Posttranscriptional regulation of gene expression has become a popular method for studying gene function and elucidating networks of gene expression. A number of tools are available that allow investigators to regulate gene expression posttranscriptionally, including RNAi, antisense oligonucleotides, DNAzymes, and ribozymes (1). Each of these methods relies on complementary basepairing between the inhibitory agent and the target mRNA. In addition, these methods require efficient delivery of short nucleic acids to cells through either carrier molecules or the introduction of genes that encode the inhibitory molecules for expression in the cells of interest (1). A unique nucleic acid inhibitor must be developed for every target of interest and, in many instances, a panel of the appropriate nucleic acid inhibitors has to be tested to achieve the desired knockdown levels of the target (2, 3).
In a recent issue of PNAS, Win and Smolke (4) describe a novel platform for posttranscriptional regulation of gene expression. Their system uses allosterically regulated, cis-cleaving ribozymes. Their system is devised such that cleavage takes place within the 3′ UTRs of the mRNAs. The premise is that cis cleavage results in separation of the poly(A) tail from the body of the message, and deadenylation is an initiating event for degradation of mRNAs (5). The system of Win and Smolke is an excellent example of the application engineering principles to a biological system. Their goals were to create a portable, scaleable, tuneable platform for regulation of gene expression.
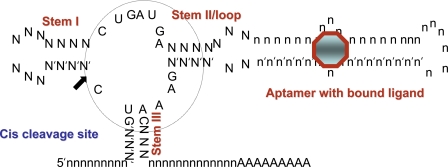
To better understand the system of Win and Smolke (4), one needs to look at each of the components. The foundation for their regulatory system is the hammerhead ribozyme, initially discovered as a cis-cleaving entity in single-stranded viroid and virusoid RNAs (6). Although the hammerhead ribozyme has been the object of extensive studies and applications over the past two decades, recent structural studies have enhanced our understanding of the critical secondary and tertiary interactions that take place between the various structural features that allow extremely rapid cleavage kinetics in the presence of physiological levels of magnesium (7). The hammerhead ribozyme essentially consists of three stems, a loop, and a catalyict core (Fig. 1). When the ribozyme folds properly, it can self-cleave via a transesterification reaction, resulting in cyclic 2′-3′ phosphate and 5′ hydroxyl cleavage products (8). Win and Smolke reasoned that insertion of the cis-cleaving ribozyme within the 3′ UTR of a message would result in self-cleavage of the transcript if the ribozyme folded properly. To ensure proper folding, they also insulated the ribozyme catalytic structure with sequences both upstream and downstream such that mRNA sequences flanking the ribozyme would not perturb the ribozyme fold. This insulation element is essential for portability because the structures of different mRNAs, even in the 3′ UTRs, could profoundly affect the folding of the ribozyme and abrogate cleavage.
Fig. 1.
The cis-cleaving hammerhead ribozyme with aptamer and ligand. The three helical stems of the ribozyme are indicated, as is the catalytic core of the ribozyme and the site of cis cleavage. The position of an aptamer is indicated. When the ligand (octagon) is bound to the aptamer, it propagates a structural change in the catalytic center of the ribozyme, allowing it to cleave in cis. The catalytic core of the ribozyme is circled.
Aptamers are in vitro-evolved nucleic acid structures that bind with high affinity to a given ligand. The high-affinity binders are selected from a random pool of nucleotides by a process called systematic evolution of ligands by exponential enrichment (SELEX) (9). The next step in the process of creating regulatable ribozyme cleavage was to insert an aptamer that binds the caffeine analogue theophylline into the ribozyme structure. Previous studies from Breaker and colleagues (10, 11) demonstrated that ribozyme cis cleavage could be controlled by such a structure. When inserted within stem/loop II or III region of the hammerhead, the aptamer structure disrupts the folding of the catalytic core such that self-cleavage of the ribozyme cannot take place. When theo phylline is added to the ribozyme self-cleavage reaction, it binds to the aptamer, propagating a conformational change from the aptamer to the catalytic core of the ribozyme, with the net result being that the catalytic core assumes a structure that allows cleavage to take place. With these two basic component parts pieced together, Win and Smolke (4) then created a series of tunable, self-cleavage modules in which the ribozyme self-cleavage could be turned on or off by graded concentrations of ligand binding. Two different RNA structural strategies were used for tuning the allosteric ribozyme activities: strand displacement and strand slippage. Strand displacement takes advantage of two different RNA conformations that are in equilibrium. This type of regulation can be engineered into the ribozyme–aptamer structure. In this case, a competing strand of RNA was appended to the 3′ end of the base of the aptamer. In one conformation, this strand binds to the 5′ end of the aptamer, resulting in disruption of the ribozyme catalytic core. Upon addition of the ligand, the conformation switches to one in which the catalytic core of the ribozyme is in an active conformation, thus resulting in a ligand “on” ribozyme. This same general strategy was used to create an allosteric ribozyme in which ligand binding triggers a conformational change that blocks the catalytic core, resulting in a ligand “off” structure. These ligand on and off strategies are both tuneable with varying concentrations of ligand.
The other regulatory approach takes advantage of strand slippage properties of RNAs (12, 13). This alternative form of structural information transfer cannot be rationally engineered into the allo steric ribozymes but requires in vitro selection. Strand slippage takes advantage of small changes in local structure that are propagated to adjacent sequences. These sequences are called communication modules because they transmit structural information from one structure to an adjoining structure. Win and Smolke (4) tested a number of published strand slippage motifs and found that several of them were very effective in regulating ribozyme activity.
To test the various approaches for allosteric regulation of the ribozyme cis-cleavage reaction in a biological system, Win and Smolke (4) inserted the insulated cassettes within the 3′ UTR of the yeast his5 gene, which is required for histidine biosynthesis when yeast are grown in minimal media. When the cis-cleaving ribozyme is active, it will trigger degradation of the his5 mRNA, resulting in histidine insufficiency and cessation of cell proliferation. With this system, they demonstrated a theophylline concentration-dependent regulation of cellular proliferation.
The second demonstration of a biological application was designed to sense the intracellular levels of a cellular metabolite, xanthine, the product of xanthosine processing. Xanthine also binds to the theophylline aptamer (albeit with 27-fold lower affinity than theophylline) but can still trigger the required conformational changes. Win and Smolke (4) used two different ligand-on aptamer–ribozyme combinations to monitor xanthine production in cells fed xanthosine. In this setting, the allosteric ribozyme was inserted in the 3′ UTR of an EGFP reporter construct. As cells were converting xanthosine to xanthine, the levels of EGPF expression increased proportionally to the amount of xanthine produced.
To demonstrate the portability of the allosteric ribozymes, Win and Smolke (4) used the strand displacement constructs for switching from a theophylline-binding aptamer to a tetracycline-binding aptamer. The tetracycline system turned out to be even more sensitive to the concentrations of drug than the theophylline system, owing to the greater cell permeability of tetracycline.
Naturally occurring riboswitches are highly evolved to exquisitely sense changes.
Overall, the clever engineering principles applied to creating the allosterically regulated ribozyme switches support this strategy as a novel one for regulating gene expression in a variety of biological systems. The initial design parameters included portability, scalability, and tuneability, making these cassettes attractive candidates for testing in mammalian systems. In principle, it should be possible to use other aptamers that bind different ligands, such as ATP or GTP, to regulate gene expression in response to changing concentrations of these important compounds (14). There are dozens of different aptamers that have been evolved to bind a large spectrum of different ligands, ranging from peptides to organic polymers (15–17). In mammalian cells, there are many possible applications for ribozyme-regulating riboswitches in basic studies and in therapeutic uses. For instance, riboswitches that control the expression of proapoptotic or antiapoptotic proteins could play a role in the treatment of diseases such as cancer.
There are now numerous examples of riboswitches in nature. These RNA structures generally assume a strong conformation upon ligand binding. In many instances, they are located in the 5′ UTR of mRNAs where they effectively block translational initiation or elongation upon ligand binding (18). Taking advantage of the design principles set forth by Win and Smolke (4), it should be possible to incorporate these naturally occurring RNA structures with ligand-regulated, self-cleaving ribozymes. Because naturally occurring riboswitches are highly evolved to exquisitely sense changes in the concentration of various cellular metabolites, including amino acids and vitamins, these could be highly effective allosteric regulators of cis-ribozyme cleavage in both prokaryotic and eukaryotic cells. An additional use of allosteric ribo zymes is as environmental sensors of levels of toxic or otherwise potentially harmful compounds via the use of ligand on aptamers for triggering expression of reporter genes. Similarly, the use of off ligands could be incorporated for monitoring reductions in the levels of various compounds or metabolites. There are potentially dozens of immediate useful applications for this system.
In conclusion, the work of Win and Smolke (4) has provided a set of design rules that should allow investigators to take advantage of the powerful approach of allosterically regulated cis-cleaving ribozymes for more widespread use. This exciting technology will find many applications in cell biology and, perhaps, even in therapeutic applications. The ability to modulate the levels of a message with varying concentrations of ligand is definitely a plus for mammalian cell culture. The use of allosterically regulated ribozymes in conjunction with gene knockdown technologies such as RNAi should allow investigators to simultaneously knock down expression of a gene in one pathway and up-regulate the expression of a gene in a different pathway. We should expect to see many applications of the allosterically regulated ribozymes in the near future.
Footnotes
The author declares no conflict of interest.
See companion article on page 14283 in issue 36 of volume 104.
References
- 1.Scherer LJ, Rossi JJ. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 2.Kalota A, Dondeti VR, Gewirtz AM. Handb Exp Pharmacol. 2006:173–196. doi: 10.1007/3-540-27262-3_9. [DOI] [PubMed] [Google Scholar]
- 3.Pan WH, Clawson GA. J Cell Biochem. 2006;98:14–35. doi: 10.1002/jcb.20790. [DOI] [PubMed] [Google Scholar]
- 4.Win MN, Smolke CD. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker R, Song H. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 6.Symons RH. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 8.Scott WG, Klug A. Trends Biochem Sci. 1996;21:220–224. [PubMed] [Google Scholar]
- 9.Tuerk C, MacDougal S, Gold L. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang J, Breaker RR. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 11.Soukup GA, Breaker RR. Structure (London) 1999;7:783–791. doi: 10.1016/s0969-2126(99)80102-6. [DOI] [PubMed] [Google Scholar]
- 12.Hall B, Hesselberth JR, Ellington AD. Biosens Bioelectron. 2007;22:1939–1947. doi: 10.1016/j.bios.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Soukup GA, Breaker RR. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z, Szostak JW. RNA. 2003;9:1456–1463. doi: 10.1261/rna.5990203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mairal T, Cengiz Ozalp V, Lozano Sanchez P, Mir M, Katakis I, O'Sullivan CK. Anal Bioanal Chem. 2007 doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 16.Nimjee SM, Rusconi CP, Sullenger BA. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich H. Handb Exp Pharmacol. 2006:305–326. doi: 10.1007/3-540-27262-3_15. [DOI] [PubMed] [Google Scholar]
- 18.Tucker BJ, Breaker RR. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]