Abstract
During adipogenesis, CCAAT/enhancer binding protein α (C/EBPα) serves as a pleiotropic transcriptional activator of adipocyte genes. Previously, we identified dual repressive elements in the C/EBPα gene and a putative transacting factor (C/EBPα undifferentiated protein, or CUP) expressed by preadipocytes, but not adipocytes, that bind to these elements. In the present investigation, CUP was purified 17,000-fold from nuclear extracts of 3T3-L1 preadipocytes. Amino acid sequence and mass spectral analysis of tryptic peptides derived from purifed CUP (molecular mass ≈50 kDa) revealed that the repressor is (or contains) an isoform of the transcription factor, AP-2α. Electrophoretic mobility shift and Western blot analysis on purified CUP and preadipocyte nuclear extracts confirmed the identity of CUP as AP-2α. Both AP-2α protein and CUP binding activity are expressed by preadipocytes and then decrease concomitantly during differentiation of 3T3-L1 preadipocytes into adipocytes. Consistent with a repressive role of AP-2α/CUP, an AP-2α1 expression vector, cotransfected with a C/EBPα promoter-reporter construct into 3T3-L1 adipocytes, inhibited reporter gene transcription. Taken together with previous results, these findings suggest that in preadipocytes the C/EBPα gene is repressed by AP-2α/CUP, which, upon induction of differentiation, is down-regulated, allowing expression of the gene.
Keywords: 3T3-L1 cells, preadipocyte, adipocyte, differentiation, C/EBPα undifferentiated protein
CCAAT/enhancer binding protein α (C/EBPα) functions as a pleiotropic transcriptional activator of adipocyte genes during adipogenesis (1–3). Indeed, compelling evidence shows that expression of C/EBPα is not only required for (4, 5), but is sufficient to trigger (6, 7), adipocyte differentiation without the use of hormonal stimuli. In view of the importance of C/EBPα in the adipocyte differentiation program, a search for regulatory elements in the C/EBPα gene was undertaken in this (8) and other laboratories (9). Recent evidence (10) indicates that the gene is regulated at least in part by a repression/derepression mechanism. DNaseI footprinting of the proximal promoter of the mouse C/EBPα gene identified several binding sites for nuclear factors that are differentially regulated during adipogenesis. One of these sites was a potential repressor binding site (8). This site, located ≈250 bp 5′ of the transcriptional start site, was footprinted by a nuclear factor present in preadipocytes, but not adipocytes (8, 11), and was therefore designated CUP (C/EBPα undifferentiated protein) (11). During adipocyte differentiation, CUP binding activity seemed to be down-regulated concomitant with the transcriptional activation of the C/EBPα gene (11), raising the possibility that CUP acts as a repressor of the gene.
More recently, we discovered that, in addition to the CUP binding site (the CUP-1 site) in the proximal 5′-flanking region of the C/EBPα gene, there is another potential binding site (the CUP-2 site) in the 5′-untranslated region that contains the same core sequence (GCCGCCG) (10). Proof that both sites are required for repression was obtained by mutating the sites in C/EBPα promoter-reporter constructs and showing that only when both CUP sites were mutated was transcription of the reporter gene derepressed (10). Thus, it seemed that dual CUP sites are necessary to ensure that the C/EBPα gene is maintained in the repressed state before differentiation.
To characterize CUP and investigate its effect on transcription of the C/EBPα gene, we set out to isolate and express CUP cDNA. As several attempts to expression-clone CUP cDNA were unsuccessful, we undertook the purification of CUP from 3T3-L1 preadipocytes to obtain sufficient amino acid sequence for the design of oligonucleotide probes for library screening. In the present paper, we report the purification of CUP and partial sequencing of the protein. From this sequence information, we discovered and verified that CUP is, in fact, an isoform of the transcription factor AP-2α. We further demonstrated that in 3T3-L1 adipocytes expression of AP-2α/CUP trans-inhibits reporter gene expression mediated by the C/EBPα gene promoter.
EXPERIMENTAL PROCEDURES
Human recombinant AP-2α1 was from Promega, and rabbit anti-hAP-2α antibody and rabbit anti-hSp1 antibody were from Santa Cruz Biotechnology. CMV-hAP-2α1 expression vector was provided by R. Buettner (University of Regensberg, Germany). Protein was determined by the Bradford method (12).
Cell Culture and Induction of Differentiation.
3T3-L1 preadipocytes were maintained and propagated in DMEM containing 10% (vol/vol) calf serum. Two-day postconfluent (designated day 0) cells were induced to differentiate (13) with DMEM containing 10% (vol/vol) fetal bovine serum, 1 μg of insulin per ml, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methyl-xanthine until day 2. Cells were then fed DMEM supplemented with 10% fetal bovine serum and 1 μg insulin per ml for 2 days, after which they were fed every other day with DMEM containing 10% fetal bovine serum. Expression of adipocyte genes and acquisition of the adipocyte phenotype begins on day 3 and is maximal by day 8.
Electrophoretic Mobility-Shift Assay (EMSA).
EMSA was performed as previously described (10). For competition experiments, a 50-fold excess of unlabeled competitor oligonucleotide was added before addition of radiolabeled oliogonucleotide. The sequence (slash indicating the delimiting linker sequence) of the CUP-1 site oligonucleotide of the mouse C/EBPα promoter is GATC/GGAGGCCGCCGAGG/. The sequence of the CUP-1 site oligonucleotide of the human C/EBPα promoter (14) used for EMSA is GGCGACGGCCGGGCCGGGGGCGGAGT (Fig. 1). Consensus wild-type and mutant AP-2 binding site oligonucleotides, i.e., GATCGAACTGACCGCCCGCGGCCCGT and GATCGAACTGACCGCTTGCGGCCCGT, were obtained from Santa Cruz Biotechnology, respectively. Nuclear extracts were prepared by a modification of the NUN protocol as previously described (10). Protein complex formation was quantitated by PhosphorImager in the linear response range; an arbitrary unit of CUP binding activity was established for each purification run (for example, the run illustrated in Table 1) to facilitate activity comparisons between purification steps.
Figure 1.
Nucleotide sequence in the CUP-1 binding site region of the mouse and human C/EBPα gene promoters. Underlined bases in the mouse C/EBPα gene promoter indicate the region that is DNaseI-footprinted by preadipocyte nuclear extract (8) or rAP-2α1 (M.-S.J. and M.D.L., unpublished results). Bases overlined indicate the AP-2 consensus sequence (-GCCNNNGGC or G). Letters in bold indicate bases in the human C/EBPα gene promoter (14) that differ from those footprinted in the mouse gene.
Table 1.
Purification of CUP from 3T3-L1 preadipocytes
| Step | Protein, μg | Activity, arbitrary units | Specific activity, units/μg | Yield, % | Purification, -fold |
|---|---|---|---|---|---|
| Cells* | 80,000 | — | — | — | — |
| Nuclear extract | 17,900 | 10,500 | 0.59 | 100 | 1 |
| DNA-cellulose | 200 | 7,350 | 37 | 70 | 63 |
| CUP-1 affinity matrix | 0.4 | 4,200 | 10,500 | 40 | 17,800 |
200 10-cm monolayers of confluent 3T3-L1 preadipocytes.
Purification of CUP from 3T3-L1 Preadipocytes.
All procedures were conducted at 4°C. Synthetic CUP-1 binding site oligonucleotide (GATC/CAGCGCCGCCGGGG/) was oligomerized to produce 5 to 10 mers, which were conjugated to CNBr-activated Sepharose 4B (Pharmacia), i.e., 22 μg of oligonucleotide/ml matrix, as described in the Pharmacia manual. Nuclear extract (see above) from 200 10-cm monolayers of 2-day postconfluent (day 0) 3T3-L1 preadipocytes was adjusted to 100 mM KCl (in buffer D containing 40 mM Tris-HCl/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride, pH 7.5) and centrifuged at 100,000× g for 45 min; the supernatant was adsorbed to 8 ml of calf thymus DNA-cellulose (Sigma) overnight, poured into a 1 × 8-cm column, and washed with 20 column volumes of buffer D containing 100 mM KCl. Fractions were monitored for CUP activity by quantitative EMSA. CUP binding activity was eluted as a single peak at ≈450 mM KCl by gradient elution. Fractions containing maximal CUP activity were pooled, desalted on a PD-10 column (Pharmacia), mixed with poly[d(A-C)⋅d(G-T)] (0.1 μg/μg protein), and then equilibrated overnight with 0.5 ml of CUP binding site affinity matrix in buffer D containing 100 mM KCl. The mixture was poured into a 0.5 × 1-cm column, washed with the same KCl-containing buffer, and then subjected to gradient elution (100–700 mM KCl), with CUP activity eluting at ≈400 mM KCl. Fractions from affinity chromatography were subjected to EMSA or SDS/PAGE (10% acrylamide) for 14–16 h at 40 V, after which SDS gels were silver-stained (Fig. 2B).
Figure 2.
Purification of CUP by binding site oligonucleotide affinity chromatography. (A) CUP purified by chromatography on DNA-cellulose was applied to a CUP-1 binding site oligonucleotide-Sepharose column, and elution was performed with a KCl gradient (□). CUP binding activity (•) in the eluted fractions was monitored by quantitative EMSA. (B) 50 μl of selected fractions were subjected to SDS/PAGE, after which the gels were silver-stained. Wf refers to final wash of the column, S refers to starting material (derived from the pooled fractions from DNA-cellulose chromatography), F refers to the flow-through volume of the CUP-1 binding site affinity column, and STD refers to molecular mass standards. The arrow identifies the 50-kDa band.
Amino Acid Sequence and Mass Analysis of CUP Tryptic Peptides.
Fractions containing peak CUP binding activity (Fig. 2A, fractions 27–31) were pooled, subjected to SDS/PAGE, blotted onto poly(vinylidene difluoride) (PVDF) membranes (ProBlott, Applied Biosystems), and stained with Coomassie Blue. The 50-kDa band was excised and reduced and alkylated with isopropylacetamide (15) followed by digestion for 17 h at 37°C with 0.2 μg of trypsin (Frozen Promega Modified) in 20 μl of 0.05 M ammonium bicarbonate containing 0.5% Zwittergent 3–16 (Calbiochem) (16). Peptides were separated on a C18 capillary column (LC Packings) using a prototype capillary gradient HPLC (Waters) with a small delay volume to facilitate hand collection of fractions (17). Solvent A was 0.1% trifluoroacetic acid, and solvent B was acetonitrile containing 0.08% trifluoroacetic acid. Peptides were eluted using a linear gradient of 0–80% B in 60 min and detected at 195 nm. Well-resolved peaks were sequenced on a model 494 CL PE Applied Biosystems sequencer using 6-mm micro cartridges and equipped with an on-line capillary PTH analyzer (model 140D). Peaks were integrated with justice innovation software using Nelson Analytical 760 interfaces. Sequence interpretation was performed on a DEC Alpha (18).
Mass analysis was performed on 0.2 μl of the isolated HPLC fractions, which were applied to a premade spot of matrix (5 μl of 20 mg/ml α-cyano-4-hydroxycinammic acid + 5 mg/ml nitrocellulose in 50% acetone/50% 2-propanol) (19) on the target plate. Ions were formed by matrix-assisted laser desorption/ionization with a nitrogen laser (337 nm). Spectra were acquired with a PerSeptive Biosystems Voyager Elite time-of-flight mass spectrometer, operated in reflector-delayed extraction mode.
Western Blot Analysis.
SDS-polyacrylamide gels were electroblotted onto PVDF membranes (Millipore). Membranes were probed with anti-hAP-2 antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma) as secondary antibody, which was detected by chemiluminescence (Amersham Corp.).
Transfection.
Differentiated 3T3-L1 adipocytes were transiently cotransfected (calcium phosphate precipitation method; ref. 20) on day 3 or 4 with 1 μg of promoter-reporter construct containing 343 bps of 5′-flanking sequence and the entire 5′-untranslated region of the C/EBPα gene in pGL3-BA luciferase or the promoterless vector (10), along with 10 μg of a CMV-hAP-2α1 expression vector (21) or a control vector lacking the AP-2α1 cDNA insert. After 48 h in culture, cell extracts were prepared and assayed for luciferase activity.
RESULTS AND DISCUSSION
Purification of CUP and Sequence Analysis of Tryptic Peptides.
Quantitative EMSA with an oligonucleotide probe corresponding to the CUP-1 binding site in the C/EBPα gene was used to monitor CUP activity during purification. Nuclear extract from 3T3-L1 preadipocytes, typically 200 10-cm cell monolayers, was subjected to chromatography on DNA-cellulose. Binding of CUP to this matrix was unexpectedly tight, requiring ≈450 mM KCl for elution and resulting in ≈60-fold purification (Table 1). Fractions from the DNA-cellulose column containing the highest CUP activity were pooled, applied to a CUP-1 binding site oligonucleotide affinity matrix, and eluted with a KCl gradient elution. A typical elution profile is shown in Fig. 2A. CUP was eluted as a single peak of activity at ≈400 mM KCl. The activity peak corresponded exactly to a major silver-stained protein of ≈50 kDa (Fig. 2B). Fractions with highest activity (fractions 27–31) were pooled, subjected to SDS/PAGE, blotted onto a PVDF membrane, and the section of the membrane containing the 50-kDa protein was excised and subjected to digestion with trypsin. Following separation by HPLC, several well-resolved tryptic peptides were subjected to Edman amino acid sequencing and mass spectral analysis.
The sequences of two peptides, i.e., QSQESGLLHTHR and SNSNAVSAIPIN, were used to search a combined nonredundant protein sequence database containing GenBank, Swiss Prot, and the Protein Identification Resource. Exact matches were obtained to sequences in mouse AP-2α, i.e., the predicted tryptic cleavage products derived from amino acids 101–112 and 185–196, respectively, of mouse AP-2α1 (22, 23). Neither of these peptide sequences is found in AP-2β (21). Mass spectral analyses of seven other CUP tryptic peptides corresponded exactly to the masses of predicted tryptic cleavage products derived from amino acids 113–124, 218–226, 229–236, 272–280, 316–323, 324–334, and 347–356 of mouse AP-2α1. Thus, it was concluded that the fractions of peak CUP activity from affinity chromatography contained an isoform of mouse AP-2α, but not AP-2β. Moreover, because the approximate molecular mass of CUP is ≈50 kDa and the molecular masses (based on amino acid sequence) of mouse AP-2α-1, -2, -3, and -4 are 48, 32, 47, and 52 kDa (23), respectively, the 50-kDa CUP/AP-2α isoform purified by affinity chromatography seems to be AP-2α-1, -3, or -4.
As the molecular mass of AP-2α2, a dominant-negative isoform of AP-2α lacking the transactivation domain, is 32 kDa (23), CUP cannot be AP-2α2. However, it should be noted that frequently an additional silver-stained band of molecular mass ≈32 kDa was detected by SDS/PAGE that coeluted with the 50-kDa CUP band from the CUP binding site affinity matrix (results not shown). It is possible, therefore, that another nuclear protein, possibly AP-2α2, may be present in the CUP complex. Investigations are underway to determine which isoforms of AP-2α are expressed during the adipocyte differentiation program.
Verification that CUP Is an AP-2α Isoform.
Several lines of evidence verify that CUP is an isoform of AP-2α. Western blot analysis showed that purified CUP is recognized by an antibody directed against a peptide corresponding to the C-terminal amino acid sequence (amino acids 420–437) of human AP-2α1 (Fig. 3A), the sequences of mouse and human AP-2α1 differing only by a single amino acid in this region. As shown in Fig. 3A, the molecular masses of CUP and recombinant AP-2α1, estimated by SDS/PAGE-Western blotting, are similar, i.e., ≈50 kDa. Interaction with this antibody, however, does not distinguish among the AP-2α isoforms (AP-2α-1, -2, -3, or -4), as the C-terminal sequences of all of the isoforms are identical (23).
Figure 3.
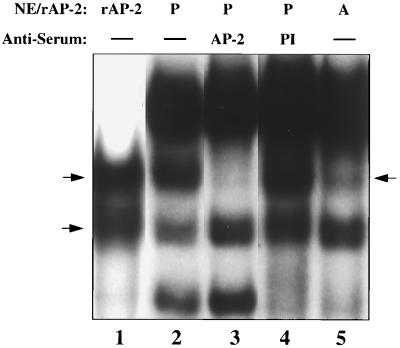
Verification that CUP is an AP-2α isoform. (A) Immunoblot of recombinant hAP-2α1 and purified CUP. 50 μl of purified CUP (from fraction 29; Fig. 2A) or 10 ng of human rAP-2α1 were subjected to SDS/PAGE (10% acrylamide), after which the gel was transblotted onto a PVDF membrane and probed with rabbit anti-hAP-2α antibody. (B) EMSA of human rAP-2α1 (rAP2), day 7 adipocyte nuclear extract (A), or day 0 preadipocyte nuclear extract (P) with 32P-labeled CUP-1 binding site oligonucleotide probe in the presence or absence of preimmune serum (PI), anti-hAP-2α antiserum (AP-2), anti-hSp1 antiserum (Sp-1), or unlabeled wild type (AP-2) or mutated (AP-2M) AP-2 binding site oligonucleotide. NE, nuclear extract. The single arrow identifies the CUP–oligonucleotide probe complex, and the double arrow identifies the position of the band supershifted by anti-hAP-2α antibody.
EMSA confirmed that the CUP/CUP site oligonucleotide complex formed with preadipocyte nuclear extract contains an AP-2α isoform. Both preadipocyte nuclear extract and recombinant AP-2α1, but not adipocyte nuclear extract (which lacks CUP), bind specifically to both the CUP-1 binding site (Fig. 3B, lanes 1–3) and consensus AP-2 (results not shown) oligonucleotides. Recombinant AP-2α1 gives rise to two protein complexes (lane 1), the slower of which corresponds to full-length AP-2α1 and the faster of which is derived from a proteolytic cleavage product also detectable on Western blots (results not shown). Anti-AP-2α antibody, but not irrelevant antibody (anti-Sp1; lane 6) or preimmune serum (lane 4), supershifts the CUP site oligonucleotide–protein complex produced by preadipocyte nuclear extract (lane 5) or by purified CUP or recombinant AP-2α1 (results not shown). Specificity of binding is indicated by the fact that the CUP site oligonucleotide-protein complex is competed away by unlabeled consensus AP-2 site oligonucleotide, but not by mutated consensus AP-2 site oligonucleotide (lanes 7 and 8).
An earlier study (8) showed that the CUP-1 site within the proximal promoter of the C/EBPα gene is DNaseI footprinted by nuclear extract from 3T3-L1 preadipocytes, but not adipocytes. Consistent with evidence presented above that AP-2α is the DNA binding protein in preadipocytes that binds at this site, rAP-2α1 also footprints this site (results not shown).
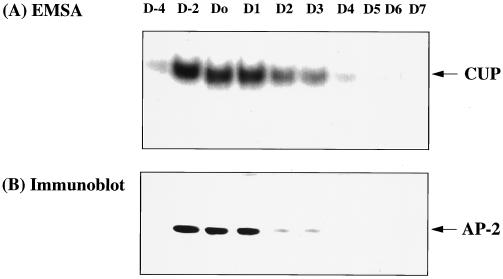
Previous investigations in this laboratory (10, 11) revealed an inverse kinetic relationship between the decrease in CUP binding activity and the increase in the rate of transcription of the C/EBPα gene during adipocyte differentiation. To determine whether decreased expression of CUP/AP-2α protein (possibly through decreased transcription of the CUP/AP-2α gene) could account for the decrease in CUP binding activity, both of these processes were monitored during adipocyte differentiation. As shown in Fig. 4 A and B, respectively, the CUP binding activity and levels of AP-2α protein declined rapidly and coordinately after induction of differentiation (day 0), becoming virtually undetectable by day 4. Moreover, rapidly dividing 3T3-L1 preadipocytes (day −4) lack both CUP protein and binding activity, but binding activity increased dramatically when the cells become growth arrested at confluence (day −2). Taken together, these findings provide compelling evidence that CUP is (or contains) AP-2α-1, -3, or -4.
Figure 4.
Changes in AP-2α and CUP binding activity during adipocyte differentiation. (A) CUP binding activity of nuclear extracts during differentiation of 3T3-L1 preadipocytes. Nuclear extracts prepared on the days indicated were subjected to Western blot analysis or EMSA using the CUP-1 binding site oligonucleotide as labeled probe. D-4 refers to proliferating 3T3-L1 preadipocytes; D-2 and D0 refer to confluent and 2-day postconfluent preadipocytes; and D1, D2, D3, D4, D5, D6, and D7 refer to cells 1, 2, 3, 4, 5, 6, and 7 days, respectively, after induction of differentiation. (B) AP-2α protein was detected by Western blot analysis of nuclear extracts during differentiation of 3T3-L1 preadipocytes.
The Human C/EBPα Gene Promoter Contains an AP-2/CUP-1 Binding Site.
It was previously reported (14) that the human C/EBPα gene lacks a functional CUP-1 binding site comparable to that in the proximal promoter (between nucleotide −251 and nucleotide −236) of the mouse C/EBPα gene, although a consensus AP-2 binding site sequence was identified (ref. 14; see also Fig. 1). In view of our finding that CUP is an AP-2α isoform, this issue was reinvestigated. EMSA was performed with an oligonucleotide probe corresponding to the potential AP-2 binding site in the human C/EBPα promoter; however, the oligonucleotide extended further 5′ than the consensus sequence tested previously without success (14). This probe gave rise to protein complexes with rAP-2α1 and preadipocyte nuclear extract (Fig. 5, lanes 1 and 2). It is evident that one of the complexes formed with preadipocyte nuclear extract contains CUP/AP-2α, as it was supershifted by antibody to hAP-2 (lane 3), was specifically competed away by unlabeled CUP-1 site oligonucleotide (results not shown) and was not formed with adipocyte nuclear extract (lane 5). It should be recalled that the two complexes formed with hAP-2α (Fig. 5, lane 1) are both supershifted by antibody to hAP-2α (results not shown) and that the faster-moving complex is due to a proteolytic cleavage product of recombinant hAP-2α. This cleavage product is not detected in preadipocyte nuclear extracts. Taken together, these results indicate that, like the mouse promoter, the human C/EBPα promoter possesses a functional CUP/AP-2 binding site in the same region (Fig. 1).
Figure 5.
EMSA of preadipocyte and adipocyte nuclear extracts with the oligonucleotide sequence in the human C/EBPα gene that corresponds to the CUP-1 binding site in the mouse gene. EMSA was performed with nuclear extracts from 3T3-L1 preadipocytes (P; day 0) or adipocytes (A; day 7) with an oligonucleotide sequence in the human C/EBPα gene that corresponds to the CUP-1 binding site in the mouse C/EBPα gene. Where indicated, nuclear extract was treated with antibody against hAP-2α (AP-2) or preimmune serum (PI) before incubation with the labeled oligonucleotide probe. Two arrows on left indicate complexes formed with rAP-2; arrow on right the complex formed with preadipocyte nuclear extract.
Trans-Inhibition by AP-2α1of Reporter Gene Expression Mediated by the C/EBPα Gene Promoter.
Previous studies showed that dual CUP binding sites in the C/EBPα gene promoter function repressively (10). Thus, mutation of the CUP binding sites in C/EBPα promoter-luciferase constructs derepressed reporter gene expression in 3T3-L1 preadipocytes, but not 3T3-L1 adipocytes. To determine whether AP-2α is capable of repressing transcription mediated by the C/EBPα gene promoter, a C/EBPα promoter-luciferase construct was cotransfected into day 3 3T3-L1 adipocytes (or 3T3-L1 preadipocytes) along with an AP-2α1 expression vector or a control vector lacking the AP-2α1 cDNA insert. As shown in Fig. 6, expression of AP-2α1 markedly “trans-inhibited” transcription directed by the C/EBPα promoter, whereas the control vector had no effect. In five different experiments, the extent of trans-inhibition varied from 55 to 90%. Importantly, the AP-2α1 expression vector had no effect on expression of the C/EBPα promoter-luciferase construct in 3T3-L1 preadipocytes (results not shown). It can be concluded, therefore, that AP-2α1 is capable of repressing transcription driven by the C/EBPα gene promoter and may be responsible, at least in part, for maintenance of the gene in a repressed state before differentiation. Although it is evident that AP-2 can act as a repressor of the C/EBPα gene, the repression mechanism remains unclear. Although AP-2 is generally considered to be a transcriptional activator, it has also been shown to function as a repressor of a number of genes including the genes for stellate type 1 collagen (24), K3 keratin (25), acetylcholinesterase (26), prothymosin (27), ornithine decarboxylase (27), retinal-fatty acid binding protein (28), retinoic acid receptor β (29), and cartilage-derived retinoic acid-sensitive protein (30). Studies are underway to determine the mechanism of repression of the C/EBPα gene by AP-2α and to assess its effect on the differentiation program.
Figure 6.
Transinhibition by AP-2α1 of reporter gene expression mediated by the promoter of the C/EBPα gene in 3T3-L1 adipocytes. Day 3 3T3-L1 adipocytes were transiently cotransfected with an AP-2α1 or control (lacking an AP-2α1 cDNA insert) expression vector and either a promoterless (lacking the C/EBPα promoter) or C/EBPα promoter-luciferase construct. After 48 h, luciferase assays were conducted on cell extracts. Results are from five independent experiments.
Acknowledgments
We thank Dr. R.Buettner (University of Regensberg, Germany) for providing the AP-2α expression vector and Ms. K. Anuzis for assistance with cell culture. This work was supported by National Institutes of Health Research Grant DK 38418.
ABBREVIATIONS
- EMSA
electrophoretic mobility-shift assay
- CUP
C/EBPα undifferentiated protein
- C/EBPα
CCAAT/enhancer binding protein α
- PVDF
poly(vinylidene difluoride)
References
- 1.MacDougald O A, Lane M D. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 2.Mandrup S, Lane M D. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 3.Hwang C-S, Loftus T M, Mandrup S, Lane M D. Annu Rev Cell Biol. 1997;13:231–259. doi: 10.1146/annurev.cellbio.13.1.231. [DOI] [PubMed] [Google Scholar]
- 4.Lin F-T, Lane M D. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 5.Samuelsson L, Stromberg K, Vikman K, Bjursell G, Enerback S. EMBO J. 1991;10:3787–3793. doi: 10.1002/j.1460-2075.1991.tb04948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin F-T, Lane M D. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freytag S O, Paielli D L, Gilbert J D. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 8.Christy R J, Kaestner K H, Geiman D E, Lane M D. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legraverend C, Antonson P, Flodby P, Xanthopoulos K G. Nucleic Acids Res. 1993;21:1735–1742. doi: 10.1093/nar/21.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Q-Q, Jiang M-S, Lane M D. Proc Natl Acad Sci USA. 1997;94:13571–13575. doi: 10.1073/pnas.94.25.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasseur-Cognet M, Lane M D. Proc Natl Acad Sci USA. 1993;90:7312–7316. doi: 10.1073/pnas.90.15.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Student A K, Hsu R Y, Lane M D. J Biol Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 14.Timchenko N, Wilson D R, Taylor L R, Abdelsayed S, Wilde M, Sawadogo M, Darlington G J. Mol Cell Biol. 1995;15:1192–1202. doi: 10.1128/mcb.15.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krutzsch H C, Inman J K. Anal Biochem. 1993;209:109–116. doi: 10.1006/abio.1993.1089. [DOI] [PubMed] [Google Scholar]
- 16.Lui M, Tempst P, Erdjmuent-Bromage H. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 17.Henzel W J, Stults J T. Curr Protocols Protein Sci. 1995;1:11.6.1–11.6.14. doi: 10.1002/0471140864.ps1106s24. [DOI] [PubMed] [Google Scholar]
- 18.Hensel W J, Rodriguez H, Watanabe C. J Chromatogr. 1987;404:41–45. doi: 10.1016/s0021-9673(01)86835-7. [DOI] [PubMed] [Google Scholar]
- 19.Schevehenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 20.Hwang C-S, Mandrup S, MacDougald O A, Geiman D E, Lane M D. Proc Natl Acad Sci USA. 1996;93:873–877. doi: 10.1073/pnas.93.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Beuttner R. Development (Cambridge, UK) 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 22.Moser M, Pscherer A, Bauer R, Imhof A, Seegers S, Kerscher M, Buettner R. Nucleic Acids Res. 1993;21:4844. doi: 10.1093/nar/21.20.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier P, Koedood M, Philipp J, Fontana A, Mitchell P J. Dev Biol. 1995;169:1–14. doi: 10.1006/dbio.1995.1121. [DOI] [PubMed] [Google Scholar]
- 24.Chen A, Beno D W A, Davis B H. J Biol Chem. 1996;271:25994–25998. doi: 10.1074/jbc.271.42.25994. [DOI] [PubMed] [Google Scholar]
- 25.Chen T-T, Wu R-L, Castro-Munozledo F, Sun T-T. Mol Cell Biol. 1997;17:3056–3064. doi: 10.1128/mcb.17.6.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Getman D K, Mutero A, Inoue K, Taylor P. J Biol Chem. 1995;270:23511–23519. doi: 10.1074/jbc.270.40.23511. [DOI] [PubMed] [Google Scholar]
- 27.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisgrove D A, Monckton E A, Godbout R. Mol Cell Biol. 1997;17:5936–5945. doi: 10.1128/mcb.17.10.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollberg, T. M., Osborn, N. & Liu, Y. (1997) Mol. Biol. Cell 8, Suppl., 429a.
- 30.Xie, W. F., Kondo, S. & Sandell, L. J. (1997) Mol. Biol. Cell 8, Suppl., 430a.