RNAP can overcome the tedious steps of removing primers and 2′-dNTP, thus allowing in great reduction of time and labor to enable preparation of much larger numbers of sequencing samples.
MATERIALS AND METHODS
Synthesis of Rhodamine Dye Attached to 3′dNTP.
The N-hydroxysuccinimidyl ester of 6-carboxytetramethylrhodamine (TMR), 6-carboxy-X-rhodamine (ROX), 5-carboxyrhodamine-6G (R6G), and 5-carboxyrhodamine-110 (R110) were purchased from Molecular Probes. Rhodamine-labeled 3′dNTPs were synthesized according to the method reported by Prober et al. (13) with minor modification.
Construction and Enzyme Purification of Mutant T7 RNA Polymerases (RNAP).
Mutant polymerase genes were constructed by PCR-mediated site direct mutagenesis (14). Mutant enzyme expression and large-scale purification have been described (15).
Enzyme Purification of Yeast Pyrophosphatase.
Baker’s yeast inorganic pyrophosphatase (Sigma) was further purified by liquid chromatography by using SP-Sepharose and Q-Sepharose (Pharmacia).
Template Preparation for Sequencing.
PCR was carried out in a 10-μl volume containing 1 pg of human thyroid-stimulating hormone (TSH) β subunit cDNA inserted into Bluescript II (designated pBS750) (16), 0.1 M forward primer (5′-ACGTTGTAAAACGACGGCCAGT-3′) and reverse primer (5′-TAACAATTTCACAGGAAACA-3′), 200 μM of each 2′-dNTP, and 0.5 units of EXTaq polymerase and EXTaq buffer (Takara Shuzo, Kyoto, Japan). In the case of p53 exon 8 amplification, 100 ng of genomic DNA prepared from human colon carcinoma line HT29 (American Type Culture Collection) or human placenta, 0.1 M T7 promoter appended primer (5′-GTAATACGACTCACTATAGGGCACCTACCTGGAGCTGGAGC-3′), and G2 primer (5′-CCAAGACTTAGTACCTGAAG-3′) were used (17). The first cycle was at 96°C for 1 min, 55°C for 30 sec, and 72°C for 1 min. This was followed by 24 cycles of 96°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min.
Transcriptional Sequencing.
Direct sequencing reactions were carried out in 10 μl containing 10 ng of unpurified PCR products, 40 mM Tris⋅HCl (pH8.0), 8 mM MgCl2, 5 mM DTT, 2 mM spermidine-(HCl)3, 500 μM GTP and UTP, 250 μM ATP and CTP, 0.1 μM R6G-3′-dATP, 0.1 μM R110–3′-dGTP, 1 μM TMR-3′-dUTP, 5 μM ROX-3′-dCTP, 10 units of yeast PPase, and 25 units of wt T7 RNAP or mutant T7 RNAP F644Y. To use 7-deaza-GTP, 500 μM 7-deaza-GTP was substituted for GTP and 1 mM GMP for the initiator. These reaction mixtures were incubated at 37°C for 30 min. Next, the unincorporated dye terminators were separated from the sequencing products with Sephadex G25 (Pharmacia), dried, and suspended with 4 μl of formamide dye (95% formamide, 10 mM EDTA). Samples were heated to 90°C for 2 min and 2 μl was applied to the sequencing gel. The sequencing reaction was run on ABI 377 DNA sequencer and analyzed by using version 2.0 xl software with a semiadaptive basecaller and DT 4% correction file. Incorporation rate ratios of the dye terminator were determined by comparing the average length of sequencing fragments generated using various dye-3′-dNTP concentrations using the ABI 377 gel image. To compare transcriptional sequencing with dye-terminator cycle sequencing with respect to various template amount on sequence accuracy, 10- to 500-ng of plasmid (pBS750) were sequenced by using both methods. Transcriptional sequencing reactions were performed at 37°C for 3 hr and dye-terminator cycle sequencing reactions were performed using the T7 primer according to the manufacturer’s instructions (Amersham). Individual quality of the sequences was assessed by calculating the percentage of correct base calls within 100-base long sections from the first signal in transcriptional sequencing.
RESULTS
Fluorescent Dye Terminator.
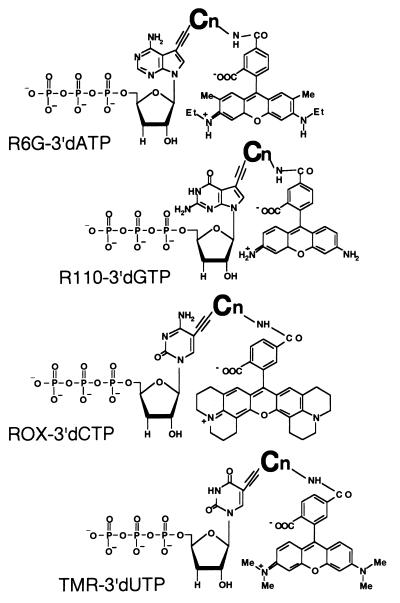
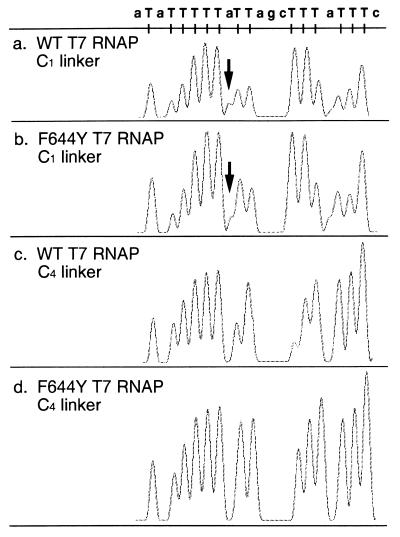
To subject the sequencing product to an automated sequencer, we synthesized the four color fluorescent dye terminators shown in Fig. 1. These four color dye-3′-dNTPs, TMR, ROX, R6G, and R110, are connected to 3′-dUTP, 3′-dCTP, 3′-dATP, and 3′-dGTP to function as terminators of the RNAP sequencing reaction. In the first trial, we synthesized four dye-3′-dNTPs with one carbon chains (C1) as spacers and assayed all of them to examine the incorporation rate. These four dye terminators showed various incorporation rates. 3′-dATP (C1) and 3′-dCTP (C1) especially were poorly incorporated by wt T7 RNAP (data not shown). We then connected three of them, TMR, R6G, and R110, to 3′-dCTP. However, none of these was incorporated efficiently, indicating that the poor incorporation rate is not due to the combination of dyes with 3′-dNTP. If the incorporation rate is due to steric hindrance caused by the short distance between 3′-dNTP and the fluorescent dye, changing the length of the spacer should change the incorporation rate. We synthesized 3′-dUTP and 3′-dCTP carrying 3-, 4-, and 6-carbon chains as spacers (C3, C4, and C6). Wild-type RNAP and mutated RNAP (F644Y; described below) were used for the assay. C1 and C3 did not cause any improvement in the incorporation rate (data not shown); however, C4 and C6 were very effective. Fig. 2 shows the result in the case of 3′-dUTP with C4. The overall effect of C4 was an improvement in the electropherogram with the disappearance of false peaks (see arrow) and an increase in the incorporation rate of these terminators, as can be seen from the peak uniformity (compare Fig. 2a with 2c, and Fig. 2b with 2d). C6 also showed a similar improvement. These data suggest that the distance of ≈6 Å obtained with C4 with a carbon connection was enough to separate the bulky rhodamine dye residues from bases.
Figure 1.
Structures of four rhodamine-labeled 3′-deoxynucleoside 5′-triphosphates. Cn indicates the number of CH2 in the linker arm between rhodamine dye and base of 3′dNTP.
Figure 2.
Effects of linker length and mutated T7 RNAP in the termination pattern with TMR-3′-dUTP. Sequencing reactions were performed with human thyroid-stimulating hormone β-subunit unpurified PCR products for the template. Wild-type or mutated F644Y T7 RNAP for enzyme, C1 or C4 linker arm for dye-3′dNTP were tested, respectively. PPase was added to all reactions. All peaks are shown on the same scale. (a) Combination of C1 linker and wt T7 RNAP. (b) Combination of C1 linker and mutant T7 RNAP. (c) Combination of C4 linker and wt T7 RNAP. (d) Combination of C4 linker and mutant T7 RNAP. Arrows indicate false peaks.
Development of Mutant T7 RNAP.
For further improvement of the uniformity of incorporation in each dye-3′-dNTP, T7 RNAP was mutated. The site of the amino acid residue involved in the recognition of 3′-dNTP is influenced by the following factors. First, three-dimensional crystallography of T7 RNAP predicts the putative NTP binding domain (18). The hydrogen in amino acid has been reported to play an important role as the site recognizing hydroxyl groups at 3′ of ribose in the nucleotide (5). Phenylalanine was chosen as a replacement residue because it can be converted to tyrosine by changing hydrogen in the side chain to hydroxyl residue without changing the main chain conformation. From the three-dimensional crystallography data, the recognition site(s) could be estimated to be in “Helix Y(625–634)”, “Helix Z(649–658)”, and the vicinity of both. Based on these findings, we introduced the mutation from phenylalanine to tyrosine at F644Y, F646Y, and F667Y. All of these mutated T7 RNAPs were examined for their processivity and the incorporation rate of 3′-dNTP using eight types of dye-3′-dNTPs, such as TMR-3′-dUTP (n = 1, 3, 4, and 6) and ROX-3′dCTP (n = 1, 3, 4, and 6). As shown in Fig. 2, F644Y and F667Y were among three mutated T7 RNAPs that showed a severalfold increase in incorporation of 3′-dUTP (data not shown). Both mutants increased the height of the peak and also reduced the false peak (Fig. 2d). This improvement of incorporation was confirmed for the remaining three nucleotides (Fig. 1). Also in the case of mutated T7 RNAP, a carbon spacer longer than four that connected the nucleotide and the fluorescent dye was confirmed to be very effective for preventing the steric hindrance of the dye. Thus, the high efficiency of incorporation of four dye-3′-dNTPs by developing the mutated T7 RNAP and the dye-3′-dNTPs with the C4 spacer allowed reduction of the total amount of the four dye terminators to decrease greatly the background and enable long reading of the DNA sequence.
Modification of Transcriptional Sequencing.
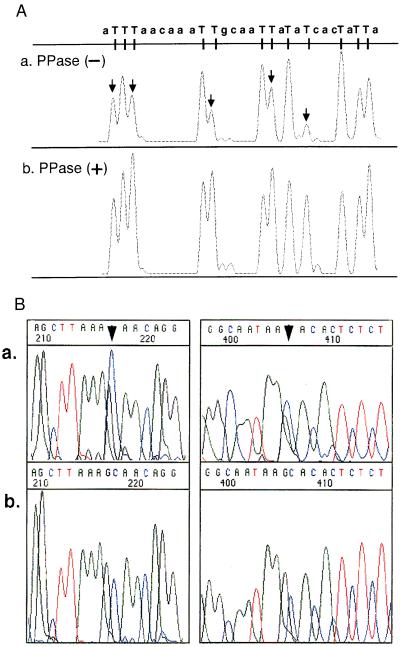
In the Sanger sequencing reaction, pyrophosphorolysis has been reported to cause degradation of specific ddNTP-terminated fragments to produce ddNTP from inorganic pyrophosphate and dideoxymononucleotide, as the reverse reaction of DNA polymerization (19). This reverse reaction can be seen also in transcriptional sequencing, resulting in impairment of the uniformity and sharpness of the peaks of the electropherogram. The impairment can be prevented by PPase in a DNA polymerase-based system. In the light of the above findings, we expected that PPase could improve further the sequencing pattern of transcriptional sequencing, and therefore, we purified the RNase-free yeast PPase. As can be seen from Fig. 3A, the shape of the peak of electropherogram was improved greatly and was more uniform, indicating that PPase is very effective for transcriptional sequencing (see Fig. 3A, arrow).
Figure 3.
(A) Improvement of peak uniformity by use of PPase. Sequencing reactions were performed in the absence or presence of PPase. Minus or plus indicates the absence or presence of PPase. All peaks indicating termination patterns with TMR-3′-dUTP are shown on the same scale. The arrows indicate sites relatively sensitive to pyrophosphorolysis a. Peak degradations in sensitive sites were prevented by PPase b. (B) Elimination of peak compression by use of 7-deaza-GTP in place of GTP. The arrows indicate two compression sites in human TSH cDNA using GTP as the substrate a. Compressions in the corresponding sequence were eliminated by use of 7-deaza-GTP for the substrate b.
In the DNA polymerase-based reaction, several types of helix-base destabilizing nucleotide analogs, such as 7-deaza-2′-dGTP and deoxyinosine triphosphate, have been used to solve the problem of peak compression (20, 21). Peak compression is considered to be more severe in transcriptional sequencing because single strand RNA has a larger repertoire of secondary structures because it has a higher Tm value than DNA (22). For additional improvement, we employed 7-deaza-GTP in place of GTP as substrates. The resulting sequencing pattern is shown in Fig. 3B. As indicated by the arrowhead, the addition of this analog eliminated the compression. Thus, impairment of the sequencing pattern because of gel compression could be further improved by employing the nucleotide analog 7-deaza-GTP.
Dual End Sequencing.
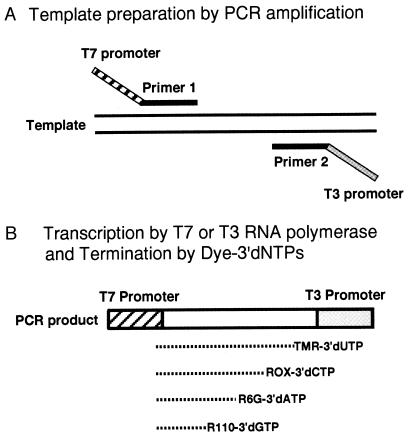
We developed a mutated hybrid T7/T3 RNAP that could recognize the T3 RNAP promoter. This hybrid T3 RNAP was designed to replace the T7 RNAP sequence with that of T3 RNAP at the 674–752 amino acid residue that recognizes the promoter sequence (23). The remaining region of the hybrid T7/T3 RNAP was identical to the sequence of T7 RNAP F644Y. Using T3 and T7 RNAPs, the insert DNA could be sequenced from both ends when the T7 and T3 promoters were designed at both flanking regions (data not shown). The principle is shown in Fig. 4.
Figure 4.
Schematic representation of direct sequencing by transcriptional sequencing. (A) Thin lines are double strand templates whereas thick lines are primers. The phage promoter sequence can be appended to one or both of the PCR primers and incorporated into the PCR product. Alternatively, because most cloning vectors contain two different opposite-oriented phage promoters flanking a multiple cloning site, an inserted DNA can be amplified by using flanking primers and transcribed directly. (B) The remaining primers and 2′-dNTPs need not be removed from PCR products prior to sequencing because the transcription reaction is independent of residual primers and dNTPs. RNA fragments are transcribed under a phage promoter from unpurified PCR reactant and terminated by four kinds of dye-3′dNTPs. Sequencing products are purified and subjected to electrophoresis on a DNA sequencer. This procedure simply requires the addition of one aliquot to the PCR product.
Effect of Template Quantity on Sequence Accuracy.
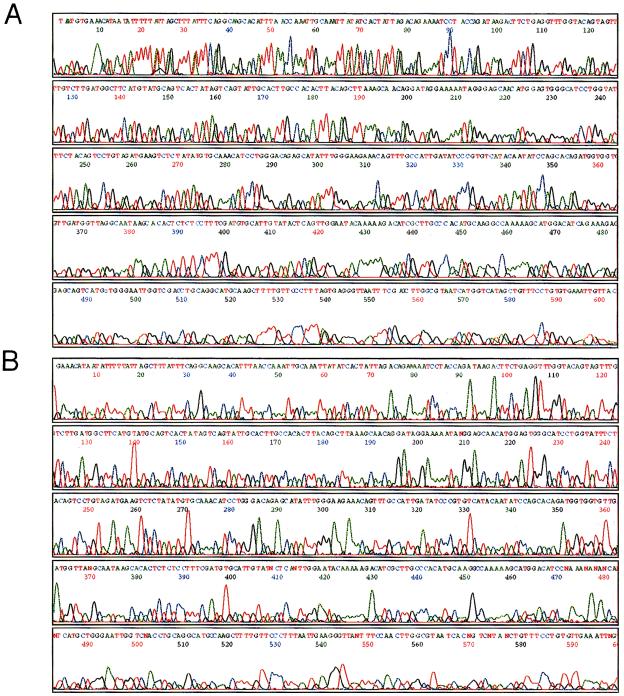
We predicted that transcriptional sequencing would produce stronger signals than that of cycle sequencing in a given time because a large amount of products can be effectively transcribed from a small amount of DNA template by phage RNAPs. We compared transcriptional sequencing with dye-terminator cycle sequencing by using various amounts of template DNA. The accuracy of the resultant sequencing data for <600 bases in transcriptional sequencing was equivalent to that of dye-terminator cycle sequencing using an optimal amount of template (200 and 500 ng) (Table 1). With <100 ng of template, transcriptional sequencing produced much better data than that of cycle sequencing (Fig. 5). With cycle sequencing, sequence accuracy was reduced seriously with 50 ng of plasmid DNA, and 25 ng of template gives >90% accuracy only, from 1 to 100 bp. Sufficient signal for basecall was not obtained with <25 ng (Table 1). On the other hand, with transcriptional sequencing, excellent data were obtained with 25- to 500-ng templates from 1 to 500 bases. Even in the case of 10 ng of template DNA, >93% accuracy could be obtained from 1 to 400 bases. In this experiment, sequencing reactions were done by incubation for 3 hr with any amount of templates. Sequencing accuracy was almost constant and independent of the template amount from 25 to 500 ng (Table 1). This property is ideal for large scale sequencing because it is too time-consuming to adjust template concentrations to optimum in cycle sequencing reactions when using a large number of clones. Although high quality sequencing data were produced until 600 bases, several bases were miscalled using ABI basecaller software. There is no deletion and insertion of the signal in raw data, and the reason for these errors was caused by the mobility correction program for DNA polymerase-based method. Adenines especially were sometimes overlapped with the next bases because the R6G-3′-dATP-terminated fragment has slower mobility than the other sequencing fragments in the RNAP-based method. Therefore, to improve the quality of sequencing data, it will be necessary to adjust the mobility correction program for transcriptional sequencing.
Table 1.
Effect of template amount on sequence accuracy
| Sequencing protocol | Amount of plasmid, ng | Bases
|
|||||
|---|---|---|---|---|---|---|---|
| 1–100 | 101–200 | 201–300 | 301–400 | 401–500 | 501–600 | ||
| Dye-terminator cycle sequencing | 500 | 99 | 99 | 100 | 100 | 94 | 85 |
| 200 | 100 | 99 | 100 | 99 | 94 | 82 | |
| 100 | 99 | 99 | 100 | 96 | 86 | 56 | |
| 50 | 98 | 96 | 98 | 88 | 63 | 45 | |
| 25 | 96 | 79 | 67 | 51 | 39 | 42 | |
| 10 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| Transcriptional sequencing | 500 | 97 | 98 | 97 | 100 | 94 | 82 |
| 200 | 97 | 96 | 97 | 97 | 94 | 74 | |
| 100 | 96 | 96 | 96 | 97 | 95 | 76 | |
| 50 | 96 | 96 | 97 | 95 | 95 | 83 | |
| 25 | 96 | 92 | 98 | 97 | 95 | 71 | |
| 10 | 96 | 92 | 97 | 93 | 77 | 50 | |
n.d., not determined.
Figure 5.
Comparison of transcriptional sequencing (A) with dye-terminator cycle sequencing (B) on the ABI 377 sequencer. 100 ng of human TSH cDNA (plasmid clone:pBS750) was sequenced with both methods as described in Materials and Methods.
PCR-Direct Sequencing.
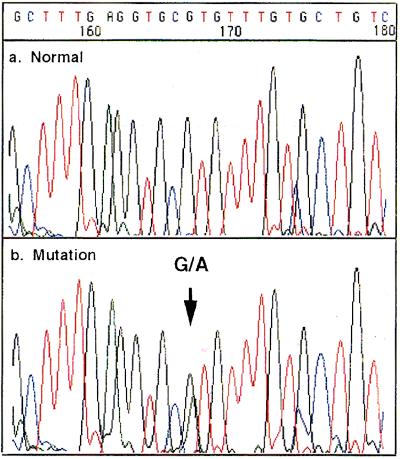
One of the most useful applications of transcriptional sequencing is that it allows direct sequencing by using a PCR-amplified DNA template without purification. In DNA polymerase-based sequencing, PCR primers and 2′-dNTP, which perturb the optimal sequencing conditions and impair the sequencing pattern, must be eliminated prior to the sequencing reaction. On the other hand, the group of promoter-dependent RNAP requires NTP and 3′dNTP (nucleotide having the hydroxyl group at the 2′ position) and a promoter for initiation of the transcription, in place of 2′-dNTP (nucleotide with “deoxy” at the 2′ position) and primers. Transcriptional sequencing is not perturbed by the presence of 2′-dNTP and primers for PCR amplification previously used to prepare the DNA template. Our protocol for PCR-direct transcriptional sequencing is shown in Fig. 4. The promoter sequence for T7 RNAP is designed in one of the PCR primers (Primer 1), and template DNA is amplified by the cooperation of Primer 2 with the T3 promoter. After PCR amplification, T7 RNAP or T3 RNAP is added to the reaction mixture without the elimination of primers and 2′dNTP. The resulting pattern of PCR-direct transcriptional sequencing using T7 RNAP is shown in Fig. 6. In this study, we employed the PCR-amplified DNA of the exon 8 of p53 prepared from human colon carcinoma cell line HT29. This cell line is known to carry the mutation R274H in which the G nucleotide is changed to A. The heterozygous alleles could be detected by the two peaks at the same position. This implies that this transcriptional sequencing produces an equalized peak height because of equalized incorporation of the four color dye-3′dNTPs as mentioned above. The ability of heterozygotic detection indicates the usefulness of this method as a diagnostic tool.
Figure 6.
Detection of heterozygotes by direct transcriptional sequencing. The top electropherogram shows sequence of the p53 exon 8 PCR product by amplification of wt DNA. The lower electropherogram shows the heterozygous point mutation from 274R (CGT) to 274H (CAT) in human colon carcinoma cell line HT29.
DISCUSSION
We have described a sequencing reaction based on the transcription of promoter-dependent RNAP. Our aim was to develop a transcriptional sequencing reaction using the rhodamine dye-3′-dNTPs with C4 linker, mutated RNAP, PPase, and 7-deaza-GTP. This method improves previous dye-terminator chemistry because it produces equalized peak heights and requires less template. This method makes possible a high throughput, longer read length, and high quality application to clinical diagnosis and genome sequencing. It is also advantageous for PCR direct sequencing. Moreover, the reaction can be performed isothermally. We predict that our method using isothermal reaction and rapid reaction without purification of PCR reactants will enhance the automation and miniaturization using robot and microfabrication technologies (24).
Acknowledgments
We thank Mari Itoh for technical assistance and Dr. Masayoshi Itoh and Kazuhiro Shibata for helpful discussions. N.S. was supported in part by Core Research for Evolutional Science and Technology from the Japan Science and Technology Corporation. This study was supported by special coordination funds and a research grant for the Genome Exploration Research Project from the Science and Technology Agency of the Japanese Government and by a Grant-in-Aid for Scientific Research on Priority Areas and Human Genome Program from the Ministry of Education and Culture of Japan to Y.H.
ABBREVIATIONS
- 3′-dNTP
3′-deoxynucleoside triphosphate
- RNAP
RNA Polymerase
- dd
2′,3′-dideoxy
- wt
wild type
- pyrophosphatase
PPase
- TMR
6-carboxytetramethylrhodamine
- ROX
6-carboxy-X-rhodamine
- R6G
5-carboxyrhodamine-6G
- R110
5-carboxyrhodamine-110
References
- 1.Guyer M S, Collins F S. Proc Natl Acad Sci USA. 1995;92:10841–10848. doi: 10.1073/pnas.92.24.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang X C, Quesada M A, Mathies R A. Anal Chem. 1992;64:2149–2154. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- 3.Lee L G, Connell C R, Woo S L, Cheng R D, McArdle B F, Fuller C W, Halloran N D, Wilson R K. Nucleic Acids Res. 1992;20:2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju J, Ruan C, Fuller C W, Glazer A N, Mathies R A. Proc Natl Acad Sci USA. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 7.Khorana S, Gagel R F, Cote G J. Nucleic Acids Res. 1994;22:3425–3426. doi: 10.1093/nar/22.16.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werle E, Schneider C, Renner M, Volker M, Fiehn W. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova O V, Makarov E M, Sousa R, Dreyfus M. Proc Natl Acad Sci USA. 1995;92:12250–12254. doi: 10.1073/pnas.92.26.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Innis M A, Myambo K B, Gelfand D H, Brow M A. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham P R, Ofengand J. BioTechniques. 1990;9:713–714. [PubMed] [Google Scholar]
- 12.Axelrod V D, Kramer F R. Biochemistry. 1985;24:5716–5723. doi: 10.1021/bi00342a005. [DOI] [PubMed] [Google Scholar]
- 13.Prober J M, Trainor G L, Dam R J, Hobbs F W, Robertson C W, Zagursky R J, Cocuzza A J, Jensen M A, Baumeister K. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 14.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 15.Zawadzki V, Gross H J. Nucleic Acids Res. 1991;19:1948. doi: 10.1093/nar/19.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashizaki Y, Miyai K, Kato K, Matsubara K. FEBS Lett. 1985;188:394–400. doi: 10.1016/0014-5793(85)80409-9. [DOI] [PubMed] [Google Scholar]
- 17.Murakami Y, Hayashi K, Hirohashi S, Sekiya T. Cancer Res. 1991;51:5520–5525. [PubMed] [Google Scholar]
- 18.Sousa R, Chung Y J, Rose J P, Wang B C. Nature (London) 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 19.Tabor S, Richardson C C. J Biol Chem. 1990;265:8322–8328. [PubMed] [Google Scholar]
- 20.Mizusawa S, Nishimura S, Seela F. Nucleic Acids Res. 1986;14:1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar G, Yoon H S, Sommer S S. Nucleic Acids Res. 1992;20:871–878. doi: 10.1093/nar/20.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joho K E, Gross L B, McGraw N J, Raskin C, McAllister W T. J Mol Biol. 1990;215:31–39. doi: 10.1016/S0022-2836(05)80092-0. [DOI] [PubMed] [Google Scholar]
- 24.Burke D T, Burns M A, Mastrangelo C. Genome Res. 1997;7:189–197. doi: 10.1101/gr.7.3.189. [DOI] [PubMed] [Google Scholar]