Abstract
The identification of cross-neutralizing antibodies to HIV-1 is important for designing antigens aimed at eliciting similar antibodies upon immunization. The monoclonal antibody (mAb) F425-B4e8 had been suggested previously to bind an epitope at the base of V3 and shown to neutralize two primary HIV isolates. Here, we have assessed the neutralization breadth of mAb F425-B4e8 using a 40-member panel of primary HIV-1 and determined the epitope specificity of the mAb. The antibody was able to neutralize 8 clade B viruses (n=16), 1 clade C virus (n=11), and 2 clade D viruses (n=6), thus, placing it is among the more broadly neutralizing anti-V3 antibodies described so far. Contrary to an initial report, results from our scanning mutagenesis of the V3 region indicate that mAb F425-B4e8 interacts primarily with the crown/tip of V3, notably Ile309, Arg315, and Phe317. Despite the somewhat limited neutralization breadth of mAb F425-B4e8, the results presented here, along with other cross-neutralizing anti-V3 mAbs, may facilitate the template-based design of antigens that target the V3 region and permit neutralization of HIV-1 strains in which the V3 is accessible to antibodies.
Keywords: HIV-1, V3 antibody, F425-B4e8, cross-clade neutralization, epitope mapping, scanning mutagenesis
INTRODUCTION
The envelope spike is the major constituent of the outer surface of human immunodeficiency virus type 1 (HIV-1) and a prominent target for neutralizing antibodies during HIV-1 infection (Haigwood and Stamatatos, 2003; Haynes and Montefiori, 2006; Nabel and Sullivan, 2000; Pantophlet and Burton, 2006; Wyatt and Sodroski, 1998; Zolla-Pazner, 2004). However, HIV has evolved to shield conserved portions on the two subunit glycoproteins –gp120 and gp41—that comprise the envelope spike from antibody. Despite an extensive regimen of viral resistance mechanisms, the existence of a handful of monoclonal antibodies (mAbs) that can neutralize a fairly broad array of primary viruses demonstrates that there are a few chinks in the viral defensive armor (Burton, Stanfield, and Wilson, 2005). A molecular understanding of regions on the HIV-1 envelope that are conserved and sufficiently exposed on the viral spike so as to be recognized by antibodies is an important aid in the design of immunogens that are aimed toward the elicitation of cross-neutralizing antibodies (Burton et al., 2004; Haynes and Montefiori, 2006; Zolla-Pazner, 2004).
Although conserved regions on gp120, such as the CD4-binding site or the heavily glycosylated 'silent face' (Wyatt et al., 1998), are generally accepted as targets that should be pursued for HIV vaccine design (Burton et al., 2004), some debate exists with regard to targeting the variable loops (V1-V5) on gp120 (Burton et al., 2004; Srivastava, Ulmer, and Barnett, 2004; Zolla-Pazner, 2004). The V3 loop in particular has been a highly discussed possible target (Hartley et al., 2005; Zolla-Pazner, 2005), ever since it was recognized as a target for neutralization on so-called T-cell line-adapted viruses (Pantophlet and Burton, 2006). However, the inability of many anti-V3 antibodies to neutralize HIV-1 primary isolates, coupled with the lack of an anti-V3 mAb that is able to neutralize primary viruses as good as the broadly neutralizing anti-gp120 mAbs b12 and 2G12, has severely diminished interest in the V3 region as a possible vaccine target.
The unattractiveness of the V3 region also stems from the notion that, as a variable region, escape from antibody neutralization will occur rapidly. Portions of the V3 region can indeed vary considerably in sequence among viral isolates (Catasti et al., 1995; Korber et al., 1994; LaRosa et al., 1990; LaRosa et al., 1991), but the sequence of V3 is relatively conserved at its center, displaying a GPGR motif in many clade B viruses and a GPGQ sequence in nearly all non-clade B viruses. This relatively high degree of conservation likely results from the need to preserve an important structural conformation of the V3 region so juxtaposed residues can efficiently interact with coreceptor molecules on the cell surface (Hartley et al., 2005; Wang et al., 1999). Based on a recent crystal structure of a V3-containing gp120 core complexed to CD4 and an antibody V3 can be subdivided into 3 structural domains (Huang et al., 2005): (1) the base (residues 296-300 (N-terminal portion) and 326-331 (C-terminal portion)), (2) the stem (residues 301-305 (N-terminal portion) and 321-325 (C-terminal portion)), and (3) the tip or crown (residues 306-320). This nomenclature differs somewhat from previous designations in the literature, in which the base and stem regions together were denoted as the stem (Catasti et al., 1995; Cormier and Dragic, 2002). Notably, in the crystal structure of the V3-containing gp120 core, the V3 crown adopts the same β-turn conformation that has been observed in the structures of anti-V3 antibodies in complex with V3 peptides (Huang et al., 2005; Rosen et al., 2006; Stanfield et al., 1999; Stanfield et al., 2003), thus, supporting the notion that certain structural elements in V3 are preserved in many different viruses despite sequence variability in other portions of V3.
Although many anti-V3 antibodies neutralize only a few HIV-1 strains, it has also become evident that some anti-V3 antibodies are able to neutralize a somewhat broader range of viruses (Binley et al., 2004; Gorny et al., 2002; Gorny et al., 2006). These include mAbs such as 447-52D and 2219, which were isolated from subtype B-infected individuals, as well as a group of more recently described anti-V3 mAbs that were isolated from patients infected with subtype A (Gorny et al., 2002; Gorny et al., 2006). The crystal structures of mAbs 447-52D and 2219 in complex with V3 peptides demonstrate how these antibodies may be able to neutralize a somewhat larger subset of viruses compared to many other anti-V3 antibodies (Rosen et al., 2005; Stanfield et al., 2004; Stanfield et al., 2006). For example, mAb 447-52D contacts the N-terminal portion of the V3 stem via main-chain interactions and only residues in the V3 tip via their side chains, thus, being able to tolerate some sequence variability in the N-terminal part of the stem of different clade B viruses without a substantial diminution in neutralizing activity (Stanfield et al., 2006). MAb 2219, in contrast, interacts mostly with conserved residues within the N-terminal half of the stem and little with residues in the V3 tip, which allows the antibody to tolerate sequence variability within the tip (Stanfield et al., 2006). A panel of recently described anti-V3 mAbs isolated from non-subtype B-infected individuals appear to neutralize clade B viruses as good as mAbs isolated from subtype B-infected individuals, but appear capable of neutralizing non-B viruses somewhat better (Gorny et al., 2006).
Pinpointing segments in V3 that are relatively conserved and also accessible to antibody is desirable for rational antigen design; although the neutralization breadth of anti-V3 mAbs is somewhat limited relative to other anti-gp120 antibodies such as b12 and 2G12 (Binley et al., 2004), it may be possible to elicit anti-V3 antibodies with greater neutralization breadth by targeted antigen engineering (Pantophlet and Burton, 2003). Here, we have investigated the neutralization breadth and epitope fine specificity of mAb F425-B4e8, which in a previous publication was shown to neutralize 2 primary viruses and reported to bind to a region at the base of V3 (Cavacini et al., 2003). We show that mAb F425-B4e8 neutralizes a range of viruses that place it in the same category as mAbs 447-52D and 2219. Furthermore, mAb F425-B4e8 does not appear to bind to the base of V3 but, rather, seems to bind to an epitope within the V3 crown; the predominant contact residues appear to be Ile309, Arg315, and Phe317. The binding of mAb F425-B4e8 is not diminished by replacing Pro313, thus, suggesting that it binds differently to the V3 tip as compared to other anti-V3 mAbs to this segment of V3 that often require the side chain of the Pro residue, or a precise β-turn conformation, for binding. The binding properties of mAb F425-B4e8 may allow it to neutralize as well as mAbs 447-52D and 2219, which makes F425-B4e8 an interesting candidate to pursue as a template for targeted vaccine design.
RESULTS
Evaluation of the neutralization breadth of mAb F425-B4e8
To assess the neutralization breadth of mAb F425-B4e8, the antibody was tested against a panel of 40 viruses in a robust and highly sensitive pseudovirus-based single-round infectivity assay with luciferase readout (Table 1). Assays such as these have been shown useful for the assessment of neutralization breadth (Binley et al., 2004; Li et al., 2005; Li et al., 2006; Richman et al., 2003). The tested viruses were picked from a previously published 90-member panel (Binley et al., 2004) so that they would encompass (a) different HIV clades, (b) various geographic origins, and (c) a diverse sensitivity to the previously tested anti-V3 mAbs 447-52D and 58.2. To enable a comparison between neutralization results obtained here and the results obtained previously with the same viruses, mAb F425-B4e8 was titrated beginning at the same starting concentration of 50 μg/ml and IC50 values were determined (Binley et al., 2004). As noted before (Binley et al., 2004), IC50 values allow for a more reliable comparison between the potency and cross-reactivity of different antibodies. Furthermore, a 1-hour incubation time of virus plus antibody was used as before.
TABLE 1.
Neutralization of a panel of 40 pseudotyped HIV-1 by mAb F425-B4e8.
| Virusa | Cladeb | IC50 (μg/ml)c |
|---|---|---|
| 92RW021 | A | >50 |
| 93UG077 | A | >50 |
| 94UG103 | A | >50 |
|
| ||
| 92TH006 | AE | >50 |
| 92TH022 | AE | >50 |
| CMU02 | AE | >50 |
| CMU06 | AE | >50 |
|
| ||
| QH0515 | B | >50 |
| QH0692 | B | 4.1 |
| BG1168d | B | >50 |
| SS1196d | B | 34.6 |
| 5768-p27 | B | >50 |
| 6101-p15 | B | >50 |
| 91US056 | B | >50 |
| 92BR020d | B | 0.4 |
| 92BR021 | B | >50 |
| 92HT594d | B | 4.3 |
| 92US712d | B | >50 |
| 93TH305 | B | 37.4 |
| JR-CSFd | B | 7.2 |
| JR-FLd | B | 2.4 |
| ME1 | B | >50 |
| NL4.3d | B | 14.0 |
|
| ||
| 11246-3 | C | >50 |
| 21068 | C | >50 |
| 93IN905 | C | >50 |
| 93IN999 | C | >50 |
| 93MW959 | C | >50 |
| 93MW960 | C | 49.7 |
| 97ZA012 | C | >50 |
| 98BR004 | C | >50 |
| 98CN006 | C | >50 |
| 98CN009 | C | >50 |
| 98IN022 | C | >50 |
|
| ||
| 92UG001 | D | >50 |
| 92UG005 | D | 22.9 |
| 92UG024 | D | >50 |
| 92UG035 | D | >50 |
| 92UG046 | D | 35.1 |
| 94UG114 | D | >50 |
|
| ||
| aMLV | N/A | >50 |
All viruses are quasispecies pools except where denoted. Virus designations are generally denoted as year (of isolation), country (of isolation), and name/number. aMLV, amphotrophic murine leukemia virus.
AE, circulating recombinant form_AE01. N/A, not applicable.
IC50 values below 50 μg/ml are denoted in bold type.
Molecular clone
Under these conditions mAb F425-B4e8 neutralized 8 clade B viruses, 1 clade C virus and 2 clade D viruses. Overall, the neutralization potency of the antibody was low, which contrasts with the higher potencies observed with certain other anti-V3 antibodies (Binley et al., 2004; Li et al., 2005). Interestingly, mAb F425-B4e8 was able to neutralize virus 92BR020, which was not neutralized previously by mAb 447-52D (Binley et al., 2004). However, virus 92BR020 was tested previously as a quasispecies pool, whereas in the current study it was tested as a molecular clone. Although this clone was selected because sensitivity of the virus to neutralization by mAbs b12, 2G12, 2F5, 4E10 and HIV+ serum N16 generally resembled that of the quasispecies pool (T. Wrin, R. Pantophlet, C. Petropoulos, D.R. Burton, unpublished results), the clone has not been tested with mAb 447-52D. Thus, it is possible that the selected clone differs in its sensitivity to anti-V3 antibodies as compared to the quasispecies pool.
We next assessed the neutralization breadth of mAb F425-B4e8, by comparing the neutralization results obtained in this study with mAb F425-B4e8 to those obtained previously with mAb 447-52D in the study by Binley et al. (Binley et al., 2004); direct comparison of the antibodies in one assay was not possible in this study due to the inability to procure sufficient 447-52D antibody for neutralization assays. For the comparison between studies, we first evaluated assay reproducibility using mAb b12. For most viruses, the results obtained here were in good agreement with the results obtained in the Binley et al. study (Table 2); i.e., there was less than a 1-dilution step difference between IC50 titers. The most noticeable difference was observed with virus SS1196, which was 5x more resistant to b12 in this study than in the Binley et al. study (Table 2). Like virus 92BR020 described above, virus SS1196 was also used here as a molecular clone, whereas it was used previously as a quasispecies pool. The SS1196 clone is similarly sensitive to neutralization by mAbs 2G12, 2F5, and 4E10, but 5–6x more resistant to mAb b12 and at least 5x more resistant to serum N16 (T. Wrin et al., unpublished results). Whether these variations in neutralization sensitivity are due to sequences differences is currently being investigated and will be presented elsewhere.
TABLE 2.
Evaluation of pseudovirus neutralization assay variability using mAb b12.
| Viruses | Clade | IC50 (μg/ml)a |
|
|---|---|---|---|
| b12 (this study) | b12 (Binley et al. study)b | ||
| 92RW021 | A | 24.6 | 13.7 |
| 93UG077 | A | >50 | 41.1 |
| 94UG103 | A | 4.0 | 3.5 |
|
| |||
| 92TH006 | AE | 39.3 | 22.4 |
| 92TH022 | AE | >50 | >50 |
| CMU02 | AE | 3.5 | 4.3 |
| CMU06 | AE | >50 | >50 |
|
| |||
| QH0515 | B | 1.6 | 0.7 |
| QH0692 | B | 0.4 | 0.2 |
| BG1168c | B | >50 | >50 |
| SS1196c | B | 15.3 | 2.7 |
| 5768-p27 | B | 2.2 | 1.3 |
| 6101-p15 | B | >50 | >50 |
| 91US056 | B | 0.3 | 0.5 |
| 92BR020c | B | 48.7 | 27.5 |
| 92BR021 | B | 3.3 | 2.2 |
| 92HT593 | B | 0.4 | 0.2 |
| 92HT594c | B | 0.08 | 0.2 |
| 92US712c | B | 19.0 | 9.2 |
| 93TH305 | B | 1.0 | 1.2 |
| JR-CSFc | B | 0.3 | 0.2 |
| JR-FLc | B | 0.04 | 0.09 |
| ME1 | B | >50 | >50 |
| NL4.3c | B | 0.1 | 0.06 |
|
| |||
| 11246-3 | C | 0.9 | 0.4 |
| 21068 | C | 1.3 | 2.3 |
| 93IN905 | C | 23.7 | 34.2 |
| 93IN999 | C | 4.4 | 4.1 |
| 93MW959 | C | >50 | >50 |
| 93MW960 | C | 0.2 | 0.2 |
| 97ZA012 | C | >50 | >50 |
| 98BR004 | C | >50 | >50 |
| 98CN006 | C | 15.2 | 8.4 |
| 98CN009 | C | 1.0 | 0.6 |
| 98IN022 | C | 0.4 | 0.04 |
|
| |||
| 92UG001 | D | 0.7 | 1.2 |
| 92UG005 | D | >50 | >50 |
| 92UG024 | D | >50 | 49.1 |
| 92UG035 | D | >50 | >50 |
| 92UG046 | D | 0.1 | 0.2 |
| 94UG114 | D | >50 | >50 |
|
| |||
| aMLV | N/A | >50 | >50 |
IC50 values below 50 μg/ml are denoted in bold type.
Information taken from (Binley et al., 2004).
Molecular clone.
Given these discrepancies and to allow for a proper comparison between mAbs F425-B4e8 and 447-52D, we did not compare the neutralization titers of the four viruses that had been used previously as a quasispecies pool and that were used here as molecular clones (i.e., SS1196, BG1168, 92BR020, and 92US712). Comparison of the remaining viral isolates shows that the neutralization breadth of mAb F425-B4e8, though limited, is comparable to that of mAb 447-52D (Table 3). However, their respective potencies often differ depending on the virus. Of note is that mAb F425-B4e8 neutralized the primary virus JR-CSF (IC50 ~7 μg/ml), whereas 447-52D failed to neutralize the same virus in the previous study (Binley et al., 2004). As noted in that study, the inability of mAb 447-52D to neutralize JR-CSF in the U87 target cell-based pseudovirus assay was perhaps surprising, given that the antibody has been reported to neutralize JR-CSF virus in a single-round PBMC-based assay (Gorny et al., 2004). However, mAb 447-52D notably failed to neutralize JR-CSF in recent pseudovirus and PBMC-based assays (Eda et al., 2006a; Eda et al., 2006b). Regardless of the underlying reason for these discrepancies, the potency with which mAb F425-B4e8 is able to neutralize JR-CSF demonstrates that this moderately neutralization-resistant virus –and the closely related virus JR-FL—can be neutralized well by select anti-V3 antibodies.
TABLE 3.
Comparison of the neutralization breadth of mAbs F425-B4e8 and 447-52D.
| Viruses | Clade | IC50 (μg/ml)a |
|
|---|---|---|---|
| F425-B4e8 | 447-52Db | ||
| 92RW021 | A | >50 | >50 |
| 93UG077 | A | >50 | >50 |
| 94UG103 | A | >50 | >50 |
|
| |||
| 92TH006 | AE | >50 | >50 |
| 92TH022 | AE | >50 | >50 |
| CMU02 | AE | >50 | >50 |
| CMU06 | AE | >50 | >50 |
|
| |||
| QH0515 | B | >50 | >50 |
| QH0692 | B | 4.1 | 40.3 |
| 5768-p27 | B | >50 | >50 |
| 6101-p15 | B | >50 | >50 |
| 91US056 | B | >50 | >50 |
| 92BR021 | B | >50 | >50 |
| 92HT593 | B | 22.1 | >50 |
| 92HT594 | B | 4.3 | 7.9 |
| 93TH305 | B | 37.4 | 8.5 |
| JR-CSF | B | 7.2 | >50 |
| JR-FL | B | 2.4 | 32.6 |
| ME1 | B | >50 | >50 |
| NL4.3 | B | 14.0 | 0.07 |
|
| |||
| 11246-3 | C | >50 | >50 |
| 21068 | C | >50 | >50 |
| 93IN905 | C | >50 | >50 |
| 93IN999 | C | >50 | >50 |
| 93MW959 | C | >50 | >50 |
| 93MW960 | C | 49.7 | >50 |
| 97ZA012 | C | >50 | >50 |
| 98BR004 | C | >50 | >50 |
| 98CN006 | C | >50 | >50 |
| 98CN009 | C | >50 | >50 |
| 98IN022 | C | >50 | >50 |
|
| |||
| 92UG001 | D | >50 | >50 |
| 92UG005 | D | 22.9 | 32.2 |
| 92UG024 | D | >50 | >50 |
| 92UG035 | D | >50 | >50 |
| 92UG046 | D | 35.1 | >50 |
| 94UG114 | D | >50 | >50 |
|
| |||
| aMLV | N/A | >50 | >50 |
IC50 values below 50 μg/ml are denoted in bold type.
Results taken from (Binley et al., 2004).
Effect of single-residue substitutions in the V3 region of gp120JR-CSF on binding of mAb F425-B4e8 to monomeric gp120
We next sought to define the epitope specificity of mAb F425-B4e8. Defining the epitopes recognized by anti-V3 antibodies has traditionally been accomplished with V3 peptides in ELISA binding assays (e.g. Pepscan) and competition studies, or by determining the ability of the antibodies to bind a series of recombinant gp120s with varying V3 regions (for example (Eda et al., 2006b; Gorny et al., 1992; Gorny et al., 1998; Laisney and Strosberg, 1999; Moore et al., 1994; Moore et al., 1995; Schreiber et al., 1997; Seligman et al., 1996; White-Scharf et al., 1993)). In some instances, the results derived from these studies have been confirmed by analyzing peptide sequences selected with antibody from peptide phage display libraries (Keller et al., 1993) or by comparing the ability of antibodies to neutralize viruses with various V3 sequences (e.g. (Moore et al., 1995; Zolla-Pazner et al., 2004)). The results of mapping studies with peptides have generally agreed with the results from X-ray crystallography and NMR studies (Rosen et al., 2005; Rosen et al., 2006; Sharon et al., 2003; Stanfield et al., 2003; Stanfield et al., 2004; Stanfield et al., 2006). However, it is not possible to assess the energetic contribution of a given residue side chain to binding affinity from structural results alone (Cunningham and Wells, 1993; Dall'Acqua et al., 1998; Seligman et al., 1996) and results from antibody binding assays with peptides as solid-phase antigens are not always good predictors of antibody binding to V3 regions in the context of gp120 (Gorny et al., 2002; Moore, 1993; Moore et al., 1994).
Here, we opted for a scanning mutagenesis approach to identify specific side chains that are required for antibody binding. Site-directed mutagenesis has been used extensively within the HIV field as well as numerous other systems to analyze the interaction between ligands (for example, (Calarese et al., 2003; Cunningham et al., 1989; Cunningham and Wells, 1989; Cunningham and Wells, 1993; Dall'Acqua et al., 1998; Darbha et al., 2004; Lu et al., 2001; Olshevsky et al., 1990; Pantophlet et al., 2003; Rizzuto et al., 1998; Scanlan et al., 2002; Thali et al., 1992; Thali et al., 1991; Vajdos et al., 2002; Wang et al., 1999; Weiss et al., 2000; Zwick et al., 2005; Zwick et al., 2004; Zwick et al., 2003)). For our approach, single residues were mutated individually to Ala. Alanine is a good replacement residue because it eliminates the side chain beyond the β carbon but, in general, does not substantially alter the main chain protein conformation aside from only small and very local structural perturbations, cause steric effects, or impose major electrostatic changes (Cunningham and Wells, 1989; Cunningham and Wells, 1993; Pal, Kossiakoff, and Sidhu, 2003). More importantly, similar alanine mutagenesis approaches have been used to identify residues in V3 that are involved in coreceptor binding and viral infectivity (Suphaphiphat, Essex, and Lee, 2007; Wang et al., 1999). The Arg residue at position 315 was also mutated to Gln, so as to generate a GPGQ sequence in V3 akin to non-subtype B viruses and, thus, to better assess the possible requirement of the Arg at position 315 for the binding interaction of mAb F425-B4e8.
Ideally, we would have preferred to perform our analysis in the context of the virus, by investigating the ability of the mAb to neutralize single-mutant viruses. This type of approach has been employed for structure-function analyses of interhelical interactions in gp41 (Lu et al., 2001) and, more recently, for studying the epitopes of the anti-gp41 antibodies 2F5 and 4E10 (Zwick et al., 2005) and a panel of anti-V3 mAbs (Krachmarov et al., 2006). However, we were not surprised to observe that not all alanine mutants were infectious (Fig. 1), as several of the mutated residues, such as Arg298, Lys305, Ile307, and Phe317, have been shown to be important for viral infectivity and binding to the CCR5 coreceptor on target cells (Rizzuto et al., 1998; Suphaphiphat, Essex, and Lee, 2007; Wang et al., 1999). Discrepancies between the infectivity of mutant viruses in this study compared to the studies by Wang et al. and Suphaphiphat et al. may be related to the use of different viruses and systems. For example, in the Wang et al. study a virus was used that expresses a consensus B gp120 (Wang et al., 1999) and virus JR-FL was used in the study by Suphaphiphat et al. (Suphaphiphat, Essex, and Lee, 2007), whereas virus JR-CSF was used here; although viruses JR-FL and JR-CSF have very similar env sequences, the minor sequence differences may be sufficient to affect the infectivity of mutant viruses. Furthermore, pseudoviruses were incubated for 3 days with target cells in our study, whereas viruses were cultured for 7 days with target cells in the two other studies.
Fig. 1. Influence of V3 mutations on viral infectivity of U87. CD4. CCR5-positive cells.
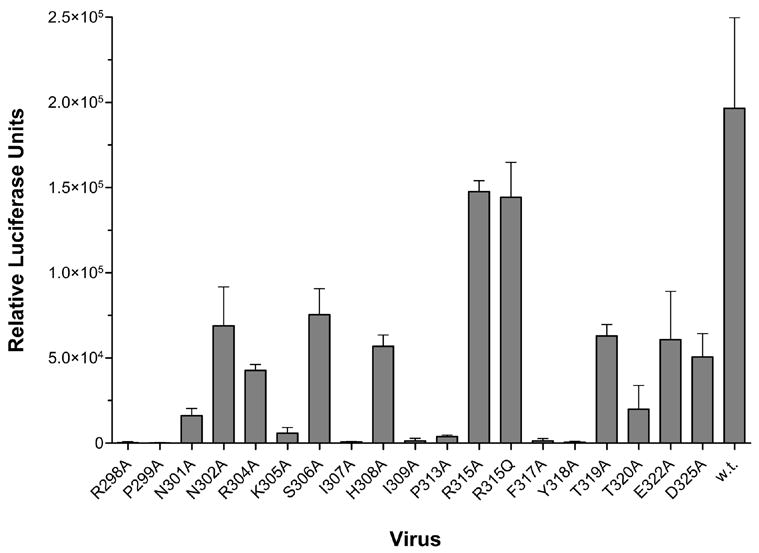
Pseudovirions were generated by transient transfection of 293T cells with an env-deficient, luciferase-expressing HIV-1 provirus (pNL4.3Luc) along with a plasmid expressing either mutant or wild-type (w.t.) JR-CSF envelope glycoprotein. The recombinant viruses were used to infect target cells with equal volumes of virus supernatant. The assay was done in triplicate (w.t.) or sextuplicate (mutants). Mean relative luciferase units and ranges are shown.
Given our inability to use all mutant viruses in infectivity assays, we opted to perform our analyses on solubilized gp120 derived from pseudotyped viruses, as was done in previous studies using anti-gp120 antibodies (Pantophlet et al., 2003; Scanlan et al., 2002). However, it is well-known that disruption of binding does not unequivocally prove that a residue is part of the antibody epitope (Cunningham and Wells, 1989); the alanine scan only identifies side chains that most affect the binding interaction, whereas crystallographic analysis of the antibody-antigen complex would be required to determine whether this indeed is the case (Cunningham and Wells, 1993). Gp120 from virus JR-CSF was used as a template for mutagenesis, given that mAb F425-B4e8 was able to neutralize this virus well (Table 1) and binds monomeric gp120JR-CSF with high affinity (data not shown). Based on the ELISA binding curves, apparent antibody binding affinities were determined and related to the apparent antibody affinity for wild-type gp120JR-CSF. Substitutions that decreased antibody binding 10-fold or more (≤ 10% apparent affinity relative to wild type) were considered indicative of having removed a side chain that is critical for antibody binding, whereas mutations that enhanced binding 2-fold or more (≥ 200% apparent affinity relative to wild type) were considered indicative of having removed a side chain that, directly or allosterically, hinders antibody binding. Intermediate values were recorded as having either moderate (11–50% apparent affinity relative to wild type) or no considerable influence on antibody binding (51%-200% apparent affinity relative to wild type). These parameters were derived from previous studies that have used mutagenesis to assess the energetic contribution of side chains in protein-protein interactions, including the binding of antibodies to antigen, by measuring the loss (or gain) in binding free energy (ΔΔG) upon residue substitution (Cunningham and Wells, 1989; Dall'Acqua et al., 1996; Pal, Kossiakoff, and Sidhu, 2003). In these studies, a loss (or gain) of at least 1 kcal/mol is considered significant and changes of 2.5 kcal/mol or greater are considered critical (also termed 'hot spots') (Dall'Acqua et al., 1996). These ΔΔG values correspond to a change in monovalent binding affinity of approximately 3-fold and 12-fold, respectively. However, to correct for possible bivalent binding of IgG in our ELISA system used here, 2-fold and 10-fold changes in affinity were chosen as the cut-off parameters. Our parameters were purposely set conservatively; bivalent interactions can greatly increase the antibody:antigen binding interaction, thus, leading to an apparent binding affinity when measured by ELISA that is several folds greater than the true binding affinity (Azimzadeh, Pellequer, and Van Regenmortel, 1992; Stevens, 1987).
A total of 18 mutants containing single Ala substitutions and 1 mutant containing a Gln substitution were generated. The locations of the substitutions spanned nearly the entire V3 region (residues 298-325, Table 4), though most altered residues encompass the N-terminal portion of the stem and V3 tip. The two Gly residues located within the β hairpin turn of V3 were not altered, to avoid a more substantial alteration of the conformation of the V3 tip upon their replacement by the larger Ala residue. As noted previously by Cunningham and Wells (Cunningham and Wells, 1989), the contribution of glycine residues to ligand binding cannot be properly assessed by mutagenesis except with larger or more conformationally disruptive substitutions. However, given that most antibody-antigen interactions are dominated by side-chain interactions (Davies and Cohen, 1996), we considered it unlikely that the side chains of the two glycine residues would contribute significantly to the interaction with antibody.
TABLE 4.
Epitope mapping of mAb F425-B4e8.
| Apparent binding affinity relative to wild-type gp120 (%) for mutant:a |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAb | R298A | P299A | N301A | N302A | R304A | K305A | S306A | I307A | H308A | I309A | P313A | R315A | R315Q | F317A | Y318A | T319A | T320A | E322A | D325A |
| F425-B4e8 | 79 | 96 | 124 | 85 | 88 | 83 | 87 | 74 | 88 | 23 | 91 | 3 | 4 | 49 | 76 | 81 | 127 | 77 | 117 |
| 447-52D | 54 | 125 | 118 | 62 | 23 | 37 | 44 | 36 | 23 | 35 | 40 | 0.1 | 0.3 | 149 | 66 | 42 | 74 | 40 | 76 |
Apparent binding affinities relative to wild-type gp120 were calculated with the formula: (apparent affinity for wild type/apparent affinity for mutants) ×100%. Red/Bold: apparent binding affinity relative to wild-type gp120 <10%; Blue: apparent binding affinity relative to wild-type gp120 between 10–50%. Amino acid numbering is based on the HXB2 reference sequence (http://www.hiv.lanl.gov/). The mutants are arranged from the N- to C-terminal portion of the V3 region (left to right).
The binding profile of mAb F425-B4e8 showed that the binding affinity of the antibody is not affected by mutating Pro313 to Ala (Table 4). However, F425-B4e8 binding is dependent on the presence of the Arg315 side chain; replacement of Arg315 with Ala or Gln severely diminished antibody binding (Table 4). The antibody also bound with diminished affinity to mutants I309A and F317A relative to wild-type gp120JR-CSF. Strikingly, binding of mAb F425-B4e8 was not enhanced or diminished more than 1.5-fold by any of the other mutations (Table 4), suggesting that it probably binds mainly to the tip of the V3 region with little or no interaction with residues in the stem, although the possibility of main-chain interactions cannot be excluded due to the inability of the alanine scan to assess this type of interaction (Cunningham and Wells, 1989). The map of F425-B4e8 obtained here contrasts with the previously held notion that it bound to an epitope at the base of V3, which was concluded based on the seeming ability of mAb F425-B4e8 to bind a gp120 mutant in which only a distal portion of the V3 region was deleted (Cavacini et al., 2003). However, a review of those experiments has revealed that this is not the case and follow-up experiments using immunoprecipitation assays have supported the results obtained in this study (J. Sodroski, personal communication).
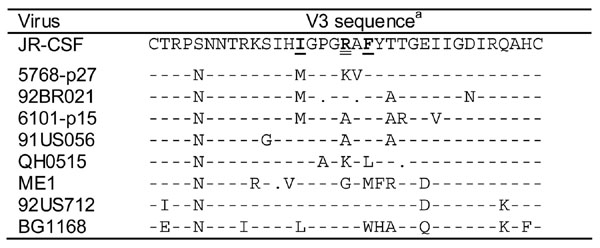
The inability of mAb F425-B4e8 to neutralize a few of the clade B viruses is in good agreement with the scanning mutagenesis, which showed that residues Ile309, Arg315, and Phe317 are critical for binding. As shown in Table 5, viruses 5768-p27, 92BR021, and 6101-p15 contain a Met instead of an Ile at position 309; the thioether group of the Met presumably disrupts the interaction with antibody. In virus 5768-p27, the Ala at position 316 is also replaced by the larger Val, which may distort the spacing between Arg315 and Phe317. In viruses 6101-p15, 91US056, and ME1, Arg315 is replaced by either an Ala or a Gly, whereas in viruses QH0515, ME1, and BG1168, Phe317 is substituted (Table 5). Only for virus 92US712 are there no substitutions in the putative antibody epitope that would readily explain the inability of F425-B4e8 to neutralize this virus well. However, we presume that the occurrence of a charged residue (Lys) at position 328 in virus 92US712 relative to the uncharged Gln residue in JR-CSF may influence the conformation of V3 and so affect the affinity of the antibody for its epitope (Table 5). However, mAb F425-B4e8 did neutralize virus 92US712 to ~40% at the highest antibody concentration tested in this study (50 μg/ml; result not shown). Whether the inability of F425-B4e8 to neutralize this virus is also related to accessibility constraints imposed by other regions of the virus, e.g. V1/V2 (Krachmarov et al., 2005; Krachmarov et al., 2006; Pinter et al., 2004), remains to be investigated. However, it has become clear that the presence of the core epitope of an antibody in the linear envelope sequence does not assure neutralization. This is not only the case for anti-V3 antibodies but has also been demonstrated with antibodies to other regions. For example, in a previous study (Binley et al., 2004), it was shown that the cross-neutralizing anti-gp41 mAb 2F5 was unable to neutralize some viruses despite the presence of the ELDKWA epitope in these viruses (the antibody core epitope DKW is underlined), indicating that the binding of mAb 2F5 to its epitope may also be prevented by quarternary constraints within the context of the viral spike.
TABLE 5.
Comparison of V3 sequences of clade B viruses that were not neutralized by mAb F425-B4e8 to the V3 sequence of JR-CSF.
Dashes denote amino acid homology to the V3 of JR-CSF. Spaces represent a deletion relative to JR-CSF. Mutation of Arg315 (double underline) to Ala severely diminishes F425-B4e8 binding whereas changing Ile309 or Phe317 (single underline) to Ala moderately decreases binding affinity. Sequence alignment was performed using ClustalW (http://www.ebi.ac.uk/clustalw/) and formatted for publication using SeqPublish (http://www.hiv.lanl.gov/content/hiv-db/SeqPublish/seqpublish.html).
For mAb 447-52D, used here as a control to gauge the effects of the amino acid substitutions, replacing Arg315 with either Ala or Gln reduced antibody binding affinity substantially (Table 4). This observation is in agreement with previous mapping studies (Gorny et al., 1992; Keller et al., 1993) as well as NMR and crystallographic analyses of this antibody in complex with a V3 peptide (Rosen et al., 2005; Stanfield et al., 2004). Alanine substitution of residues at positions 304-309, 313, and 319-320 moderately diminished the binding affinity of 447-52D. Pro313 in the V3 sequence is important for proper 447-52D binding interactions (Stanfield et al., 2004) and, thus, a reduction in binding affinity upon mutation to Ala was not surprising. The other reductions were somewhat striking, given that mAb 447-52D interacts for the most part via main-chain contacts with the V3 region; Ala substitutions would not be expected to affect main-chain interactions (Cunningham and Wells, 1989). However, in the NMR structure of mAb 447-52D with a V3 peptide (Sharon et al., 2003), the formation of the β hairpin is stabilized by an extensive network of hydrogen bonds and hydrophobic interactions between the N- and C-terminal segments of the V3 stem. Covariation within the V3 region suggests that these interactions likely play a role in stabilizing the V3 loop (Korber et al., 1993). In the case of mAb 447-52D, these interactions may also be conducive to high-affinity antibody binding, as they likely stabilize the β-strand-β-strand interaction between the antibody heavy chain and V3 (Stanfield et al., 2004); substitution of individual residues flanking the β hairpin turn (without compensatory mutations in the opposite flank) likely distorts this bonding network, thus, affecting 447-52D binding affinity.
Effect of select single-residue substitutions in the V3 region on virus susceptibility to neutralization by mAb F425-B4e8
As noted above, we would have preferred to perform our mutagenesis analyses in the context of the virus. However, this was not pursued due to the diminished and, in some instances, severely impaired infectivity of the mutant viruses relative to wild-type virus (Fig. 1); we reasoned that the lowered infectivity of the viruses might obscure the effects of the mutations on antibody neutralization. In the case of mAb F425-B4e8 in particular, we could not test mutant viruses I309A and F317A due to their substantially reduced level of infectivity. However, because mAb F425-B4e8 bound gp120 with mutations at position 315 (Arg) poorly, we did test the corresponding viruses (R315A and R315Q) for neutralization sensitivity to the antibody, given also that the infectivity of these viruses was least impaired by the introduced mutations. Both viruses were indeed more resistant to neutralization by mAb F425-B4e8; virus R315A was neutralized to only 30% and virus R315Q was neutralized to 19% at the highest concentration of antibody tested here (50 μg/ml), whereas wild-type virus was neutralized at >80% at the same concentration (data not shown). This change in neutralization sensitivity seemed to be limited to the V3 region, given that the neutralization sensitivity of the viruses to mAb b12 remained unchanged (data not shown).
DISCUSSION
Variation in the neutralization breadth of anti-V3 antibodies to HIV-1 is influenced by the fine specificity of the respective antibody vis-à-vis overlap of the antibody epitope with conserved portions of the V3 region that are accessible to the antibody on virions. The aim of this study was to comprehensively assess the neutralization breadth and epitope fine specificity of mAb F425-B4e8, so as to further identify residues within the V3 region that are accessible to antibody on primary viruses and that, thus, might pinpoint segments in V3 that can become targets for rational HIV vaccine design. Although mAb F425-B4e8 had been shown previously to neutralize 2 primary viruses, its neutralization breadth had not been analyzed to date. Here, we determined that this antibody is able to neutralize a modest fraction of viruses (Table 1) and that the neutralizing activity seems at least as good as that of mAb 447-52D, which is considered among the more broadly neutralizing anti-V3 antibodies (Binley et al., 2004; Zolla-Pazner et al., 2004). However, other anti-V3 mAbs have been described more recently that seem to neutralize as well as or even more broadly than mAb 447-52D. MAb 2219, for example, initially did not appear to neutralize as broadly as mAb 447-52D (Gorny et al., 2004; Gorny et al., 2002; Li et al., 2005), but in a more recent study by Gorny et al., using a different selection of viruses, seems to neutralize equivalently to mAb 447-52D (Gorny et al., 2006). In this latter study by Gorny et al., a panel of anti-V3 mAbs is also described that were isolated from subtype A-infected individuals and that seem to neutralize somewhat better than mAb 447-52D (Gorny et al., 2006). However, these mAbs have only been tested on a relatively small panel of viruses so far (in the Gorny et al. study only 11 viruses were tested); thus, it is possible that if other or additional virus strains were included, such as the viruses described in this study, then the neutralization breadth of 447-52D and these antibodies would be similar. In this regard it is worth noting that statistical significance was not achieved in the Gorny et al. study between the neutralization breadth of mAb 447-52D and the newly described panel of non-B anti-V3 mAbs (Gorny et al., 2006).
The ability of mAb F425-B4e8 to neutralize a relatively larger number of viruses may stem from its apparent independence of a precise β-turn conformation at the V3 tip, as evidenced by its ability to bind mutant P313A. Furthermore, the apparent binding affinity of mAb F425-B4e8 relative to wild-type gp120 was not substantially enhanced by any of the Ala substitutions. At present it is not possible to exclude the possibility that, like mAb 447-52D, F425-B4e8 interacts with the N-terminal portion of V3 via main-chain interactions which cannot be revealed by alanine mutagenesis (Cunningham and Wells, 1989).
The vulnerability of HIV to some anti-V3 antibodies that seem to bind mainly the tip segment of V3 is supported by the recently described mAb KD-247, which interacts mainly with residues IGPGR and is not influenced much by mutations in flanking residues (Eda et al., 2006b), and by mAb 447-52D, which has been shown to interact with V3 in a manner that encompasses mainly the tip of V3 (Stanfield et al., 2004). The moderate decrease in binding of mAb 447-52D to several mutants along the N-terminal portion of the V3 stem (Fig. 2) suggests that it is affected more by the destabilization of the hydrogen-bonding network between the N- and C-terminal segments of the V3 stem than mAb F425-B4e8.
Fig. 2. Effects of single-residue substitutions on binding of mAbs F425-B4e8 and 447-52D.
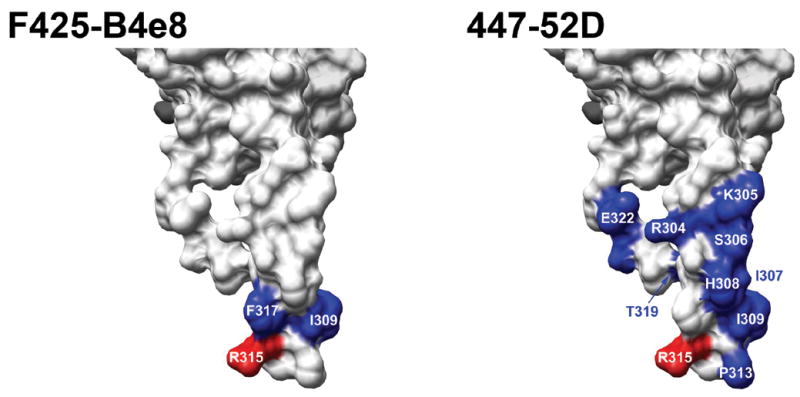
Results from mutagenesis analyses (Table 4) were mapped onto the structure of gp120JR-FL-core+V3 (Huang et al., 2005) (only the V3 portion of the structure is depicted). Only substitutions that substantially affected antibody binding are colored and labeled (i.e., <50% affinity relative to wild type). As in Table 4, substitution of residues colored blue had a moderate effect on binding (10–50% affinity relative to wild type), and those colored red caused a strong decrease in binding (<10% affinity relative to wild type). Molecular graphics images were produced using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. The resulting images were labeled and compiled with Adobe Photoshop CS (v. 8.0).
MAb F425-B4e8 did not neutralize most non-clade B viruses with a Gln instead of an Arg at position 315, as would be expected based on the mutagenesis. However, although the affinity of the antibody for mutant R315Q was >20-fold lower than for wild-type gp120, a high concentration of antibody (>10 μg/ml) did achieve an absorbance that was equal to wild type (data not shown), thus, explaining at least in part its ability to weakly neutralize viruses 93MW960 (IGPGQTF) and 92UG046 (IGLGQAY), which both have GPGQ sequences in V3, as well as mutant virus R315Q. Of the non-clade B viruses that contain an Arg at position 315, only one, virus 92UG005 (IGPGRAY) was neutralized. These observations support the hypothesis that in non-B viruses the conformation of V3 may be somewhat different than in clade B viruses (Gorny et al., 2006; Liao et al., 2006; Stanfield et al., 2006).
The V3 region of HIV-1 gp120 is arguably a potential target for HIV vaccine design (Zolla-Pazner, 2005). It is often presumed that the inability of anti-V3 antibodies to neutralize broadly is largely due to limited accessibility of the V3 region to antibody when V3 is in the context of the functional envelope spike on the viral surface. Indeed, recent studies have shown that V3 may be shielded –at least in part and on certain viruses—by the V1/V2 region (Krachmarov et al., 2006; Pinter et al., 2004), thus, supporting the results from earlier investigations into the functional properties of V1/V2 (Cao et al., 1997; Wyatt et al., 1993). However, it should also be noted that sequence variability in the regions flanking the conserved β-turn in the V3 crown (GPGR sequence) may alter the stability of the turn or alter its surface accessibility (Catasti et al., 1995). The influence of V1/V2 and sequence variation in V3 on the binding interaction of mAb F425-B4e8 with V3 will be subject to further investigation.
Whether an antigen can be designed that elicits antibodies that bind exclusively to the tip of V3, similarly to mAb F425-B4e8 but perhaps with greater neutralization breadth, cannot be answered currently. We are seeking to solve the structure of this antibody in complex with V3 peptides and gp120 to gain further insight into its mode of binding at the atomic level. The crystallographic results will be useful in generating variants of mAb F425-B4e8 that (a) are able to bind clade B viruses more potently, and/or (b) are able to bind V3 sequences with a GPGQ motif with better affinity than the parent antibody. The latter should allow for a continuation of the assessment of the accessibility of the V3 regions in non-subtype B viruses. Broadening the specificity of mAbs has been demonstrated with antibodies to the BAFF (B-cell activating factor of the tumor necrosis factor family) receptor BR3 using phage display (Lee et al., 2006) and, somewhat more recently, Garcia-Rodriguez et al. used yeast display and a co-selection strategy to increase the cross-reactivity of a single chain Fv antibody to botulinum neurotoxin type A (Garcia-Rodriguez et al., 2007). Given a recent report in which immunization with a group M consensus envelope glycoprotein induced a somewhat broad neutralizing antibody response in which ~50% of the average serum neutralizing activity was attributable to anti-V3 antibodies (Liao et al., 2006), information gained from our current and future analyses may positively influence the rational design of V3-based antigens and be valuable in general to further define neutralizing epitopes on the HIV-1 envelope spike.
MATERIALS AND METHODS
MAbs
The anti-V3 mAb F425-B4e8 was isolated from an HIV-1 infected patient by fusion of mononuclear splenocytes with HMMA human myeloma cells (Posner et al., 1989; Posner, Elboim, and Santos, 1987); mAb F425-B4e8 resulted from a fusion of splenocytes stimulated with Epstein Barr virus for 48 h (Cavacini et al., 2003). Hybridomas were selected based on their specific reactivity with HIV-infected cells and ability to capture a relatively high degree of virions in a virus capture assay (Cavacini et al., 2003). The general specificity of mAb F425-B4e8 for the V3 loop was determined based on its binding profile with variable-loop deleted gp120 mutants (Cavacini et al., 2003).
MAb 447-52D, an anti-V3 mAb that is capable of neutralizing a number of mainly subtype B viruses (Binley et al., 2004; Conley et al., 1994), was included in the epitope mapping analysis. This antibody was isolated from an HIV-1 infected individual and selected based on its reactivity with a V3 peptide (Gorny et al., 1992; Gorny et al., 1993). The core epitope of the antibody has been mapped to the GPxR motif at the tip of the V3 region, although some preference is exhibited for certain residues in the N-terminal portion of the V3 stem, as judged by its reactivity profile with phage-displayed peptides (Keller et al., 1993).
Neutralization assays
MAbs were evaluated for their neutralization breadth by employing a high-throughput luciferase-based single-round of infectivity assay with pseudotyped viruses and U87 target cells expressing CD4, CCR5, and CXCR4 (Richman et al., 2003). Serial dilutions of mAb, starting at 50 μg/ml, were incubated for 1 hour with virus after which the mixture was added to target cells. The tested viruses (n=40) are shown in Table 1; they included 3 clade A viruses, 4 CRF01_AE circulating recombinant forms, 16 clade B viruses, 11 clade C viruses, and 6 clade D viruses. All but one of the viruses (NL4.3) was a primary viral strains. The viruses were selected from a 90-member panel used in a recent study by Binley et al. (Binley et al., 2004). Most viruses were from quasispecies derived from primary virus cultures. However, eight of the tested viruses were molecular clones (Table 1); four of these clones were the same as used previously in the Binley et al. study (Binley et al., 2004), whereas the remaining four clones (BG1168, SS1196, 92BR020, 92US712) were new (T. Wrin et al., unpublished results); the neutralization sensitivity of these four latter clones largely reflected the neutralization sensitivity of the viral quasispecies to mAbs 2F5, 2G12, 4E10, b12, and a cross-neutralizing HIV+ plasma termed N16 (T. Wrin et al., unpublished results). To allow for comparison between the results obtained in this study and the results obtained in the Binley et al. study, we included mAb b12 in our analyses (Barbas et al., 1992; Roben et al., 1994) because it neutralizes a broad range of viruses (Binley et al., 2004; Li et al., 2005; Li et al., 2006). MAb F425-B4e8 was also assessed for its ability to neutralize mutant viruses with single-point substitutions in V3 (see below), using an in-house luciferase-based assay with U87 target cells expressing CD4 and CCR5 as described previously (Pantophlet et al., 2003).
Quikchange mutagenesis
For epitope mapping, single amino acid substitutions in V3 were generated using relevant complementary primers by Quikchange II mutagenesis (Stratagene) of the plasmid pSVIII-JR-CSF, which expresses the entire env gene of the primary isolate JR-CSF (Pantophlet et al., 2003). Amino acid substitutions were verified by DNA sequencing.
Generation of V3 mutant pseudovirions
JR-CSF pseudovirions were obtained by transient co-transfection of 293T cells with wild-type or mutant env plasmid and the luciferase reporter plasmid pNL4.3.Luc.R−E− (obtained from the NIH AIDS Research and Reference Reagent Program and contributed by Nathaniel Landau (Connor et al., 1995; He et al., 1995)) using FuGENE (Roche) or polyethylenimine (Kirschner et al., 2006). The culture media was replaced with fresh media ~6 h after transfection when polyethylenimine was used. Supernatants were collected 3 days post-transfection and used immediately for neutralization assays or detergent was added (Empigen; 1% v/v final concentration) and the viral lysates used for ELISA (see below).
ELISAs
To compare the apparent binding affinities of the antibodies for JR-CSF wild-type virus relative to the V3 mutants, ELISA binding assays were performed as described before using detergent-treated supernatants collected from transiently-transfected 293T cells (Pantophlet et al., 2003). Briefly, detergent-containing supernatants, diluted so as to equalize the amount of gp120 in each preparation, were added to ELISA plate wells (Costar, #3690) coated at 5 μg/ml with a monospecific sheep antibody preparation which binds to the C5 region of gp120 (Cliniqa). Anti-V3 mAbs were added to the ELISA plate wells in 5-fold serial dilutions. MAb binding was detected with a peroxidase-conjugated secondary antibody and TMB substrate (Pierce). Absorbances were measured at 450 nm after stopping the color reaction with sulfuric acid (2 M concentration). Apparent affinities were determined as the antibody concentration at half-maximal binding based on ELISA binding curves using the program Graphpad Prism (v. 4.0); antibody affinity for each mutant gp120 relative to wild-type gp120 was calculated as: (apparent affinity for wild-type gp120/apparent affinity for mutant gp120) ×100.
Acknowledgments
We thank Susan-Zolla Pazner for providing mAb 447-52D for initial ELISA binding studies and comments on early drafts of this manuscript. This work was supported by the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium and NIH grant AI33292 (to D.R.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azimzadeh A, Pellequer JL, Van Regenmortel MH. Operational aspects of antibody affinity constants measured by liquid-phase and solid-phase assays. J Mol Recognit. 1992;5(1):9–18. doi: 10.1002/jmr.300050103. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Bjorling E, Chiodi F, Dunlop N, Cababa D, Jones TM, Zebedee SL, Persson MA, Nara PL, Norrby E, et al. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci U S A. 1992;89(19):9339–43. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci U S A. 2005;102(42):14943–8. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–71. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71(12):9808–12. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catasti P, Fontenot JD, Bradbury EM, Gupta G. Local and global structural properties of the HIV-MN V3 loop. J Biol Chem. 1995;270(5):2224–32. doi: 10.1074/jbc.270.5.2224. [DOI] [PubMed] [Google Scholar]
- Cavacini L, Duval M, Song L, Sangster R, Xiang SH, Sodroski J, Posner M. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS. 2003;17(5):685–9. doi: 10.1097/00002030-200303280-00006. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Gorny MK, Kessler JA, 2nd, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68(11):6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–44. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76(17):8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham BC, Jhurani P, Ng P, Wells JA. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243(4896):1330–6. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244(4908):1081–5. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234(3):554–63. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- Dall'Acqua W, Goldman ER, Eisenstein E, Mariuzza RA. A mutational analysis of the binding of two different proteins to the same antibody. Biochemistry. 1996;35(30):9667–76. doi: 10.1021/bi960819i. [DOI] [PubMed] [Google Scholar]
- Dall'Acqua W, Goldman ER, Lin W, Teng C, Tsuchiya D, Li H, Ysern X, Braden BC, Li Y, Smith-Gill SJ, Mariuzza RA. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry. 1998;37(22):7981–91. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- Darbha R, Phogat S, Labrijn AF, Shu Y, Gu Y, Andrykovitch M, Zhang MY, Pantophlet R, Martin L, Vita C, Burton DR, Dimitrov DS, Ji X. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry. 2004;43(6):1410–7. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci U S A. 1996;93(1):7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda Y, Murakami T, Ami Y, Nakasone T, Takizawa M, Someya K, Kaizu M, Izumi Y, Yoshino N, Matsushita S, Higuchi H, Matsui H, Shinohara K, Takeuchi H, Koyanagi Y, Yamamoto N, Honda M. Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J Virol. 2006a;80(11):5563–70. doi: 10.1128/JVI.02095-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda Y, Takizawa M, Murakami T, Maeda H, Kimachi K, Yonemura H, Koyanagi S, Shiosaki K, Higuchi H, Makizumi K, Nakashima T, Osatomi K, Tokiyoshi S, Matsushita S, Yamamoto N, Honda M. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J Virol. 2006b;80(11):5552–62. doi: 10.1128/JVI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25(1):107–16. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66(12):7538–42. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Mascola JR, Israel ZR, VanCott TC, Williams C, Balfe P, Hioe C, Brodine S, Burda S, Zolla-Pazner S. A human monoclonal antibody specific for the V3 loop of HIV type 1 clade E cross-reacts with other HIV type 1 clades. AIDS Res Hum Retroviruses. 1998;14(3):213–21. doi: 10.1089/aid.1998.14.213. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol. 2004;78(5):2394–404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80(14):6865–72. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150(2):635–43. [PubMed] [Google Scholar]
- Haigwood NL, Stamatatos L. Role of neutralizing antibodies in HIV infection. Aids. 2003;17(Suppl 4):S67–71. doi: 10.1097/00002030-200317004-00008. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21(2):171–89. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5(4):579–95. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69(11):6705–11. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PM, Arnold BA, Shaw AR, Tolman RL, Van Middlesworth F, Bondy S, Rusiecki VK, Koenig S, Zolla-Pazner S, Conard P, et al. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology. 1993;193(2):709–16. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr Purif. 2006;48(1):61–8. doi: 10.1016/j.pep.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Korber BT, Farber RM, Wolpert DH, Lapedes AS. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc Natl Acad Sci U S A. 1993;90(15):7176–80. doi: 10.1073/pnas.90.15.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, MacInnes K, Smith RF, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68(10):6730–44. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79(2):780–90. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–35. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laisney IL, Strosberg AD. Dual specificity of a human neutralizing monoclonal antibody, specific for the V3 loop of GP120 (HIV-1) Immunol Lett. 1999;67(3):185–92. doi: 10.1016/s0165-2478(99)00010-3. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, et al. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990;249(4971):932–5. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Weinhold K, Profy AT, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, Bolognesi DP, Matthews TJ, Emini EA, et al. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant: further clarifications. Science. 1991;253(5024):1146. doi: 10.1126/science.1887238. [DOI] [PubMed] [Google Scholar]
- Lee CV, Hymowitz SG, Wallweber HJ, Gordon NC, Billeci KL, Tsai SP, Compaan DM, Yin J, Gong Q, Kelley RF, DeForge LE, Martin F, Starovasnik MA, Fuh G. Synthetic anti-BR3 antibodies that mimic BAFF binding and target both human and murine B cells. Blood. 2006;108(9):3103–11. doi: 10.1182/blood-2006-03-011031. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of acute and early subtype C human immunodeficiency virus type 1 molecular env clones from heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, Ma BJ, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353(2):268–82. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Stoller MO, Wang S, Liu J, Fagan MB, Nunberg JH. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J Virol. 2001;75(22):11146–56. doi: 10.1128/JVI.75.22.11146-11156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP. The reactivities of HIV-1+ human sera with solid-phase V3 loop peptides can be poor predictors of their reactivities with V3 loops on native gp120 molecules. AIDS Res Hum Retroviruses. 1993;9(3):209–19. doi: 10.1089/aid.1993.9.209. [DOI] [PubMed] [Google Scholar]
- Moore JP, Cao Y, Conley AJ, Wyatt R, Robinson J, Gorny MK, Zolla-Pazner S, Ho DD, Koup RA. Studies with monoclonal antibodies to the V3 region of HIV-1 gp120 reveal limitations to the utility of solid-phase peptide binding assays. J Acquir Immune Defic Syndr. 1994;7(4):332–9. [PubMed] [Google Scholar]
- Moore JP, Trkola A, Korber B, Boots LJ, Kessler JA, 2nd, McCutchan FE, Mascola J, Ho DD, Robinson J, Conley AJ. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69(1):122–30. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel GJ, Sullivan NJ. Antibodies and resistance to natural HIV infection. N Engl J Med. 2000;343(17):1263–5. doi: 10.1056/NEJM200010263431711. [DOI] [PubMed] [Google Scholar]
- Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64(12):5701–7. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal G, Kossiakoff AA, Sidhu SS. The functional binding epitope of a high affinity variant of human growth hormone mapped by shotgun alanine-scanning mutagenesis: insights into the mechanisms responsible for improved affinity. J Mol Biol. 2003;332(1):195–204. doi: 10.1016/s0022-2836(03)00898-2. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. Immunofocusing: antigen engineering to promote the induction of HIV-neutralizing antibodies. Trends Mol Med. 2003;9(11):468–73. doi: 10.1016/j.molmed.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(1):642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–15. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner MR, Barrach HJ, Elboim HS, Nivens K, Santos DJ, Chichester CO, Lally EV. The generation of hybridomas secreting human monoclonal antibodies reactive with type II collagen. Hybridoma. 1989;8(2):187–97. doi: 10.1089/hyb.1989.8.187. [DOI] [PubMed] [Google Scholar]
- Posner MR, Elboim H, Santos D. The construction and use of a human-mouse myeloma analogue suitable for the routine production of hybridomas secreting human monoclonal antibodies. Hybridoma. 1987;6(6):611–25. doi: 10.1089/hyb.1987.6.611. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100(7):4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68(8):4821–8. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen O, Chill J, Sharon M, Kessler N, Mester B, Zolla-Pazner S, Anglister J. Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization. Biochemistry. 2005;44(19):7250–8. doi: 10.1021/bi047387t. [DOI] [PubMed] [Google Scholar]
- Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion. Proc Natl Acad Sci U S A. 2006;103(38):13950–5. doi: 10.1073/pnas.0606312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Wachsmuth C, Muller H, Odemuyiwa S, Schmitz H, Meyer S, Meyer B, Schneider-Mergener J. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J Virol. 1997;71(12):9198–205. doi: 10.1128/jvi.71.12.9198-9205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman SJ, Binley JM, Gorny MK, Burton DR, Zolla-Pazner S, Sokolowski KA. Characterization by serial deletion competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol Immunol. 1996;33(9):737–45. doi: 10.1016/0161-5890(96)00044-2. [DOI] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure (Camb) 2003;11(2):225–36. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Srivastava IK, Ulmer JB, Barnett SW. Neutralizing antibody responses to HIV: role in protective immunity and challenges for vaccine design. Expert Rev Vaccines. 2004;3(4 Suppl):S33–52. doi: 10.1586/14760584.3.4.s33. [DOI] [PubMed] [Google Scholar]
- Stanfield R, Cabezas E, Satterthwait A, Stura E, Profy A, Wilson I. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs. Structure Fold Des. 1999;7(2):131–42. doi: 10.1016/s0969-2126(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Ghiara JB, Ollmann Saphire E, Profy AT, Wilson IA. Recurring conformation of the human immunodeficiency virus type 1 gp120 V3 loop. Virology. 2003;315(1):159–73. doi: 10.1016/s0042-6822(03)00525-7. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure (Camb) 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens FJ. Modification of an ELISA-based procedure for affinity determination: correction necessary for use with bivalent antibody. Mol Immunol. 1987;24(10):1055–60. doi: 10.1016/0161-5890(87)90073-3. [DOI] [PubMed] [Google Scholar]
- Suphaphiphat P, Essex M, Lee TH. Mutations in the V3 stem versus the V3 crown and C4 region have different effects on the binding and fusion steps of human immunodeficiency virus type 1 gp120 interaction with the CCR5 coreceptor. Virology. 2007;360(1):182–190. doi: 10.1016/j.virol.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Ho DD, Robinson J, Tilley S, Pinter A, Sodroski J. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 1992;66(9):5635–41. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Olshevsky U, Furman C, Gabuzda D, Posner M, Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol. 1991;65(11):6188–93. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J Mol Biol. 2002;320(2):415–28. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- Wang WK, Dudek T, Essex M, Lee TH. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci U S A. 1999;96(8):4558–62. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97(16):8950–4. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Scharf ME, Potts BJ, Smith LM, Sokolowski KA, Rusche JR, Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993;192(1):197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67(8):4557–65. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies. 2005;14(3–4):69–72. [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–8. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol. 2005;79(2):1252–61. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Komori HK, Stanfield RL, Church S, Wang M, Parren PW, Kunert R, Katinger H, Wilson IA, Burton DR. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J Virol. 2004;78(6):3155–61. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Parren PW, Saphire EO, Church S, Wang M, Scott JK, Dawson PE, Wilson IA, Burton DR. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(10):5863–76. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]