Abstract
The structure of the proline-specific aminopeptidase (EC 3.4.11.9) from Escherichia coli has been solved and refined for crystals of the native enzyme at a 2.0-Å resolution, for a dipeptide-inhibited complex at 2.3-Å resolution, and for a low-pH inactive form at 2.7-Å resolution. The protein crystallizes as a tetramer, more correctly a dimer of dimers, at both high and low pH, consistent with observations from analytical ultracentrifuge studies that show that the protein is a tetramer under physiological conditions. The monomer folds into two domains. The active site, in the larger C-terminal domain, contains a dinuclear manganese center in which a bridging water molecule or hydroxide ion appears poised to act as the nucleophile in the attack on the scissile peptide bond of Xaa-Pro. The metal-binding residues are located in a single subunit, but the residues surrounding the active site are contributed by three subunits. The fold of the protein resembles that of creatine amidinohydrolase (creatinase, not a metalloenzyme). The C-terminal catalytic domain is also similar to the single-domain enzyme methionine aminopeptidase that has a dinuclear cobalt center.
Proline-specific peptidases have been described in a wide variety of organisms and specifically cleave either the amide bond after a proline residue or the imide bond that precedes it (1). Proline aminopeptidases (EC 3.4.11.9) specifically release the N-terminal residue from a peptide where the penultimate residue is proline. These enzymes have significant sequence homology with (i) some other amino peptidases (e.g., methionine aminopeptidase, AMPM) and (ii) the Xaa-Pro dipeptidases, prolidases, that specifically cleave Xaa-Pro dipeptides. All these enzymes are activated in the presence of divalent metal ions, though the biologically active metal is not always certain. In vitro the prolyl peptidases are most active in the presence of Mn2+. Structural comparisons between AMPM and creatine amidinohydrolase (creatinase) and sequence comparisons between these proteins and aminopeptidase P (AMPP) and prolidase have suggested that the catalytic domains of all four enzymes have a common fold (2). This work presents the crystal structure of AMPP, the product of the pepP gene in Escherichia coli. The structures of a complex of AMPP with a dipeptide inhibitor and a low-pH inactive form are used to devise a plausible mechanism for peptide hydrolysis.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of AMPP.
AMPP was originally isolated and characterized in E. coli (3) and was subsequently cloned and overexpressed (4). The details of the overexpression and purification of AMPP used in this work will be published elsewhere. Briefly, AMPP was overexpressed in the E. coli AN1459/pPL670. Lysed cells were subject to (NH4)2SO4 precipitation at 4°C followed by centrifugation. The pellet was resuspended and applied to a DEAE-Fractogel column (Merck). The AMPP-containing fraction was then passed over a ceramic hydroxyapatite column (Bio-Rad). This simple purification scheme provided pure protein ready for concentration and crystallization trials.
Crystallization of AMPP.
Crystals of AMPP were grown by using vapor diffusion in 6-μl hanging drops containing 1:1 mixtures of protein and well solutions. Hexagonal crystals, space group P6422, a = 178.4, and c = 95.6 Å, were grown at 4°C near pH 8.5, the optimum for the activity of the enzyme (4). The protein solution contained 8.7 mg of protein per ml in 50 mM Tris (pH 8.3) with 5 mM MnCl2; and the reservoir solution was 30–33% polyethylene glycol 4,000 in 100 mM Tris (pH 8.3). Small crystals (<0.02 mm, maximum dimension), which grew within 1 month, were used for macroseeding into drops that had been preequilibrated for 7 days, resulting in hexagonal rods with dimensions of 0.2 × 0.1 × 0.1 mm3. Tetragonal crystals, space group I4122 with cell dimensions a = 139.7 and c = 230.9 Å, were grown from a solution containing protein at 8.7 mg/ml in 50 mM sodium acetate (pH 4.6–5.5) with 5 mM MnCl2 and a reservoir solution with 20–30% 2-methyl-2,4-propanediol (MPD). The crystals were bipyramidal, about 0.2 × 0.2 × 0.2 mm3, and grew in several days. The pH at which these crystals grew is 3 to 4 units below the optimal pH for AMPP activity. Because these crystals were grown from MPD, no additional cryoprotectant was necessary for flash freezing.
X-Ray Data Collection.
Data were recorded with a Rigaku R-Axis IIc imaging plate detector mounted on a Rigaku RU-200 rotating anode generator with a Cu target (λ = 1.5418 Å) and focusing mirror optics (Z. Otwinowski and G. Johnson, Yale University; as marketed by Molecular Structure, The Woodlands, TX). Flash freezing was carried out in a stream of cold nitrogen gas. Data were integrated and scaled with denzo and scalepack (6).
The data used in the structure solution were recorded at room temperature because of lack of isomorphism in flash-frozen crystals of potential derivatives. The data used for the refinement of the native structures in the two space groups and of the inhibitor complex were recorded from flash-frozen crystals at −160°C. The hexagonal crystals were briefly soaked in reservoir solution before being transferred to the cryoprotectant solution, which was the reservoir solution with the addition of 20% 2-propanol.
Structure Solution and Refinement.
The structure of the native protein at high pH was solved by multiple isomorphous replacement. The locations of the heavy atoms of the di-μ-iodobis(ethylenediamine)diplatinum(II) (PIP) derivative were found by using rsps (7) and were confirmed by inspection of the appropriate Harker sections of difference Patterson maps. Heavy atom positions were refined and phases calculated with mlphare (7). The phases derived from the PIP derivative were used to locate the heavy atom sites of a number of other derivatives in cross-difference Fourier syntheses. Although only the PIP derivative provided any significant phasing power, it was found that removal of any of the other derivatives produced maps that were not as readily interpretable, particularly when density modification with dm (7) was used to improve and extend the phases. The correct space-group enantiomer P6422 was chosen by inspection of electron-density maps. The structure at room temperature was completed by using data, 50 Å ≥ d ≥ 3 Å, with cycles of manual model building using o (8) and refinement using refmac (7). The refinement was then continued by using the data recorded from flash-frozen crystals to a resolution of 2.0 Å. Rigid-body refinement was performed to allow for slight changes in unit-cell dimensions. Three cycles of manual model building and positional refinement with refmac were followed by further cycles of building and refinement with x-plor (9). arpp (10) was used for the automatic selection of potential water sites. Each of these sites was checked by the inspection of omit difference electron density maps. Bulk solvent contributions were included in all calculations, and all electron-density maps were calculated by using σA weights (11). The final model contains all 440 amino acid residues of AMPP and 742 water molecules with refined temperature factor ≤65 Å2. Two loops, residues 68–71 and 85–89, were in weak electron density. No electron density was visible for the side chain of Glu-393 or for either the main chain or side chain of His-243. Ala-37 has φ and ψ angles that are unusual in protein structures but lies in well-defined electron density. No other residue lies in a disallowed region of a Ramachandran plot calculated with procheck (12) (91.7% in most favored regions). The crystallographic analysis is summarized in Table 1. Both the temperature factors and occupancies of the water molecules were refined. This procedure was justified by a lack of correlation between the refined occupancy and temperature factors (14). The calculated volume fraction of the crystals occupied by solvent is 72%. Because this value lies outside the “normal” range reported by Matthews (15), extensive checks were made in electron-density maps for an additional protein molecule in the asymmetric unit. No additional nonsolvent atoms were found.
Table 1.
Crystallographic data and refinement statistics
| Native-1 | Native | PIP-1 | ProLeu-1 | ProLeu-2 | Native-2 | |
|---|---|---|---|---|---|---|
| Space group | P6422 | P6422 | P6422 | P6422 | P6422 | I4122 |
| pH | 8.3 | 8.3 | 8.3 | 8.3 | 8.3 | 4.6 |
| Temperature, °C | −160 | 20 | 20 | −160 | −160 | −160 |
| Resolution limit, Å | 2.0 | 3.0 | 3.0 | 2.3 | 2.7 | 2.7 |
| No. observations | 1,132,574 | 113,398 | 169,957 | 549,017 | 370,031 | 344,400 |
| Unique reflections, no. | 58,976 | 17,986 | 17,542 | 36,900 | 24,537 | 26,338 |
| Completeness, %* | 97.2 (95) | 93.9 (96) | 91.0 (92) | 91.9 (92) | 97.2 (97) | 83.1 (67) |
| Rmerge, %† | 5.1 (24.5) | 15.1 (40.8) | 12.5 (34.2) | 9.6 (37.5) | 10.0 (33.7) | 9.3 (29.0) |
| Rcentric, %‡ | — | — | 0.89 | — | — | — |
| Phasing power§ | — | — | 0.64 | — | — | — |
| Rscale, %¶ | — | — | 14.1 | 13.7 | 14.2 | — |
| Soak time | — | — | 1 h | 3 days | 5 days | — |
| R, %‖ | 16.5 | 20.0 | — | 18.4 | 19.5 | 18.2 |
| Rfree, %** | 19.2 | 24.6 | — | 22.6 | 22.5 | 22.7 |
| rmsΔ bond length, Å | 0.011 | 0.007 | — | 0.008 | 0.007 | 0.008 |
| rmsΔ bond angles, degrees | 1.7 | 1.4 | — | 1.4 | 1.3 | 1.5 |
| 〈B〉, Å2 | 25.4 | {20.0} | — | 29.2 | 31.5 | 28.3 |
| rmsΔ bonded B, Å2 | 2.2 | — | — | 3.2 | 3.7 | 3.1 |
| DPI, ҇ | 0.103 | 0.411 | — | 0.185 | 0.328 | 0.340 |
| No. protein atoms | 3,504 | 3,488†† | — | 3,504 | 3,504 | 3,504 |
| No. nonprotein atoms | 750 | 2 | — | 457 | 212 | 314 |
Values for the highest resolution shell are shown in parentheses.
Rmerge = Σ|Ii − 〈Ii〉|/Σ|〈Ii〉|, where Ii is the intensity measurement for the reflection i and 〈Ii〉 is the mean intensity calculated for refection i from multiple measurements.
Rcentric = Σ||FH(obs)|−|FH(calc)||/Σ|FH(obs)|for centric reflections, where FH is the structure factor for the heavy atom.
Phasing power = 〈FH〉/E, where 〈FH) is the rms heavy atom structure factor and E is the residual lack of closure.
Rscale = Σ||FPH|−|FP||/Σ|FP|, where FPH and FP are the derivative and native structure factors, respectively.
R = Σ||FOBS|−|FCALC||/Σ|FOBS|.
Rfree is R calculated for 5% of the reflections, which were omitted throughout the refinement.
DPI = diffraction precision indicator = 1.0(N/P)1/2C−1/3dminR, where N = no. of atoms, P = no. of degrees of freedom (no. of independent reflections − no. of variables), C = completeness of the data, dmin = resolution (13).
Model was still incomplete at the conclusion of the room-temperature refinement.
The structure of the tetragonal crystals of the native protein at low pH was solved by molecular replacement with amore (7) using normalized structure factor amplitudes and with the high-pH structure, omitting water molecules and metal ions, as the search model. The highest peak (10.3σ) in the rotation function was ultimately found to be the correct solution. The translation search was straightforward, with the lowest R value solution being correct. After rigid-body refinement, the structure was subjected to molecular dynamics simulated annealing from 3,000 K to reduce model phase bias. The refinement was completed in a manner similar to that described for the native high-pH structure. At an early stage of refinement, it was noticed that electron density in a bridging position with respect to the two metal atoms was more extended than density that had been interpreted as a bridging water molecule or hydroxide ion in the high-pH structure analysis. Given the presence of 50 mM sodium acetate in the crystallization buffer, this electron-density feature was modelled as an acetate ion. With one molecule in the asymmetric unit, the solvent content of the crystals was 80%. The two loops at residues 68–71 and 85–89, which were in weak density in the hexagonal structure, still had high B values but lay in significant-difference electron density. There was no side-chain density for the following residues: Arg-5, Arg-305, Lys-327, Glu-337, Lys-439, and Gln-440 (whole residue). A Ramachandran plot (data not shown) placed 92.4% of the residues in most favored regions. As in the high-pH structure, Ala-37 lies outside the allowed regions.
Complex with an Inhibitor.
The dipeptide Pro-Leu is known to be a competitive inhibitor of AMPP (16). Because of the insolubility of Pro-Leu in the crystallization buffer, a complex was formed by adding a few grains of Pro-Leu (Sigma) to hanging drops (33% polyethylene glycol/0.1 M Tris, pH 8.5) containing a single crystal of the native high-pH form of AMPP. The crystals were kept overnight at 4°C and then placed at room temperature for 3–5 days to aid dissolution of the peptide. The crystals were extremely fragile after soaking. They were washed briefly in cryoprotectant solution before freezing. Data sets to 2.3-Å and 2.7-Å resolution, respectively, were recorded from a crystal (ProLeu-1) soaked for 3 days and from a crystal (ProLeu-2) soaked for 5 days. The crystallographic and refinement statistics are shown in Table 1. The starting model for refinement of this structure was the refined model of AMPP at high pH with all water molecules removed. After rigid-body refinement, difference electron-density maps clearly showed the location of Pro-Leu in the active site. The ProLeu-1 and -2 data sets were used to fit and refine models independently in a manner similar to that described for the native structure. The complete chain of 440 amino acids was readily traceable for both ProLeu-1 and -2. Because of the lower resolution of the two data sets, the occupancies of water molecules were not refined and were set to 1.0. The occupancy of the dipeptide was lower in ProLeu-1, consistent with the shorter soaking time.
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed with a Beckman Optima XL-A analytical ultracentrifuge. Experiments were carried out with 125-μl samples of AMPP at 0.06, 0.03, and 0.015 g/liter in 100 mM Tris (pH 8.5) using six-channel centerpieces. Absorption versus radius scans were collected at 9,000 rpm at 4°C with intervals of 0.001 cm using 10-fold averaging. These data were collected at 4-h intervals until successive scans gave identical profiles, indicative of equilibrium. The experiments were repeated in the presence of (i) 1 mM MnCl2, (ii) EDTA, and (iii) about 1 mM Pro-Leu. Data were analyzed to determine molecular weight and association constants with the program origin 2.94 (Microcal, Northampton, MA).
RESULTS
The structures of AMPP at high and low pH are identical within the limits of precision (rms deviation for the 440 Cα positions is 0.3 Å). Except where stated otherwise, all descriptions refer to the higher-resolution high-pH structure. The high- and low-pH crystals contain the same homotetramer, in which the monomers are related by crystallographic 222 symmetry. The result is a dimer of dimers, the monomer–monomer interface within a dimer being different from the interface between dimers. The presence of tetramers in the crystals is consistent with the molecular mass of AMPP in solution, as estimated by gel filtration (4) and by ultracentrifugation (present work). The ultracentrifugation experiments leave little doubt that the active form of AMPP at biologically significant concentrations is a tetramer. The monomer–monomer association constant is too high to permit monomers to be detected by ultracentrifugation in even the most dilute solutions (3 × 10−7 M) used in the experiments. The dimer–tetramer association constant is >1 × 106 M−1, calculated for a fixed dimer molecular mass of 99,300 Da. The sedimentation results were not changed in the presence of Mn2+ ions or an inhibitor.
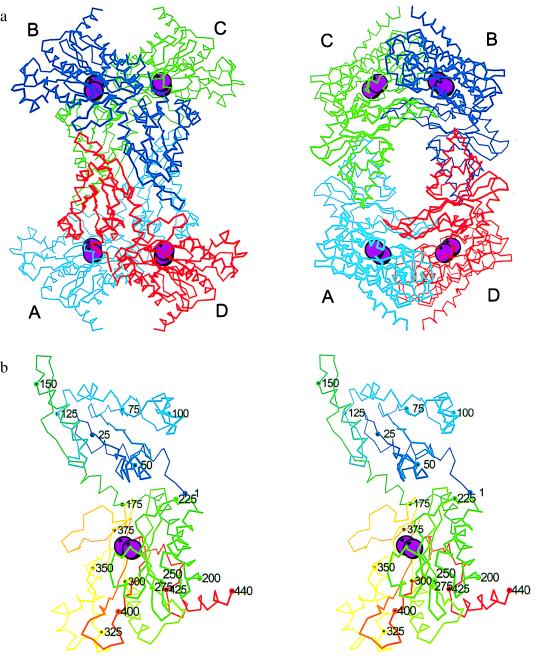
The tetramer has approximate overall dimensions 125 × 100 × 80 Å3 with a large central hole (Fig. 1a). The four subunits are labeled A, B, C, and D. These letters are used to prefix residue numbers when referring to subunits other than A. The surface area buried between subunits A and B is 650 Å2, whereas the surface between subunits A and D is 2,111 Å2. Pairwise superpositions of the AB and AD dimers in the high-pH and low-pH structures indicate that the more extensive AD interface is identical in both forms, whereas the AB interface is slightly different. These changes result in a slight expansion of the tetramer in the tetragonal low-pH structure (differences in the distances between the centroids in the tetramer in the high- and low-pH forms are as follows: A–B = 0.5 Å, A–C = 0.5 Å, and A–D = 0.04 Å).
Figure 1.
(a) Orthogonal views of the tetramer of AMPP. Chain A is cyan, chain B is blue, chain C is green, and chain D is red. Mn2+ ions in the dinuclear active site are enlarged magenta spheres. (b) Stereo Cα trace of AMPP. Residues are color ramped from blue at the N terminus to red at the C terminus.
The polypeptide fold of AMPP is shown in Fig. 1b. AMPP has two domains: domain I—residues 1–174; domain II—residues 175–440. Domain I consists of a central six-stranded mixed β-sheet flanked by four α-helices, two on each side. These β-sheets are extended to 12 strands by the subunit interfaces of type AB in the tetramer. The fourth α-helix lies on the outer edge of this long β-sheet. There are two arms (residues 31–44 and 81–92) whose ends come close to the active sites in domain II of an adjoining subunit. Domain II consists of a strongly curved mixed six-stranded β-sheet with seven α-helices on the “outer” convex face of the sheet. The four longer α-helices run parallel to the β-strands. The C-terminal α-helix lies nearly perpendicular to the strands of the sheet.
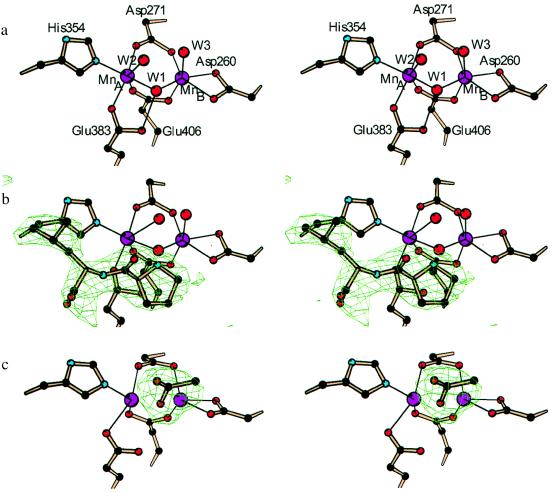
The active site of AMPP is located in a oval depression on the inner surface of the β-sheet of domain II (Fig. 1b). The ends of the depression are defined by two loops (residues 240–245 and 355–362). The central feature of the catalytic site is a dinuclear metal cluster (Fig. 2a). X-ray spectroscopic measurements have shown that the metal is manganese and that both atoms are in the +2 oxidation state (L. Zhang, M. J. Crossley, N.E.D., P. J. Ellis, M. L. Fisher, G. F. King, P.E.L., D. MacLachlan, R. J. Pace, and H.C.F., unpublished results). The presence of 5 mM MnCl2 in the crystallization buffers, and the heights of peaks in electron-density difference maps, are consistent with the presence of two Mn2+ ions in the crystals. The Mn–Mn distance of 3.3 Å is consistent with the value 3.5 Å found by electron paramagnetic resonance (EPR) measurements (L. Zhang, M. J. Crossley, N.E.D., P. J. Ellis, M. L. Fisher, G. F. King, P.E.L., D. MacLachlan, R. J. Pace, and H.C.F., unpublished results). Each of the Mn2+ ions is in a distorted octahedral environment (Table 2 and Fig. 2a). MnA is coordinated by the side chains of Asp-271, Glu-383, His-354, and Glu-406 and two water molecules. MnB is coordinated by Asp-260, Asp-271, and Glu-406 and two water molecules. Asp-271 and Glu-406 are bidentate bridging ligands of the Mn2+ ions. Asp-260 is bound in a bidentate fashion to MnB. The bridging solvent molecule W1 is identified as a hydroxide ion in view of the pH at which the crystals were grown. His-361 makes a hydrogen bond from its Nɛ2 atom to the water coordinated axially to MnA, and a hydrogen bond or salt bridge from its Nδ1 atom to Asp-D38 Oδ2 in a neighboring molecule. The dinuclear site is similar to that in AMPM, where the two Co2+ ions are coordinated by sequentially equivalent protein residues (17). In the structure analysis of AMPM, no coordinated water molecules were identified, possibly due to the lower resolution of the data, but the presence of additional water ligands was inferred (17). Also noteworthy are features that the metal site in AMPP shares with rat liver arginase (18): a MnA2+–MnB2+ separation of ≈3.3 Å and a bridging hydroxide ion that is hydrogen bonded to a carboxyl whose second oxygen coordinates to MnA2+.
Figure 2.
(a) Stereo view of the catalytic site of AMPP for native crystals at pH 8.3. Residues are colored by atom type. (b) Stereo view of the catalytic site of AMPP for crystals at pH 8.3 showing omit difference electron density for the inhibitor Pro-Leu contoured at 4σ. (c) Stereo view of the catalytic site of AMPP for crystals at pH 4.6 showing omit difference electron density for coordinated acetate ion contoured at 4σ.
Table 2.
Manganese ligand distances
| Ligand | MnA | MnB | Ligand | MnA | MnB |
| Asp-260 Oδ1 | 2.1 | Glu-406 Oɛ1 | 2.2 | ||
| Asp-260 Oδ2 | 2.4 | Glu-406 Oɛ2 | 2.2 | ||
| Asp-271 Oδ1 | 2.1 | W1 | 2.1 | 2.2 | |
| Asp-271 Oδ2 | 2.1 | W2 | 2.8 | ||
| His-354 Nɛ2 | 2.2 | W3 | 2.4 | ||
| Glu-383 Oɛ1 | 2.2 | MnA or B | 3.3 | 3.3 | |
Values in Å are for the 2.0-Å resolution refinement of AMPP at pH 8.3.
In the tetragonal low-pH crystal form the hydroxide ion bridging the two Mn2+ ions is replaced by one oxygen atom of an acetate ion (Fig. 2c). The other oxygen atom of the acetate ion forms a hydrogen bond or salt bridge with the Nɛ2 atom of His-361. The axial water molecules seen in the native structure at high pH are not seen in the low-pH structure, possibly because of the lower resolution of the structure analysis. On the other hand there is weak but significant density for His-243 that is apparently completely disordered in the high-pH structure.
The structure of the Pro-Leu complex shows that the proline residue of the dipeptide lies immediately above the bridging hydroxide ion (Fig. 2b). As in the low-pH form, omit electron density indicates that His-243 is at least partly ordered. The peptide oxygen atom of Pro-Leu forms a long hydrogen-bonded contact to Nɛ2 of His-243, and the terminal imino nitrogen atom of the proline residue forms a short contact to the bridging hydroxide ion. There is an additional hydrogen bond to a water molecule that is in an axial direction with respect to MnA but too distant to coordinate. The carboxyl group of the dipeptide forms hydrogen bonds or salt bridges with the side chains of Arg-370 and Arg-B153. Other residues that contact Pro-Leu include His-350 and Arg-404, which pack against the ring of Pro, and His-361 and His-354, which pack around the Leu side chain. Residues from a neighboring subunit that extend the rim of the active-site pocket are Asp-D38, Trp-D88, and Phe-D89.
DISCUSSION
The present work reveals a number of striking similarities for AMPP, AMPM, and creatinase. The catalytic C-terminal domains of the three proteins have the same pita-bread fold, as predicted from an earlier sequence comparison and from the structures known at the time (2). In view of the similarity between the catalytic domains, it is not surprising that the dinuclear Mn2+ center in AMPP has ligands equivalent to those at the dinuclear Co2+ center in AMPM. What was not predicted is that the N-terminal domains of AMPP and creatinase also have similar folds. In fact, the more stable dimer in the tetramer of AMPP uses the same interface as the dimer of creatinase, but the angle between the monomers is different. The structural similarities between AMPP and creatinase do not reflect a high level of sequence identity. In a clustal (19) sequence alignment (data not shown) of AMPP, AMPM, creatinase, and prolidases from Homo sapiens, E. coli, and Lactobacillus delbrueckii, there are only 13 totally conserved residues. AMPP and creatinase have the lowest level of sequence identity and yet have a common structure. Pair-wise sequence comparisons among AMPP and the prolidases yield sequence identities of 24–30% (L. Zhang, M. J. Crossley, N.E.D., P. J. Ellis, M. L. Fisher, G. F. King, P.E.L., D. MacLachlan, R. J. Pace, and H.C.F., unpublished results), making it probable that all these proteins have structures similar to AMPP.
If we assume that there is significant structural homology among the enzymes listed above, then how do these molecules distinguish between substrates of different sizes? We suggest that the aggregation of the enzyme molecules into various types of oligomer is involved. AMPM, whose active site can accommodate (and must be accessible to) large polypeptide substrates, is a monomer (17). Creatinase, which acts only on small substrates, is a dimer in which the volume and shape of the active site pocket are partly defined by specific contacts with residues in the neighboring subunit (20, 21). The same mechanism can, in principle, account for the strong preference of prolidases for di- rather than oligopeptide substrates, because the active form of these enzymes is also reported to be a dimer (22, 23). Finally, in AMPP, each active site in the dimer of dimers is defined partly by the residues of a neighboring subunit. Although the overall fold of AMPP is similar to that of creatinase, the specific interactions between the subunits leave the active sites in AMPP more exposed, consistent with a preference for oligopeptide substrates and an ability to cleave even larger polypeptides.
AMPP is one of a number of structurally characterized enzymes that contain a dinuclear metal (Mg2+, Mn2+, Fe2+/3+, Co2+, Ni2+, or Zn2+) site (24). The choice of a particular metal ion and the nature of the enzyme reaction are not readily correlated with the primary structures of the proteins. AMPP and AMPM have chemically and sequentially identical metal ligands, and yet the former is activated by Mn2+ and the latter by Co2+. Furthermore, AMPP and AMPM are aminopeptidases. On the other hand, AMPP and rat liver arginase (18, 25, 26) have dinuclear Mn2+ sites, despite the facts that there is no detectable sequence homology between them and that they have different combinations of metal-ligand residues. The fact that the ligating side chains in arginase are not implicated directly in the proposed enzyme mechanism (18) supports the view that neither the metal nor the ligating groups are directly responsible for the nature of the specific chemical reaction catalyzed by the enzyme. The requirement for a particular metal is more likely to be correlated with the optimal pH for the enzymatic reaction.
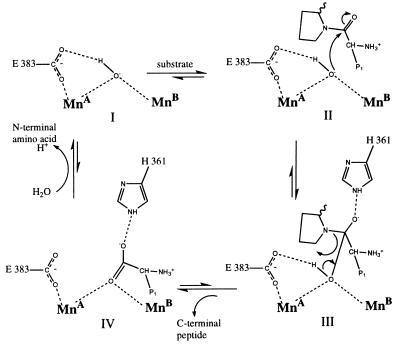
Given the similar pH rate profiles of AMPP (27) and arginase (28), it is most likely that the ligand that bridges the two Mn2+ ions in the active form of the enzymes is a hydroxide ion rather than a water molecule (18). It has been suggested that in arginase this hydroxide acts as the nucleophile (18), and our structures support this hypothesis for AMPP. The bridging hydroxide ion is perfectly placed to act as the nucleophile. The dipeptide inhibitor in the AMPP–Pro-Leu complex is precisely where the C-terminal peptide of a cleaved substrate would be if it remained bound to the enzyme (Fig. 3). When a model of a peptide substrate, Xaa-Pro-Leu, is superposed on the Pro-Leu inhibitor, the carbon atom of the scissile peptide group is 2.3 Å from the hydroxide, and the oxygen atom of the scissile peptide group is within hydrogen bonding distance of Nɛ2 of His-361. Thus the conditions for the stabilization of an oxyanion intermediate exist (Fig. 3, III). The position of His-361 is fixed partly by a hydrogen bond or salt bridge to Asp-D38 in a neighboring subunit. The close contact between Glu-383 and the bridging hydroxide ion suggests that a hydrogen bond exists between them. This would also stabilize the reaction intermediate (Fig. 2a), as has been proposed for arginase. There is no experimental evidence that the hydroxide ion is also the source of the proton (Fig. 3, III), but by reason of proximity and simplicity, it is a likely candidate. In a role analogous to that of the nonligand histidine residues in arginase (18), the conserved residues His-243, His-350, and His-361 may shuttle protons from the dinuclear center to the solvent.
Figure 3.
Proposed mechanism for the nucleophilic attack by the bridging hydroxide ion on a peptide, P1-Pro-. The leaving peptide is probably protonated by a shuttle from the solvent. Surrounding residues and other Mn ligands are omitted for clarity.
Unlike several other proposals (29, 30), the mechanism outlined in Fig. 3 does not require coordination of substrate to the metal ions. In this regard it is consistent with EPR and x-ray spectroscopic evidence (L. Zhang, M. J. Crossley, N.E.D., P. J. Ellis, M. L. Fisher, G. F. King, P.E.L., D. MacLachlan, R. J. Pace, and H.C.F., unpublished results) that three known inhibitors either do no bind directly to the Mn centers or substitute existing ligands with little change in coordination. The role of the metal ions is presumably to increase the acidity of the bridging water ligand, as has been proposed for arginase (31). The fine tuning is provided by the other metal ligands and their hydrogen bonding networks. There is no equivalent in AMPP to the MnA 190 His⋅⋅⋅Asp⋅⋅⋅Ser network of hydrogen bonds in arginase (25). The corresponding histidine ligand in AMPP His-354 forms a hydrogen bond to a carbonyl oxygen of the peptide backbone.
The basis of the specificity of AMPP and prolidases for substrates with a proline residue C-terminal to the scissile peptide bond of the substrate is not apparent from the present structure. The cis peptide configuration facilitated by proline cannot be the reason for the specificity, because it has been shown that AMPP (32) and prolidase (5) are specific for trans peptide linkages. We note that seven of the eight residues that are conserved in AMPP and prolidases but not in creatinase or in AMPM (Leu-258, His-350, Gly-385, Tyr-387, Ile-405, Gly-262, Ala-269, and Val-360) surround the dinuclear active site, suggesting that they play a role in proline specificity. However, only His-350 directly contacts the inhibitor in the structure of the Pro-Leu dipeptide–inhibitor complex.
Acknowledgments
We are grateful to Ms. Katrina Giles for many crystallization experiments. This work was supported by a grant from the Australian Research Council (A29601726) to H.C.F. and J.M.G.
ABBREVIATIONS
- AMPM
methionine aminopeptidase
- AMPP
aminopeptidase P
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference nos. 1AZ9, 1A16, and 1JAW).
References
- 1.Yaron A, Naider F. Crit Rev Biochem Mol Biol. 1993;28:31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]
- 2.Bazan J F, Weaver L H, Roderick S L, Huber R, Matthews B W. Proc Natl Acad Sci USA. 1994;91:2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaron A, Mlynar D. Biochem Biophys Res Commun. 1968;32:658–663. doi: 10.1016/0006-291x(68)90289-1. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimoto T, Murayama N, Honda T, Tone H, Tsuru D. J Biochem (Tokyo) 1988;104:93–97. doi: 10.1093/oxfordjournals.jbchem.a122430. [DOI] [PubMed] [Google Scholar]
- 5.Lin L-L, Brandts J F. Biochemistry. 1979;18:43–47. doi: 10.1021/bi00568a007. [DOI] [PubMed] [Google Scholar]
- 6.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 7.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 8.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 9.Brunger A T. x-plor, Version 3.1: A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 10.Lamzin V S, Wilson K S. Acta Crystallogr D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 11.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 12.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 13.Cruickshank D W J. In: Macromolecular Refinement, Proceedings of the CCP4 Study Weekend January 1996. Dodson E, Moore M, Ralph A, Bailey S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1966. pp. 11–22. [Google Scholar]
- 14.Sielecki A R, James M N G. In: Refinement of Protein Structures. Machin P A, Campbell J W, Elder M, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1981. pp. 78–87. [Google Scholar]
- 15.Matthews B W. In: The Proteins. Neurath H, Hill R L, editors. III. New York: Academic; 1977. pp. 468–477. [Google Scholar]
- 16.Yoshimoto T, Orawski A T, Simmons W H. Arch Biochem Biophys. 1994;311:28–34. doi: 10.1006/abbi.1994.1204. [DOI] [PubMed] [Google Scholar]
- 17.Roderick S L, Matthews B W. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 18.Kanyo Z F, Scolnick L R, Ash D E, Christianson D W. Nature (London) 1996;383:554–557. doi: 10.1038/383554a0. [DOI] [PubMed] [Google Scholar]
- 19.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 20.Hoeffken H W, Knof S H, Bartlett P A, Huber R. J Mol Biol. 1988;204:417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- 21.Coll M, Knof S H, Ohga Y, Messerschmidt A, Huber R. J Mol Biol. 1990;214:597–610. doi: 10.1016/0022-2836(90)90201-v. [DOI] [PubMed] [Google Scholar]
- 22.Mock W L, Green P C. J Biol Chem. 1990;265:19606–19610. [PubMed] [Google Scholar]
- 23.Yoshimoto T, Matsubara F, Kawano E, Tsuru D. J Biochem (Tokyo) 1983;94:1889–1896. doi: 10.1093/oxfordjournals.jbchem.a134542. [DOI] [PubMed] [Google Scholar]
- 24.Wilcox D E. Chem Rev. 1996;96:2435–2458. doi: 10.1021/cr950043b. [DOI] [PubMed] [Google Scholar]
- 25.Scolnick L R, Kanyo Z F, Cavalli R C, Ash D E, Christianson D W. Biochemistry. 1997;36:10558–10565. doi: 10.1021/bi970800v. [DOI] [PubMed] [Google Scholar]
- 26.Stemmler T L, T M, Sossong J, Goldstein J I, Ash D E, Elgren T E, D M, Kurtz J, Penner-Hahn J E. Biochemistry. 1997;36:9847–9858. doi: 10.1021/bi9702795. [DOI] [PubMed] [Google Scholar]
- 27.Yaron A, Berger A. Methods Enzymol. 1970;19:521–534. [Google Scholar]
- 28.Kuhn N J, Talbot J, Ward S. Arch Biochem Biophys. 1991;286:217–221. doi: 10.1016/0003-9861(91)90031-d. [DOI] [PubMed] [Google Scholar]
- 29.King G F, Crossley M J, Kuchel P W. Eur J Biochem. 1989;180:377–384. doi: 10.1111/j.1432-1033.1989.tb14659.x. [DOI] [PubMed] [Google Scholar]
- 30.Mock W L, Liu Y. J Biol Chem. 1995;270:18437–18446. doi: 10.1074/jbc.270.31.18437. [DOI] [PubMed] [Google Scholar]
- 31.Dismukes G C. Chem Rev. 1996;96:1909–2926. doi: 10.1021/cr950053c. [DOI] [PubMed] [Google Scholar]
- 32.Lin L-N, Brandts J F. Biochemistry. 1979;18:5037–5042. doi: 10.1021/bi00590a002. [DOI] [PubMed] [Google Scholar]