Abstract
The membrane-bound complex of the Salmonella typhimurium periplasmic histidine permease, a member of the ABC transporters (or traffic ATPases) superfamily, is composed of two integral membrane proteins, HisQ and HisM, and two copies of an ATP-binding subunit, HisP. The complex hydrolyzes ATP upon induction of the activity by the liganded soluble receptor, the periplasmic histidine-binding protein, HisJ. Here we take advantage of the modular organization of this system to show that the nucleotide-binding component can be stripped off the integral membrane components, HisQ and HisM. The complex can be reconstituted by using the HisP-depleted membranes containing HisQ and HisM and pure soluble HisP. We show that HisP has high affinity for the HisP-depleted complex, HisQM, and that two HisP molecules are recruited independently of each other for each HisQM unit. The in vitro reassembled complex has entirely normal properties, responding to HisJ and ATPase inhibitors with the same characteristics as the original complex and in contrast to those of soluble HisP. These results show that HisP is absolutely required for ATP hydrolysis, that HisQM cannot hydrolyze ATP, that HisP depends on HisQM to relay the inducing signal from the soluble receptor, HisJ, and that HisQM regulates the ATPase activity of HisP. We also show that HisP changes conformation upon exposure to phospholipids.
The superfamily of ABC transporters (also called traffic ATPases) includes both prokaryotic and eukaryotic proteins (1, 2). Its significance in prokaryotes has been recently highlighted by the finding that in the completed Escherichia coli genome sequence the largest family of paralogous proteins is that of ABC transporters (3). In eukaryotes the superfamily has acquired an increasingly growing importance as more and more proteins of medical significance are found to belong to it; among many others are the human cystic fibrosis transmembrane conductance regulator (CFTR), P-glycoprotein (multidrug resistance protein, or MDR), the heterodimeric transporter associated with antigen processing (Tap1/Tap2), and the retina-specific transporter involved in recessive Stargardt macular distrophy (4–7). All members of the superfamily have the same overall composition, with two ATP-binding domains and two transmembrane domains, all four of which in eukaryotes are usually fused into a single (or occasionally two) polypeptide chain(s). Relatively little is known as yet about the properties of the individual domains of ABC transporters: Which of the domains hydrolyzes ATP? How is ATP hydrolysis regulated? How do the four domains interact with each other in the complex? What specific roles are fulfilled through this interaction? Which domain is responsible for the specificity of translocation? How is the signal transmitted through the membrane? To study these questions it is essential to take apart such a modular structure into its building blocks and characterize them. Because the prokaryotic and the eukaryotic systems bear a high level of homology, both with respect to the sequence of the ATP-binding domains and to the predicted tertiary organization of the entire structure, the prokaryotic systems provide an excellent model for studying the mechanism and function of these transporters in general. Among several advantages, most important is their modular composition allowing the various domains to be separated and characterized. Several prokaryotic ABC transporters have been analyzed and provide possible model systems for the superfamily; among them, the Salmonella typhimurium histidine permease and the E. coli maltose permease are the most notable (8).
The histidine permease, which is responsible for the high-affinity uptake of histidine in S. typhimurium and E. coli, has been characterized extensively by using biochemical, genetic, and physiological tools (9–13). The permease includes a membrane-bound complex consisting of two integral membrane components, HisQ and HisM, and two copies of an ATP-binding component, HisP. Although HisP has a hydrophilic sequence (14), it does not behave like a typical peripheral membrane protein, because it is only partly released by urea, Triton X-114, and alkaline pH; neither does it behave like a typical integral membrane protein because it is partially extracted with the same agents (15). In addition, although HisP is accessible on the cytoplasmic aspect of the membrane (16), it is also accessible at the periplasmic membrane surface (13), indicating that it penetrates the membrane deeply. Such an organization has also been reported for MalK (17), KpsT (18), and the isolated ATP-binding domain of CFTR (19, 20). ATP hydrolysis, which supplies the energy for transport, is strictly dependent on a soluble receptor, the periplasmic substrate-binding protein, HisJ, which binds histidine with high affinity and stimulates ATP hydrolysis by a complex containing HisQ, HisM, and HisP in 1:1:2 proportions (HisQMP2) (12). The ATP-binding subunit, HisP, has been produced free of the transmembrane components, purified in soluble form, and its ATP-hydrolyzing properties characterized (10). The intact HisQMP2 complex has also been purified and is functional in both ATP hydrolysis and histidine transport (ref. 21 and unpublished results).
Here we show that HisP can be separated from HisQ and HisM and reassembled back into an active HisQMP2 complex. The dimeric form of HisP is shown to be the enzymatically active form of HisP in the complex, whereas the HisP-depleted complex (HisQM) by itself does not hydrolyze ATP. HisQ and HisM are shown to be required to relay the ATPase-inducing signal from HisJ and to regulate ATP hydrolysis by HisP, by a combination of suppression and stimulation. The availability through genetic screens of individual building blocks endowed with specialized properties provides a powerful tool for investigating how these components collaborate to form a properly functioning complex.
MATERIALS AND METHODS
The bacterial strains used in this study are: TA1889, which is E. coli carrying, in an unc− background, plasmid pFA17 containing the hisQ, hisM, and hisP genes from S. typhimurium, under the control of the phage λ PL promoter (22); TA5093, which is E. coli carrying chromosomal deletions uncΔ702 and hisΔ700, which eliminates genes hisJ, hisQ, hisM, and hisP; and GA298, isogenic with TA1889, but lacking plasmid pFA17. Protein was assayed by using a modified Lowry (23). Phospholipids were obtained from Avanti Polar Lipids.
RESULTS
Disassembly of HisQMP2 into HisP and HisQM and in Vitro Reassembly into a Functional Complex.
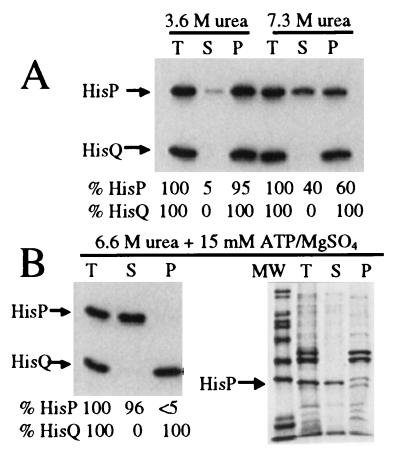
Using the histidine permease, the first step in this study has been the release of the ATP-binding subunit from the membrane-bound complex and its reconstitution into the depleted structure containing only HisQ and HisM. Among various agents tried, urea was found to be effective in both removing HisP and not damaging permanently the remaining components, such that they would be capable of being reconstituted into an active complex. Fig. 1A shows that 3.6 M urea releases ≈5% of the HisP present in a preparation of inside-out membrane vesicles (first three lanes), whereas 7.3 M urea extracts 40% of HisP (last three lanes). These results confirm the notion that the association of HisP with HisQ and HisM is tighter than that of a classic peripheral membrane protein (15). Fig. 1B, shows that the addition of 15 mM ATP and 15 mM MgSO4 to urea (6.6 M) allows the extraction of essentially all of HisP, while all of HisQ remains in the membrane.† Although several proteins are released, HisP is the dominant one (Fig. 1B, Right). Thus, the presence of ATP/Mg2+ increases the ability of urea to remove HisP from membrane vesicles, yielding a membrane preparation that is essentially free of HisP.
Figure 1.
Disassembly of HisQMP2. Inside-out membrane vesicles were prepared as described (12), with minor modifications, from E. coli strain TA1889. Intact cells and the final membrane preparation were resuspended in buffer A [50 mM 4-morpholinepropanesulfonic acid/K+, pH 7.0/100 mM NaCl/complete protease inhibitor cocktail (Boehringer Mannheim)/1 mM DTT]. For the urea extraction bacterial membranes (10 μl, total protein concentration ≈10 mg/ml) were diluted into 100 μl of a urea solution in buffer A to give final urea concentrations as indicated. The mixture was incubated on ice for 1 h, and subsequently ultracentrifuged at 200,000 × g for 1 h at 4°C. The top 40% of the supernatant was analyzed further. The pellets were resuspended in a volume of buffer A that corresponds to the volume subjected to centrifugation. Samples were diluted 2.5-fold in Laemmli sample buffer (33) and a 5 μl aliquot was analyzed without heating by 10% (unless indicated differently) SDS/PAGE. Detection was either by immunoblotting by using polyclonal antibody raised against HisP and HisQ and visualization with enhanced chemiluminescence (Amersham), or by Coomassie blue staining. Quantitation was performed by densitometry as described (34) and is indicated as percentages of HisP and HisQ present in the total fraction. T, total mixture prior to centrifugation; S, supernatant; P, pellet; MW, molecular weight standard. (A) The final concentrations of urea were 3.6 M (leftmost three lanes) and 7.3 M (rightmost three lanes). (B) The final concentration of urea was 6.6 M; in addition, 15 mM ATP and 15 mM MgSO4 were present in the extraction mixture. The proteins in the four lanes on the right were resolved on 12% acrylamide SDS/PAGE and visualized with Coomassie blue.
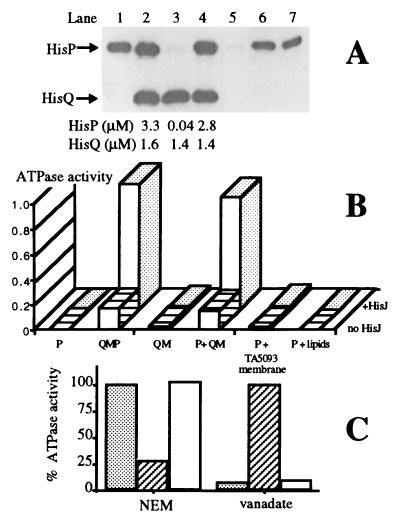
The ability of such a depleted preparation to accept pure HisP and reassemble into a functional complex was tested. Soluble HisP(his6), a HisP protein with a carboxyl-terminal extension of eight amino acids (Leu-Glu-His-His-His-His-His-His), was purified from cells unable to produce HisQ and HisM (10) (Fig. 2A, lane 1). It was then added to HisQM membranes (lane 3) in the same proportion relative to HisQ as in native membranes (lane 2). Essentially all of HisP(his6) becomes associated with the sedimentable material (lane 4), with only trace amounts remaining in the supernatant (lane 5). Accurate quantitation of the untreated and reconstituted samples shows that HisP(his6) is recruited into a complex that has essentially the same proportions of HisQ to HisP as the original sample in lane 2. Thus, a complex with the correct proportions of all subunits is formed. The possibility that the presence of HisP(his6) in the membrane fraction is artifactual, being due to its precipitation into sedimentable material, was considered. This is an especially relevant concern because soluble HisP(his6) is known to be particularly prone to precipitating (10). However, under the conditions of this experiment, no HisP is present in the pellet if membrane vesicles are absent from the reaction mixture (data not shown). The ability of soluble HisP to associate unspecifically with membranes prepared from a strain that lacks all permease components (TA5093) was also tested; about half of HisP becomes associated with such a membrane, presumably nonspecifically (data not shown). Similarly, about half of HisP(his6) becomes associated with phospholipids liposomes (lanes 6 and 7). These results are not surprising in view of the fact that HisP produced in vivo in the absence of HisQ and HisM is partially associated with the membrane (15).
Figure 2.
Reassembly of HisP with HisQM. Membrane vesicles, prepared as described in the legend to Fig. 1, were diluted 10-fold into a solution of 8 M urea, 15 mM ATP, and 15 mM MgSO4 in buffer A, and incubated on ice for 1 h with occasional mixing; they were then centrifuged at 45,000 rpm (TLA-45 rotor, Beckman) for 1 h at 4°C. The pellet was washed once in buffer A and resuspended in 50 mM 4-morpholinepropanesulfonic acid/K+, pH 7.0, by using a homogenizer, and stored in aliquots at −80°C. This preparation is stable for at least 6 months. The amount of HisQ and HisP present in each sample of either pure HisP(his6) or HisQM membranes or intact membranes was quantitated by immunoblotting with antibodies raised against HisQ and HisP, respectively. Standard curves (six points) were made for each SDS/PAGE, by using known concentrations of pure HisP(his6) (10) and pure HisQMP2 complex (21). Pure HisQMP2 in proteoliposomes has been shown to have the same stoichiometry as in membrane vesicles (15). For reassembly, pure HisP(his6) (final concentration: 2.8 μM) and HisQM membranes (containing final concentration: 1.4 μM HisQ) were incubated on ice for 30 min in a buffer containing 50 mM 4-morpholinepropanesulfonic acid/K+, pH 7.0, 2 mM ATP, and 4% glycerol (final total volume: 100 μl) and centrifuged at 45,000 rpm (TLA45 rotor, Beckman) for 1 h at 4°C. The pellet was resuspended in 100 μl of the same buffer. The supernatant and pellet were analyzed for protein content as described in Fig. 1. (A) 10% SDS/PAGE of various samples containing the indicated amounts of protein. Lane 1, pure soluble HisP(his6); lane 2, intact membrane vesicles; lane 3, HisQM membranes; lanes 4 and 5, pellet and supernatant from reassembly reaction by using HisQM membranes; lanes 6 and 7, pellet and supernatant respectively, from a reassembly reaction by using E. coli phospholipids (2 mg/ml). (B) Initial rate of ATP hydrolysis by samples from a reassembly experiment. The ATPase activity (10, 12) was assayed in the presence or absence of 18 μM l-histidine-liganded HisJ, in a total volume of 50 μl, taking 10-μl samples at different time intervals. The activity is expressed as nmol Pi released per min per 10 μl sample, containing 25 pmol of pure HisP(his6) and/or 13 pmol of HisQ (quantitated by densitometry by using purified HisQMP as standard). Columns 1–4 correspond to lanes 1–4 in A; column 5 is the pellet from a reassembly reaction by using membranes prepared from control strain TA5093. Column 6 is the pellet from a reassembly reaction with E. coli phospholipids replacing membranes. (C) Inhibitor profile of the ATPase activity (in the presence of 18 μM l-histidine-liganded HisJ) after preincubation with 200 μM N-ethylmaleimide for 10 min at 37°C (Left) or 200 μM of vanadate for 30 min on ice (Right). The activity is expressed as percentage of the activity of the respective untreated samples. ░⃞, Intact membranes (same material as in lane 2 of A); ▨, soluble HisP(his6) (same material as in lane 1 of A); □, reassembled complex (same material as in lane 4 of A).
An important question is whether the reassociated material is correctly reassembled. Fig. 2B shows the ATPase activity of equal relative volumes of samples displayed in Fig. 2A. The second column shows the intrinsic activity of untreated membrane vesicles (0.17 nmol/min), which increases to a rate of 0.99 nmol/min in the presence of liganded HisJ (as expected; see ref. 12). HisQM membranes are completely inactive in either the presence or absence of HisJ. In addition, proteoliposomes reconstituted from solubilized HisQM membranes are unable to transport (data not shown). Soluble HisP(his6) has a very low ATPase activity (0.006 nmol/min), and the presence of HisJ has no effect.‡ Both the intrinsic and HisJ-stimulated ATPase activities of membranes reconstituted with an equivalent amount of soluble HisP(his6) (P+QM) are restored essentially to the same levels (0.15 and 0.89 nmol/min, respectively) as those of the untreated sample. HisP that was released from the complex with urea is active in ATP hydrolysis after the removal of urea by slow dialysis and can be reassembled back into HisQM membranes giving a fully active complex (data not shown). That HisP specifically requires HisQM for its activity is also shown by the fact that HisP associated either with membranes that do not contain HisQ and HisM (from TA5093) or with phospholipids, has no ATPase activity, either in the presence or absence of HisJ (Fig. 2B, fifth and sixth columns). Additional evidence that the complex has been reassembled correctly is the fact that it transports histidine upon solubilization and reconstitution into propteoliposomes (11) (data not shown). In conclusion, not only are the relative amounts of HisP quantitatively restored upon reassembly, but the ATP-hydrolyzing activity, both intrinsic and HisJ-induced, and transport activity are also restored.
Soluble HisP has several properties different from those of complex-associated HisP (10, 12). Therefore, in another approach to determine whether the reassembled HisP is “normal” in all respects, these properties were tested. Soluble HisP is insensitive to HisJ, while the reassembled HisP responds to HisJ in the same way as the untreated complex (as already pointed out in Fig. 2B, first and fourth columns, respectively). Soluble HisP is sensitive to N-ethylmaleimide and resistant to vanadate (Fig. 2C, hatched bars); this profile of inhibitor sensitivity is the opposite of that of the complex (stippled bars). Upon reassembly, the inhibitor profile of HisP changes from that of the soluble protein to that of the complex, becoming inhibitable by vanadate and insensitive to N-ethylmaleimide (empty bars). The urea extractability of HisP(his6) from the reassembled complex is also the same as that of HisP in the original complex (data not shown). Thus, upon reassembly, HisP loses the characteristic properties of the soluble form and is indistinguishable from HisP that has never left the complex.
The reassembly reaction is fast. All of HisP(his6) disappears from the soluble fraction within 3 min of incubation on ice, with concomitant appearance in the complex of the level of activity expected for that amount of HisP. The reassembly reaction does not require Mg2+ or glycerol and proceeds in the absence of ATP. Thus, there appears to be no energy requirement. The pH and NaCl concentration optima for reassembly are 7.5 and 50 mM, respectively; higher concentrations of NaCl inhibit reassembly, indicating that ionic interactions may be involved (data not shown).
Disassembly and reassembly can also be performed by using proteoliposomes reconstituted with purified HisQMP2, indicating that membrane vesicles do not contain additional unknown assembly factors. Proteoliposomes are not routinely used because of poor recovery.
HisP Molecules Reassemble into the Complex Independently of Each Other, but Function as Dimers.
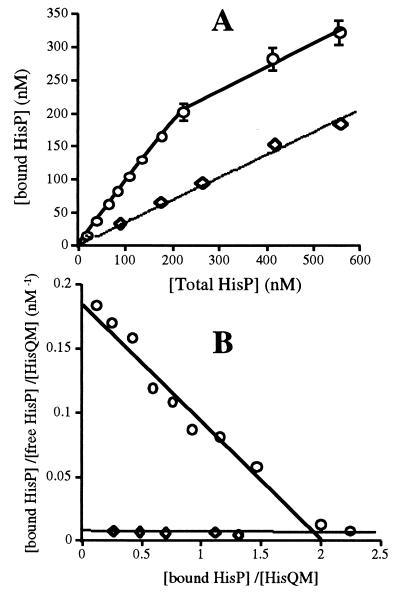
It has been shown (12) that the two HisP subunits in the native complex display positive cooperativity for ATP, presumably reflecting the presence of a dimer. The properties of HisP during reassembly and in the final reassembled complex were analyzed in this respect. The affinity of HisP for HisQM and the number of binding sites involved in the reassembly process were determined by varying the concentration of HisP(his6) between 0 and 600 nM (at which concentrations, at least 99.5% of soluble HisP would be monomeric), while the HisQM concentration was maintained constant (determined to be 140 nM). Fig. 3A shows that the amount of HisP(his6) bound increases linearly (up to 200 nM HisP); there is no indication of cooperativity. Thus, it appears that monomeric HisP molecules are recruited independently of each other. Membranes from strain GA298, which is isogenic with TA1889, but lacks plasmid pFA17 and thus does not overproduce HisQ, HisM, or HisP, were used as control. Fig. 3A shows that HisP can be bound nonspecifically, without reaching saturation. The amount of HisP(his6) bound specifically (after correcting for nonspecific binding by GA298 membranes) reaches saturation when its concentration equals 2-fold the concentration of HisQM. If there were an equilibrium between association and dissociation of HisP(his6) and HisQM, a Scatchard plot analysis of these data would yield a Kd value. Because an equilibrium has not been demonstrated, the value obtained from the Scatchard plot is referred to as “apparent Kd.” Such an analysis (Fig. 3B) shows that HisP has a high affinity for HisQM (apparent Kd 10 nM) and that there are two HisP-binding sites for each HisQM molecule (n = 2). This is in agreement with the previously established stoichiometry of the native HisQMP2 complex (15) . The presence of ATP (0–15 mM), Pi (0–50 mM), or glycerol (0–4%) during the reassembly reaction does not affect the affinity of HisP for HisQM (data not shown).
Figure 3.
Scatchard plot for reassembly. Reassembly was performed as described in the legend to Fig. 2, except that incubation on ice was for one h to insure having reached equilibrium, and by using variable concentrations of HisP(his6) and either HisQM membranes containing 140 nM HisQM (circles) or membranes from control strain GA298 (diamonds). Accurate estimate of bound and free HisP(his6) was achieved by a more sensitive immunoblot quantitation that uses CDP-Star (Boehringer Mannheim and Tropix, Bedford, MA), a chemiluminescence substrate for alkaline phosphatase. (A) Amount of HisP(his6) bound vs. total HisP(his6). (B) Scatchard plot of data in (A) Such an analysis has been performed for several independent experiments, giving apparent Kd values between 3 and 20 nM and n = 2 ± 0.3. The concentration range of HisP(his6) is such that it is never <99.5% monomeric.
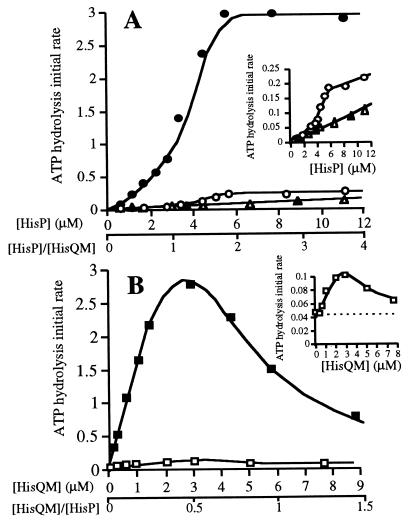
The emergence of ATPase activity in the complex as it is being reassembled with varying amounts of HisP and a constant amount of HisQM membranes was also measured. Because it appears that HisP molecules enter the complex independently of each other, two species, HisQMP1 and HisQMP2, would be formed in a continuously changing ratio, as dictated statistically by the varying proportions of HisP versus HisQM. If HisQMP1 were capable of hydrolyzing ATP, the initial appearance of reconstituted activity should be linearly correlated to the concentration of HisP. Fig. 4A shows that the ATPase activity is sigmoidally dependent on the concentration of HisP (0–12 μM soluble HisP(his6) added to 3 μM HisQM), being strongly cooperative, both in the presence (•) and in the absence (○; see also Inset) of HisJ, with a Hill coefficient of 2. This result suggests that if the HisQMP1 form exists, it has no, or relatively little, ATPase activity and that HisQMP2 is the enzymatically active form. In agreement with this notion is the finding that a saturation point is reached when the molarity of HisP is twice that of HisQM (6 and 3 μM, respectively), which agrees with the number of binding sites in HisQM being 2, as previously determined (Fig. 3) and as expected from the known stoichiometry of the native complex.
Figure 4.
Dependence of ATPase activity on HisQM and HisP concentrations. The initial rate of ATP hydrolysis was measured as described in the legend to Fig. 2, in the absence (open symbols) or presence of 18 μM l-histidine-liganded HisJ (solid symbols). The activity is expressed as nmol Pi released per min per 10 μl sample, containing various concentrations of pure HisP(his6) and/or HisQ, as indicated. All ATPase assays involving membranes were corrected for a low level contaminating ATPase activity that is present in HisQM membranes. (A) Varying amounts of soluble HisP(his6) (0–12 μM) were incubated either with HisQM membranes containing 3 μM HisQM (○ and •) or in the absence of membranes (▵ and ▴). (B) Varying amounts of HisQM membranes (containing 0–12 μM HisQM) incubated with 6 μM of soluble HisP(his6) (□ and ▪). The dotted line is drawn through the level of activity of 6 μM pure soluble HisP(his6). Both insets are enlargements of the respective data from samples with low activity. In this experiment no ultracentrifugation to separate the bound and free HisP was performed, because the ATPase activity HisP(his6) at such low concentrations in the absence of HisQM does not interfere with the assay, also shown by the fact that similar results were obtained if the reassembly mixture containing nmolar amounts of HisP(his6) were centrifuged prior to analysis.
The notion that HisQMP1 has no or poor activity is corroborated by reassembly experiments in which the concentration of HisQM is varied (between 0 and 12 μM), keeping the amount of HisP(his6) constant (6 μM). The corresponding ATPase activity (in the presence of HisJ) increases linearly until it reaches a maximum at 3 μM HisQM (Fig. 4B, ▪) at which point, the concentration of HisQM relative to that of HisP is 1:2 and corresponds to the calculated amount of HisQM capable of binding all of the HisP present (as determined in Fig. 3B). After this point the activity decreases. The decrease is not due to residual urea or other unknown contaminating inhibitor(s) because it persists after the membranes are washed (data not shown). A likely explanation is that as HisP becomes limiting, excess HisQM increases the probability of HisQMP1 formation at the expense of HisQMP2. This result supports the notion that HisQMP1 is inactive. The level of the emerging ATPase activity matches the expected level as predicted from the probability of HisQMP2 formation. For example, at 6 μM each HisQM and HisP, the probability of forming HisQMP2 is 50%, which fits the observed ATPase activity of 1.5 nmol/min (50% of maximum). The same result is obtained in the absence of HisJ (Fig. 4B, □; see also Inset). In conclusion, the results in Figs. 3 and 4A, combined with those in Fig. 4B indicate that HisP molecules are recruited individually, that the HisQMP1 form has no or little ATPase activity, and that HisQMP2 is the active form.
The reassembled HisQMP2, in which HisP has presumably assumed the dimeric form, has an intrinsic ATPase activity higher than that of equivalent amounts of soluble HisP. For example, soluble HisP at a concentration of 6 μM, 5% of which is calculated to be in the dimeric form, has an activity of 0.05 nmol/min, while the same amount recruited into a correctly proportioned HisQMP2 complex has an activity of 0.17 nmol/min (Fig. 4A). This result indicates that HisQ and/or HisM have a stimulatory function and may act by forcing the dimerization of HisP into the HisQMP2 complex. However, if all of HisP were a dimer in the soluble form, it is calculated to have an activity of 1 nmol/min, which is considerably higher than 0.17, suggesting that either HisQ and/or HisM have a suppressing function or that the activation is incomplete.
The Conformation of HisP Changes upon Interaction with Phospholipids.
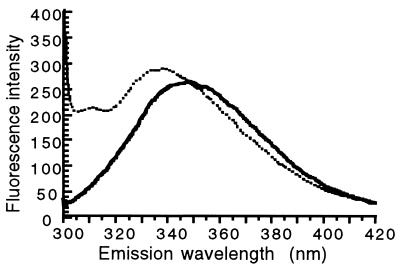
Soluble HisP(his6) binds avidly to phospholipids: maximally three molecules of HisP(his6) bind to 1,000 molecules of phospholipids. The possibility that this binding might affect the conformation of HisP was considered. The single tryptophan residue (Trp-105) present in each HisP molecule can be used as a convenient probe for analyzing conformational changes. The fluorescence emission spectrum of soluble HisP has a maximum at 348 nm (Fig. 5) (10), indicating that Trp-105 is in a polar environment. In the presence of 0.4 mg/ml of phospholipid liposomes, the fluorescence maximum shifts to 337 nm (Fig. 5). This indicates that HisP undergoes a significant conformational change and suggests that, in so doing, the molecular environment of Trp-105 becomes more hydrophobic. Such an interaction might be involved in the process of reassembly, in vivo and/or in vitro because Trp-105 is located in the so-called helical domain, which has been hypothesized to be involved in the interaction of HisP with HisQ and HisM (25) and the interaction of MalK with MalG and MalF in the maltose permease (26). Because the association of HisP with phospholipids is unproductive (Fig. 2B), it must reflect a very different organization than that in the native form or in the one reassembled in the presence of HisQM.
Figure 5.
HisP interacts with phospholipids and changes conformation. The tryptophan fluorescence spectrum of HisP(his6) was measured as described (10) in the absence (—) or the presence of 0.4 mg/ml sonicated phospholipids (⋅⋅⋅).
DISCUSSION
Using the histidine permease as a model system for ABC transporters, we have shown that it is possible to disassemble and reassemble the membrane-bound complex. It is likely that modularity of structure and function is an integral property of all members of this superfamily of transporters, even for those eukaryotic members with the individual domains fused into a single polypeptide. This opens numerous opportunities for studying their mechanism of action involving the interaction between the integral membrane and the ATP-hydrolyzing domains, especially by using complementation experiments with properly designed mutants and in vitro chemical modifications.
We show that the nucleotide-binding domain of the histidine permease is released as an undamaged soluble protein capable of hydrolyzing ATP and of being reassembled with the remaining urea-treated hydrophobic components into a normally functioning complex, indicating that the urea treatment is gentle with respect to all parts of the complex. Because HisP has a very high affinity for HisQM, but also a relatively high affinity for phospholipids, the process of reassembly might involve an early interaction between HisP and phospholipids followed by a two-dimensional movement of HisP in the lipid bilayer in search of the hydrophobic proteins, HisQ and HisM. Because HisP is hydrophilic (14) and spans the membrane (13), HisQ and HisM must engulf it forming a protective shell around it. Soluble HisP is inhibited by phospholipids (10); therefore it is likely that such a shell screens it also from this inhibition. It is possible that a regulated exposure of HisP to phospholipids in vivo provides one of the means of controlling its activity.
The disassembly of the complex has allowed the assignment of specific functions to the individual domains. Although isolated HisP hydrolyzes ATP (10), and similar results have been obtained in other systems (27, 28), it has never been possible to determine directly whether the hydrophobic components are also able to hydrolyze ATP. This is a particularly relevant question because it is known that HisQ binds ATP (ref. 22; V. Shyamala and G.F.-L.A., unpublished data). By separating HisP from the hydrophobic components, we show here that HisQ and HisM have no ATPase activity, and thus, that their function must be structural and/or regulatory. One regulatory function is that of acting as intermediates in the signaling mechanism because HisP responds to signaling by HisJ only when complexed with HisQ and HisM. Another regulatory function appears to be the control of the rate of hydrolysis of ATP, as demonstrated by the ability of HisQM to raise the activity of the isolated, soluble HisP. This is clearly shown, for example, in Fig. 4 in which the ATPase activity of HisP (6 μM) is increased 3-fold (from 0.05 to 0.17 nmol/min) when HisQM (3 μM) is present and 60-fold (to 3 nmol/min) when HisJ is also present.
What is a likely scenario for this reactivation process? Because soluble HisP at low concentration contains little active dimer (10) and because the monomers are recruited individually into the complex (Fig. 3), they must be subsequently reorganized to form a dimer and be activated.§ Thus, it is reasonable to postulate that the activation of HisP is due to a combination of (i), the two high-affinity HisP-binding sites present in each HisQM complex, which allow the efficient scavenging of HisP out of solution or from the lipidic phase; (ii), the subsequent coercion of HisP to assemble as a dimer within the functional complex, HisQMP2; and (iii), some form of activation by HisQM. The conclusion that HisQM activates HisP does not conflict with previous evidence that HisQM suppresses the ATPase activity of HisP in the complex (12). The soluble dimeric form of HisP has a turnover rate of 2 s−1 (10), which is higher than that of the equivalent amount of dimeric complex-bound HisP in the absence of HisJ (intrinsic activity, 0.5 s−1), but still lower than that of the fully induced complex in the presence of liganded HisJ (8 s−1) (12). Therefore, it appears that the hydrolytic activity of HisP in the complex is constrained by HisQM from needlessly pursuing a rampant hydrolysis. In conclusion, although HisQ and HisM do not participate directly in ATP hydrolysis, they have an important role in regulating the activity of HisP, through an equilibrium between the competing forces of stimulation and repression.
How could the two HisP molecules coordinate their activity within the complex? That the active form of reassembled HisP is a dimer involving physical contact between the two subunits is in agreement with the evidence previously presented that both in native HisQMP2 (12) and in the soluble form (10), dimeric HisP is the active species. In addition, chemical cross-linking between two HisP molecules has been observed (K. Nikaido and G.F.-L.A., unpublished observations). Dimeric HisP appears to be the active form also in the absence of HisJ (Fig. 4), indicating that the dimer is the basic functional ATP-hydrolizing unit under all conditions. It cannot be completely excluded that HisQ and HisM control the activity of HisP by holding it in a conformation that does not involve physical contact between the two molecules. The positive cooperativity displayed by HisQMP2 suggests that the two HisP subunits are different (12). It is possible that they are organized differently via the interaction with the hydrophobic components, and that one is catalytic while the other one is exclusively involved in signaling. Alternatively, the model proposed for MDR, the multidrug resistance protein, in which both nucleotide-binding sites hydrolyze ATP, but do so alternately (29), and for the maltose permease (30), in which both ATP-binding subunits have to be intact for ATP hydrolysis, might also apply to the histidine permease.
The ease of reassembly of HisP into HisQM membranes suggests that its in vivo assembly in the complex is not, or need not be, cotranslational. Presumably HisQM and HisP undergo a refolding process as they reassemble into an active complex. In addition, a comparison can be made with SecA, a membrane-associated translocation ATPase, that spans the membrane (31) and undergoes an ATP-driven cycle of membrane insertion and deinsertion (32). Possibly HisP undergoes a similar cycle in connection with its transport function.
Acknowledgments
We thank Kishiko Nikaido, who also participated in the initial reassembly experiment, and Cheng E. Liu for providing pure soluble HisP(his6) and pure HisJ, respectively, and for invaluable extensive discussions. This work was supported by National Institutes of Health Grant DK12121 (to G.F-L.A.).
ABBREVIATIONS
- HisQMP2
complex containing HisQ, HisM, and HisP in 1:1:2 proportions
- HisQM membranes
HisP-depleted membranes containing HisQ and HisM
- HisQM
HisP-depleted complex
- HisP(his6)
HisP protein with a carboxyl-terminal extension of eight amino acids: Leu-Glu-His-His-His-His-His-His
Footnotes
Because of the high level of sequence similarity between HisQ and HisM (24), they are expected to behave similarly also with regard to extraction. HisM is not routinely tested because the available anti-HisM antibody is of low titer.
Only the dimer form of soluble HisP is active. Its formation is dependent on concentration; at the HisP concentration used in Fig. 2 (90 μg/ml) 97.3% is calculated to be in the inactive monomer form (10).
It should be noted that it cannot be excluded that small amounts of dimeric HisP also enter the complex directly.
References
- 1.Ames G F-L, Mimura C, Holbrook S, Shyamala V. Adv Enzymol. 1992;65:1–47. doi: 10.1002/9780470123119.ch1. [DOI] [PubMed] [Google Scholar]
- 2.Higgins C F, Gottesman M M. Trends Biochem Sci. 1992;17:18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 5.Doige C A, Ames G F-L. Annu Rev Microbiol. 1993;47:291–319. doi: 10.1146/annurev.mi.47.100193.001451. [DOI] [PubMed] [Google Scholar]
- 6.Dean M, Allikmets R. Curr Opin Genet Dev. 1995;5:779–785. doi: 10.1016/0959-437x(95)80011-s. [DOI] [PubMed] [Google Scholar]
- 7.Allikmets R, Singh N, Sun H, Shroyer N F, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 8.Boos W, Lucht J M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1175–1209. [Google Scholar]
- 9.Ames G F-L. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido K, Liu P-Q, Ames G F-L. J Biol Chem. 1997;272:27745–27752. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 11.Liu C E, Ames G F-L. J Biol Chem. 1997;272:859–866. doi: 10.1074/jbc.272.2.859. [DOI] [PubMed] [Google Scholar]
- 12.Liu C E, Liu P-Q, Ames G F-L. J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- 13.Baichwal V, Liu D, Ames G F-L. Proc Natl Acad Sci USA. 1993;90:620–624. doi: 10.1073/pnas.90.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins C F, Haag P D, Nikaido K, Ardeshir F, Garcia G, Ames G F-L. Nature (London) 1982;298:723–727. doi: 10.1038/298723a0. [DOI] [PubMed] [Google Scholar]
- 15.Kerppola R E, Shyamala V, Klebba P, Ames G F-L. J Biol Chem. 1991;266:9857–9865. [PubMed] [Google Scholar]
- 16.Prossnitz E. Ph.D. thesis. Univ. of California at Berkeley; 1989. [Google Scholar]
- 17.Schneider E, Hunke S, Tebbe S. J Bacteriol. 1995;177:5364–5367. doi: 10.1128/jb.177.18.5364-5367.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bliss J M, Silver R P. J Bacteriol. 1997;179:1400–1403. doi: 10.1128/jb.179.4.1400-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko Y H, Delannoy M, Pedersen P L. Biochemistry. 1997;36:5053–5064. doi: 10.1021/bi9630265. [DOI] [PubMed] [Google Scholar]
- 20.Gruis D B, Price E M. Biochemistry. 1997;36:7739–7745. doi: 10.1021/bi9701585. [DOI] [PubMed] [Google Scholar]
- 21.Liu C E. Ph.D. thesis. Univ. of California at Berkeley; 1996. [Google Scholar]
- 22.Hobson A, Weatherwax R, Ames G F-L. Proc Natl Acad Sci USA. 1984;81:7333–7337. doi: 10.1073/pnas.81.23.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson G L. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 24.Ames G F-L. In: Current Topics in Membranes and Transport. Adelberg E A, Slayman C W, editors. New York: Academic; 1985. pp. 103–119. [Google Scholar]
- 25.Mimura C S, Holbrook S R, Ames G F-L. Proc Natl Acad Sci USA. 1991;88:84–88. doi: 10.1073/pnas.88.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilken S, Schmees G, Schneider E. Mol Microbiol. 1996;22:655–666. doi: 10.1046/j.1365-2958.1996.d01-1724.x. [DOI] [PubMed] [Google Scholar]
- 27.Walter C, Honer zu Bentrup K, Schneider E. J Biol Chem. 1992;267:8863–8869. [PubMed] [Google Scholar]
- 28.Koronakis V, Hughes C, Koronakis E. Mol Microbiol. 1993;8:1163–1175. doi: 10.1111/j.1365-2958.1993.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 29.Senior A E, Al-Shawi M K, Urbatsch I L. FEBS Lett. 1995;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- 30.Davidson A L, Sharma S. J Bacteriol. 1997;179:5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y J, Rajapandi T, Oliver D. Cell. 1994;78:845–53. doi: 10.1016/s0092-8674(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 32.Economou A, Wickner W. Cell. 1994;78:835–43. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli V K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Ames G F-L, Liu C E, Joshi A K, Nikaido K. J Biol Chem. 1996;271:14264–14270. doi: 10.1074/jbc.271.24.14264. [DOI] [PubMed] [Google Scholar]