Abstract
Erroneous replication of lesions in DNA by DNA polymerases leads to elevated mutagenesis. To understand the molecular basis of DNA damage-induced mutagenesis, we have determined the x-ray structures of the Y-family polymerase, Dpo4, in complex with a DNA substrate containing a bulky DNA lesion and incoming nucleotides. The DNA lesion is derived from an environmentally widespread carcinogenic polycyclic aromatic hydrocarbon, benzo[a]pyrene (BP). The potent carcinogen BP is metabolized to diol epoxides that form covalent adducts with cellular DNA. In the present study, the major BP diol epoxide adduct in DNA, BP-N2-deoxyguanosine (BP–dG), was placed at a template–primer junction. Three ternary complexes reveal replication blockage, extension past a mismatched lesion, and a −1 frameshift mutation. In the productive structures, the bulky adduct is flipped/looped out of the DNA helix into a structural gap between the little finger and core domains. Sequestering of the hydrophobic BP adduct in this new substrate-binding site permits the DNA to exhibit normal geometry for primer extension. Extrusion of the lesion by template misalignment allows the base 5′ to the adduct to serve as the template, resulting in a −1 frameshift. Subsequent strand realignment produces a mismatched base opposite the lesion. These structural observations, in combination with replication and mutagenesis data, suggest a model in which the additional substrate-binding site stabilizes the extrahelical nucleotide for lesion bypass and generation of base substitutions and −1 frameshift mutations.
Keywords: cancer, DNA replication, mutagenesis, translesion synthesis, polycyclic aromatic hydrocarbon
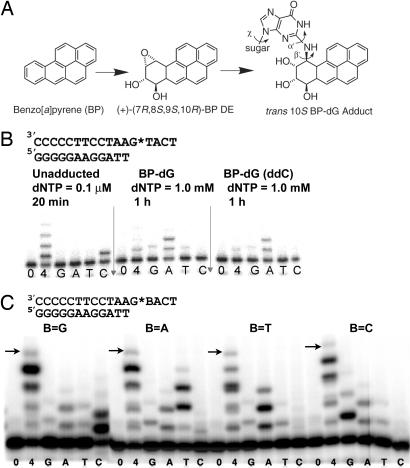
Translesion DNA synthesis (TLS), the extension of primers opposite damaged DNA templates, is a fundamental process that enables a cell to survive DNA damage (1–3). Although high-fidelity replicative DNA polymerases are generally blocked by DNA lesions, specialized DNA polymerases belonging to the Y-family are capable of replicating past DNA damage. These translesion polymerases are much less accurate than the replicative polymerases and can cause mutations by introducing errors as they replicate DNA. Thus, error-prone TLS by the Y-family polymerases provides a molecular connection between DNA damage and mutagenesis. Many mutagenic lesions in DNA are caused by environmental agents such as UV light or exogenous chemicals or their metabolic products. Polycyclic aromatic hydrocarbons (PAHs) are widespread environmental contaminants derived from incomplete combustion. They are present in cigarette smoke, automobile exhaust, smoked foods, and industrial emissions, and they enter the body through the air or through the diet (4, 5). Benzo[a]pyrene (BP) is a prototypical PAH that is highly tumorigenic and implicated in specific mutations found in lung tumors of smokers (6). BP is metabolized to BP diol epoxides (BP DEs) in mammals (7). These highly reactive DEs form covalent adducts with the purine bases of DNA (8). The predominant BP DE isomer, (+)-(7R,8S,9S,10R)-BP DE, is the most tumorigenic in animal models (7). Its reaction at N2 of guanine gives the major BP DE DNA adduct (trans 10S BP-N2-dG, Fig. 1A) in mammalian cells (9, 10). This adduct induces mainly base substitutions as well as −1 frameshift mutations in vivo (11, 12). In this study, we describe the structural basis for incorrect bypass of the BP–dG adduct by a Y-family polymerase.
Fig. 1.
Formation of a benzo[a]pyrene diol epoxide adduct (BP–dG) and its effect on DNA replication. (A) Structures of BP, its (+)-(7R,8S,9S,10R) diol epoxide (DE) metabolite and the trans 10S BP–dG adduct in DNA. The α′, β′, and χ torsion angles are labeled. (B) Incorporation of nucleotides opposite the BP–dG adduct in the template in comparison with incorporation opposite an unmodified dG residue. The BP–dG adduct is indicated as G*. Assays were performed with the dNTP concentrations and the times indicated. Lanes 0, 4, G, A, T, and C correspond to no dNTP, all four dNTPs, and each single dNTP, respectively. ddC (Right) indicates the 3′ end of the template strand is dideoxy-C, which is used in crystallization to prevent nucleotide addition at the blunt end (21). (C) Incorporation of dNTP opposite the BP–dG adduct in four sequence contexts by Dpo4. The arrows indicate the expected size of the full-length products. Reaction time was 1 hour with 100 μM dNTPs.
Y-family DNA polymerases are involved in the mutagenic bypass of BP adducts (1, 3, 13–15). There are multiple Y-family DNA polymerases in cells. The human enzymes polη and polκ can replicate past BP–dG (16). DNA polymerase IV (Dpo4) from Sulfolobus solfataricus is homologous to polκ, yet shows lesion bypass abilities akin to those of polη (17). It has become a model system for understanding of replication past DNA damage at an atomic level. Dpo4 consists of four domains, a three-domain classic polymerase core of palm, finger, and thumb domains and an additional “little finger” (LF) domain unique to the Y-family (18). The four domains are conserved in all of the Y-family DNA polymerases (18). The finger and thumb domains are unusually small and form an open active site with few DNA contacts. This spacious active site has been demonstrated to be responsible for replication slippage, mismatched primer extension, and lesion bypass (18–22). The LF domain of Y-family polymerases interacts with the major groove of the bound DNA. Attachment of this LF domain to the core domain creates a structural gap between the core and the LF domains. This structural gap harbors the extrahelical BP dG adduct at the template–primer junction in the protein–DNA–dNTP complexes reported here, solved at 2.5- and 2.25-Å resolution [supporting information (SI) Table 1]. The extrusion of the adducted base by template misalignment results in either base substitution or −1 frameshift. The present results reveal a previously unidentified structural basis for mutagenic replication by Dpo4.
Results and Discussion
Benzo[a]pyrene Intercalation Blocks G* Bypass.
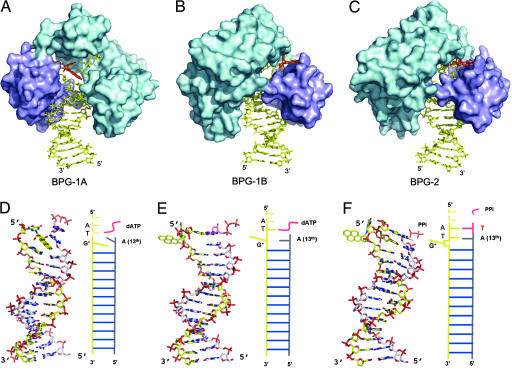
We have determined structures for the adducted DNA–Dpo4–dNTP complex in two crystal forms. In the first crystal (BPG-1), the DNA substrate has a mispaired G*:A at the template–primer junction (sequence in Fig. 1B plus an A base in the primer opposite the adducted base G*) and an incoming dATP matching the template dT 5′ to the lesion. In primer extension assays with the given template a dA residue was most efficiently incorporated opposite the dG* adduct (Fig. 1B), and this dA:dG* mispair was also most efficiently extended (Fig. 1B and SI Fig. 5A). With 1 mM dNTPs, bypass of the adduct was less efficient in the presence of all four dNTPs than with dATP alone (Fig. 1B). We suggest that the other dNTPs, which are poor substrates compared with dATP, may act as competitive inhibitors of dATP incorporation at high dNTP concentration. BPG-1 contains two ternary complexes, BPG-1A and BPG-1B, in the asymmetric unit. The structures represent intermediates for primer extension beyond the lesion site. The conformation of the Dpo4 is almost identical in the two complexes, with rmsd on Cα of 0.3 Å, but the conformation of the DNA differs in the structures.
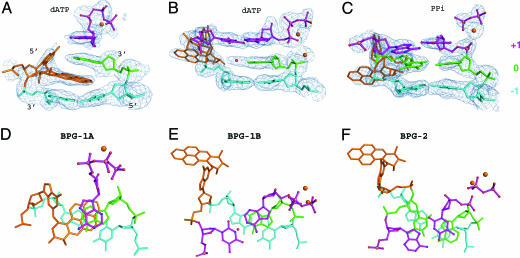
BPG-1A has the pyrene ring system intercalated into the DNA helix. The hydrocarbon occupies the space corresponding to one base pair and inserts between the last two bases at the primer strand terminus (Fig. 2). Intercalation of the BP rings displaces the G* base into the minor groove in a syn conformation (SI Table 2). Base-pairing is disrupted both at the lesion (0) position (G*:A) and at the position 5′ to the lesion (+1 position, T:dATP). By pointing toward the 3′ side of the modified G base, the BP ring system stacks with the bases surrounding the lesion site and has its long axis at an angle of ≈40° with the DNA duplex axis. Accordingly the A base at the primer strand end tilts by 40° from its ideal position and overwinds the helix by ≈15° to maximize its stacking interactions with the pyrene rings (Fig. 2D). The intercalation also disturbs the −1 base pair (A:T) that is beneath the pyrene rings, introducing 32° of buckle and −5° of opening, lengthening the N6(A)–O4(T) and N1(A)–N3(T) hydrogen bonds to 3.1 and 3.5 Å, respectively. The single-stranded template 5′ to the lesion is disordered.
Fig. 2.
Structures of BPG-1A, BPG-1B and BPG-2. (A–C) Dpo4 is represented as a molecular surface with the polymerase core in cyan and the LF domain in purple; DNA and nucleotide are shown as sticks, and BP–dG is highlighted in orange. BPG-1B and BPG-2 in B and C are rotated 180° relative to BPG-1A in A around the DNA helix axis, to show the extrahelical BP–dG in the gap between the core and LF domains. (D–F) The DNA conformations corresponding to (A–C) as stick models, all with the same orientations as in A. The primer strands are in gray, and incoming dATP is in pink. The single-stranded portion of the template DNA is not shown. Figs. 2, 3, and 4 were generated by using PYMOL (46).
Disruption of the base-pairing at the template–primer junction and distortion of the template prevents the incoming dATP from binding in the correct conformation for primer extension. Without an ordered template base with which to pair, the incoming dATP is flipped over, with its Hoogsteen edge facing the template strand to maximize its stacking with the 3′-terminal base A of the primer (Fig. 3). In contrast to a normal cis base-paired nucleotide, the trans paired dATP has the sugar ring nearly upside down and its α phosphate shifting away from the 3′ end of the primer (Fig. 3). The distance between the 3′ OH of the primer strand and Pα of the dATP is 9.0 Å, too great for the phosphoryl transfer reaction to occur. In addition, only one Ca2+ was found in the active site, occupying the “B” site of the two catalytic metal ion-binding sites (18, 20). The complex BPG-1A thus represents a nonproductive conformation in which intercalation of the hydrocarbon prevents primer extension.
Fig. 3.
The BP–dG adduct and its surrounding base pairs in the active site. The base pairs are colored according to their position relative to the lesion site: The lesion site (0) is green, +1 (above) is purple, and −1 is light cyan. The adducted nucleotide itself is orange. The DNA and dATP are shown as ball-and-stick models with Ca2+ ions and water molecules shown as orange and red spheres. (A–C) Side views of the base pairs with the simulated annealing omit map contoured at 2.5σ at 2.5 Å and 2.25 Å resolution, respectively. (D–F) Views rotated 90° relative to the corresponding image in A–C. The oxygen, nitrogen, and phosphorus atoms are colored red, blue, and pink, respectively, for the top layer.
BP–dG Flipping-Out and Base Substitutions.
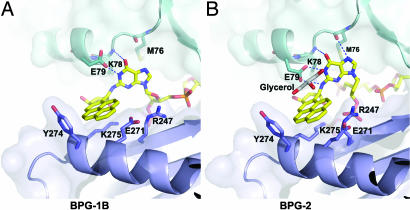
In marked contrast, the BP adducted G* is flipped from the minor groove side into the gap between the LF and the core domains in the second complex (BPG-1B) (Fig. 2). In the enzyme-substrate complex, the conformation of the adduct with χ, α′, and β′ torsion angles (Fig. 1A) of −153.0, −160.0, and 161.7, respectively, does not lie in a low-energy region as predicted for the corresponding nucleoside adduct in the absence of an enzyme (23). In this conformation, the BP–dG adduct would be exposed completely to the solvent if there were no protein present. Full exposure of the hydrocarbon rings and aromatic base G is extremely unfavorable in an aqueous environment. None of the experimentally observed DNA structures containing BP–dG adducts have shown such a fully exposed conformation (24–27). The protein-free NMR structures have the BP ring system either intercalated or bound to DNA in the minor groove to avoid full exposure of the aromatic rings to water. However, such a BP ring-exposed conformation is stabilized in BPG-1B by burying the hydrophobic moieties in the gap between the core and the LF domains of Dpo4. The interface of the core and the LF domains is near the DNA template–primer junction and thus can accommodate this otherwise unfavorable conformation. When oriented in the gap, the pyrene ring packs against the LF domain, whereas the purine contacts the catalytic core domains (Fig. 4). The BP rings contact the hydrophobic parts of Lys 275 and Glu 271 (LF domain), with the outside edge being partially shielded by Tyr 274. The adducted base is stabilized by two H bonds between N1 of the base and the side chain of Glu 79 and between O6 of G* and the main-chain N of Lys 78, as well as by hydrophobic interactions between the purine ring and Met 76 and the hydrophobic portion of Lys 78 (Fig. 4).
Fig. 4.
Close-up views of BP–dG in the structure gap between the core and the LF domains. (A) BPG-1B. (B) BPG-2. The protein is in ribbon models covered by a transparent molecular surface. The key residues interacting with the adduct G* are shown as stick models. The BP ring system is in van der Waals contact with the LF domain (purple); the adducted G base interacts with the core domain (cyan). The glycerol molecule is in gray.
Flipping-out of the BP lesion from the intrahelical space reduces the structural distortion of the DNA duplex and allows base pairs around the lesion to stack with each other as in a normal B-form DNA (Fig. 2). The replicating base pair T:dATP and the two unpaired template nucleotides 5′ to the replicating base pair are structured, in contrast to a substantial disordering of this region in the intercalated structure BPG-1A. This allows the incoming dATP to bind normally in a standard Watson–Crick base pair (Fig. 3). The dATP sugar and phosphate moieties are now in their proper positions, and the distance between the primer 3′ OH and the Pα is 3.9 Å. This distance is small enough to produce a catalytically productive conformation with only relatively minor conformational adjustments. Both metal ions were observed in the active site at their canonical binding sites (20, 28).
Extrusion of the BP from the helix leaves a noninstructional gap in the template strand. This gap is occupied by the terminal A of the primer strand, which would otherwise have been paired with the flipped-out G* (Fig. 2). A water molecule (red sphere, Fig. 3B) fills the remaining space between the A at the primer terminus and the opposite strand of the template. This noninstructional space in the template strand potentially allows the incorporation of any nucleotide opposite the lesion, which would account for base substitutions induced by the BP-adduct. The conformation in BPG-1B could arise from untemplated incorporation of dA, or it could arise from a pairing of the dA with the dT 5′ to the lesion and a subsequent rearrangement. To examine this possibility with Dpo4, a second crystal was prepared (see next section).
BP–dG Looping-Out and −1 Frameshift Mutations.
The second crystal (BPG-2) was cocrystallized Dpo4 with the same adducted DNA but with dTTP as the incoming nucleotide to match the A base that is the second template base 5′ to G*. In this structure, dA in the primer strand has skipped the dG* and paired with the dT 5′ to the lesion. This complex shows a successful extension of a misaligned adduct in the template–primer junction (Fig. 2). The dT has been incorporated into the primer strand, with PPi remaining in the active site. The remaining PPi was observed in the ternary complex of Dpo4 with a mismatched base pair (G:T) in the active site (22). The slow release of PPi has been suggested as facilitating pyrophosphorolysis, a reversal of DNA polymerization that might serve as a proofreading step for Y-family polymerases. Instead of flipping out of the adducted base as in BPG-1B, the whole BP-adducted nucleotide unit, including the sugar–phosphate backbone, is looped out of the DNA helix by template misalignment in BPG-2. The looping-out was secured by two base pairs 5′ to the lesion site. This conformation of BP–dG (with χ, α′, and β′ torsion angles of 87.0, −100.0, and 179.0, respectively) is energetically unfavorable based on molecular simulation (23). The BP–dG adduct again is extruded from the minor groove side and occupies the cleft between the core and LF domains. The BP ring system is stabilized by the same residues from the LF domain as in BPG-1B, with the solvent accessible face covered by a glycerol molecule (Fig. 4). The carboxyl group of Glu-79 and main chain N and O from Lys-78 and Met-76 in the palm domain form four H-bonds with N1, N2, O6 and N7 of the G* base. The G* stacks against Met 76 and Lys 78 as in BPG-1B. Therefore, the bulky adduct is sandwiched between the LF and the palm domains in both structures.
With the BP–dG completely looping out, the template dT 5′ to the lesion is brought down to fill the gap left by the BP–dG and to pair with the A at the 13th position of the primer strand, resulting in a −1 frameshift (Fig. 2F). The very weak top bands for the full-size product of primer extension in Fig. 1C indicate that the major product is one base shorter than the template strand as a result of template misalignment. When the 5′ dT is shifted down to fill the gap, it is pushed much farther into the center of the helix, closer to the primer strand (green base pair in Fig. 3 C and F). To retain its normal backbone position in the protein active site, the complementary dA at the 13th position in the primer strand rotates from an anti to a syn conformation and exposes its Hoogsteen edge to the shifted dT next to G*. Interestingly, the shifting of the dT 5′ to the lesion propagates to its neighboring dA at the replicating (+1) position, such that this dA also assumes a syn conformation to compensate for the shortened distances between the template and primer strands in BPG-2 (Fig. 3). As a result, the two base pairs after the looped-out adduct are Hoogsteen base pairs (Fig. 3F). The C1′–C1′ distances in the Hoogsteen base-pairs are 7.7 and 8.3 Å, respectively, shorter than the corresponding distance (≈10.5 Å;) for a Watson–Crick base pair. The Hoogsteen base pairs accommodate the distortion caused by the template misalignment and permit the primer backbone to fit into the active site of the protein for extension. The incorporation of dT into the growing primer strand in BPG-2 indicates that the loop-out conformation is productive. In this state, the polymerase is in the process of moving along the template strand to the next base.
Template Misalignment and the 5′ Rule.
The loop-out of BP–dG provides a structural basis for 5′ sequence-dependent base substitutions and −1 frameshift mutations induced by the major BP–dG adduct in vivo (11, 12, 29, 30). Once the lesion is looped out by template misalignment, primer extension proceeds by using the nucleotide 5′ to the lesion as template for incoming nucleotides. In consequence, the base 5′ determines the identity of the base incorporated opposite the lesion, which is referred as the 5′ rule (20). The mutations induced by BP–dG in Escherichia coli are >95% G → T in a 5′-TG*C sequence and >95% G → A in a 5′-AG*A sequence, respectively, which follow the 5′ rule and are Y-family polymerase-dependent (12, 29, 30). The 5′ rule is also clearly demonstrated by the present replication assays on templates that have four different bases 5′ to the BP–dG lesion (Fig. 1C). In the presence of a single dNTP, primer extension was most efficient when the incoming dNTP was complementary to the base 5′ to the adducted G* (Fig. 1C). For the 5′-TG*A-3′ sequence in the crystal structures, the preference for A incorporation opposite the lesion is a result of looping-out of the modified base (BPG-2) and a subsequent strand realignment (BPG-1B). A −1 frameshift will result if the entire adducted residue loops out, and strand realignment does not occur (BPG-2). In addition, the pronounced 5′ dependency for base incorporation by Dpo4 opposite BP–dG has not been observed for Dpo4 with other types of lesions. Bypass of the minor cis-(10R)-BP-N6-dA (BP–dA) adduct by Dpo4 strongly follows the A rule, which shows preferential insertion of dA opposite a lesion (21, 31). This is consistent with the Dpo4/BP–dA structure in which the dA* is directly paired with dA at the active site (21). With an abasic lesion, both dependences on the 5′-base and the A rule were observed, indicating possible direct insertion of dA opposite the lesion (20). In contrast, replication past the BP–dG lesion by Dpo4 strictly follows the 5′ rule, not the A rule (Fig. 1C). This mechanism clearly supports a link between base substitutions and frameshift mutations and suggests a straightforward explanation for 5′ sequence dependence in base substitutions induced by the bulky adducts in E. coli (2, 12, 30, 32).
Additional Substrate-Binding Site Provides a Previously Uncharacterized TLS Mechanism.
Previous structural analyses of Dpo4 have attributed its lesion bypass ability to its open and solvent-accessible active site that can accommodate DNA lesions in productive conformations (19–21). No direct stabilization was provided by Dpo4 to the DNA lesions in the previous studies. For instance, Dpo4 bypasses a minor cis-(10R)-BP-N6-dA adduct by placing the PAH moiety into the solvent-accessible major groove (N6 is on the major-groove side). The BP–dA adduct is stabilized by packing one face of the PAH against base pairs in the major groove (21). Loop-out bypass by Dpo4 was also observed for abasic lesions (20), but its structural basis differs from that for bypass of the present BP–dG adduct. In the structure of Dpo4 complexed with an abasic lesion (Ab-2A), the looped-out backbone is in the open active site and exposed to solvent (20). A small and hydrophilic abasic lesion (a sugar–phosphate unit without a base moiety) does not need to be shielded from water. In the absence of an enzyme, the hydrocarbon portion of the BP-N2-dG adduct either intercalates with base pairs or lies in the minor groove (N2 is on the minor groove side) in NMR structures (24–26). PAH intercalation blocks the primer extension past BP–dG in BPG-1A. The minor groove-bound bulky PAH in the NMR model would be in steric conflict with Dpo4 if it were put into the active site. A modeling study predicted either an altered Dpo4 structure with a wider gap between the palm and finger domains to accommodate the minor groove adduct or a DNA model with the PAH shifted into the major groove to fit BP–dG into the active site (33). In the present crystal structures, Dpo4 stabilizes this hydrophobic lesion in a pocket outside the active site. The stabilization of a productive conformation allows the adducted DNA and the incoming dNTP to fit into the catalytic site and permits bypass synthesis, which reveals a structural basis different from all previous observations for translesion synthesis. There is no significant difference among the Dpo4 conformations in the three structures here, which closely resemble previously determined structures of Dpo4. The rmsd on Cα superimposition is within 0.6 Å on overlapping the present three structures with the previous structures of Dpo4 in complex with undamaged or damaged DNA substrates (18–21). Dpo4 is in a ready-to-go state and does not need to undergo conformational changes to accept a variety of DNA substrates. The additional site is an intrinsic part of Dpo4 and is well positioned near the template–primer junction to harbor bulky lesions, potentially resulting in template misalignment, regardless of the specific DNA lesion. In contrast, a crystal structure of a DNA substrate containing the same major BP–dG adduct in complex with DNA polymerase I from Bacillus stearothermophilus shows that this high-fidelity polymerase has no room to accommodate a productive conformation of BP–dG-adducted DNA, so that replication was completely blocked by the lesion (34).
The LF domain is conserved in all Y-family polymerases, and the structure gap between the LF and the core domains has been observed in many Y-family polymerase structures (35). The human Y-family polκ bypasses the major BP adduct with correct insertion of C (15). Notably, efficiency, (Vmax/Km)ins, for this correct insertion is increased by ≈4 to 11-fold when the base 5′ to the adduct is a G (36). To account for this observation, a misalignment mechanism whereby G* is looped out, and the 5′-neighboring G serves as a template for C insertion was suggested (36). Structural superposition of human Y-family polκ (37) and Dpo4 indicates that polκ may use the same additional binding site to harbor the looped-out adduct, which cannot fit into the active site without intercalation. Polκ could stabilize the adducted base better than Dpo4 by stacking the G base with Phe 171, which is equivalent to Lys 78 in Dpo4 (SI Fig. 5C). Polκ also has a unique “N-clasp” to encircle the DNA for tight DNA binding (37). These two structural features of polκ may contribute to its efficient bypass of the BP adduct. Furthermore, strand misalignment is a spontaneous process that induces frameshift mutations (38, 39). The frequency of frameshift mutations in replication depends on the stability of misaligned intermediates at a template–primer junction. The additional binding site that stabilizes extrahelical bases may also facilitate generation of frameshift mutations in DNA replication. The study reported here demonstrates a mechanism for translesion synthesis by a Y-family polymerase past bulky and hydrophobic DNA adducts and extends our understanding of carcinogen-induced mutagenesis.
Materials and Methods
Adducted Oligonucleotide.
The template oligonucleotide (17-mer) used for the present crystal structures, 5′-(TC ATG* AAT CCT TCC CCC)-3′, with the trans (10S)-BP–dG adduct indicated by the asterisk and a dideoxycytidine residue at the 3′ terminus, was synthesized essentially as described (40). The adducted oligonucleotide was purified by reverse phase HPLC and annealed with a slight excess of its 13-mer primer strand, 5′-(GGG GGA AGG ATT A)-3′. The resulting duplex was separated from the excess primer by HPLC on a hydroxyapatite column (for further details see SI Materials and Methods).
Crystallization and Structure Determination.
Dpo4 was expressed and purified as described (18). Crystals were produced by vapor diffusion. The details of crystallization are described in SI Materials and Methods. The diffraction data were collected at APS beamline 19-ID and CHESS beamline F-1. The data sets were processed with HKL (41). The structures were solved by molecular replacement with CNS (42) or PHASER (43) by using the type I structure as a search model (18). The structures were iteratively adjusted with O (44) and refined by using CNS and Refmac5 (45) with all residues in the allowed regions of the Ramachandran plot.
Primer Extension.
Primer extension by Dpo4 was performed at 37°C with 10 nM DNA substrate with 5′ 32P-labeled primer and 100 μM dNTPs (or as indicated in Fig. 1). The buffer used contained 40 mM Tris·HCl (pH 8.0), with 5 mM MgCl2, 10 mM DTT, 0.35 mg/ml BSA, and 2.5% glycerol, along with 10 nM Dpo4. Oligonucleotides were prepared by preannealing of 150 nM template and 100 nM primer. Products were separated on 20% polyacrylamide 7 M urea gels and visualized by PhosphorImager analysis.
Supplementary Material
Acknowledgments
We thank Dr. R. Zhang (Structural Biology Center, Argonne National Laboratory, Argonne, IL) who collected BPG-1 data. The CHESS in which BPG-2 data were collected is supported by the National Science Foundation and the National Institutes of Health (NIH)/National Institute for General Medical Sciences under award DMR-0225180. We thank Dr. M. Cai (NIH) for some of the primer extension studies. This work was supported by the Terry Fox Foundation and the Intramural Research Program of the NIH (National Institute of Diabetes and Digestive and Kidney Diseases). H.L. acknowledges the New Investigators Award from the Canadian Institutes of Health Research.
Abbreviations
- PAH
Polycyclic aromatic hydrocarbons
- BP
benzo[a]pyrene
- BP–dG
BP diol epoxide-N2-deoxyguanosine, the major adduct of (+)-(7R,8S,9S,10R)-BP DE, formed by trans addition of N2-dG to (+)-(7R,8S,9S,10R)-BP DE
- Dpo4
DNA polymerase IV
- TLS
translesion synthesis
- LF domain
little finger domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.Y. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2IA6 (BPG-1) and 2IBK (BPG-2)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0700717104/DC1.
References
- 1.Friedberg EC, Wagner R, Radman M. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 2.Goodman MF. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR. Mutat Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 4.Committee on the Biological Effects of Atmospheric Pollutants. Washington DC: Natl Acad Sci; 1972. [Google Scholar]
- 5.Phillips DH. Mutat Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 7.Thakker DR, Yagi H, Levin W, Wood AW, Conney AH, Jerina DM. Bioactivation of Foreign Compounds. New York: Academic; 1985. [Google Scholar]
- 8.Jerina DM, Chadha A, Cheh AM, Schurdak ME, Wood AW, Sayer JM. Adv Exp Med Biol. 1991;283:533–553. doi: 10.1007/978-1-4684-5877-0_70. [DOI] [PubMed] [Google Scholar]
- 9.Vock EH, Wolfe AR, Meehan T. Mutat Res. 2001;478:199–206. doi: 10.1016/s0027-5107(01)00153-1. [DOI] [PubMed] [Google Scholar]
- 10.Koreeda M, Moore PD, Wislocki PG, Levin W, Conney AH, Yagi H, Jerina DM. Science. 1978;199:778–781. doi: 10.1126/science.622566. [DOI] [PubMed] [Google Scholar]
- 11.Lenne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP. Mol Microbiol. 2000;38:299–307. doi: 10.1046/j.1365-2958.2000.02116.x. [DOI] [PubMed] [Google Scholar]
- 12.Shukla R, Geacintov NE, Loechler EL. Carcinogenesis. 1999;20:261–268. doi: 10.1093/carcin/20.2.261. [DOI] [PubMed] [Google Scholar]
- 13.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. DNA Repair (Amsterdam) 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Chiapperino D, Kroth H, Kramarczuk IH, Sayer JM, Masutani C, Hanaoka F, Jerina DM, Cheh AM. J Biol Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- 15.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Wu X, Guo D, Rechkoblit O, Geacintov NE, Wang Z. Mutat Res. 2002;510:23–35. doi: 10.1016/s0027-5107(02)00249-x. [DOI] [PubMed] [Google Scholar]
- 17.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling H, Boudsocq F, Woodgate R, Yang W. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 19.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 20.Ling H, Boudsocq F, Woodgate R, Yang W. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 21.Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W. Proc Natl Acad Sci USA. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaisman A, Ling H, Woodgate R, Yang W. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie XM, Geacintov NE, Broyde S. Biochemistry. 1999;38:2956–2968. doi: 10.1021/bi9825605. [DOI] [PubMed] [Google Scholar]
- 24.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, Patel DJ. Proc Natl Acad Sci USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosman M, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 26.Geacintov NE, Cosman M, Hingerty BEAS, Broyde S, Patel DJ. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Lin C, Huang X, Kolbanovskiy A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. J Mol Biol. 2005;346:951–965. doi: 10.1016/j.jmb.2004.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steitz TA. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 29.Mackay W, Benasutti M, Drouin E, Loechler EL. Carcinogenesis. 1992;13:1415–1425. doi: 10.1093/carcin/13.8.1415. [DOI] [PubMed] [Google Scholar]
- 30.Yin J, Seo KY, Loechler EL. DNA Repair (Amsterdam) 2004;3:323–334. doi: 10.1016/j.dnarep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Strauss BS. DNA Repair (Amsterdam) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez H, Loechler EL. Mutat Res. 1995;326:29–37. doi: 10.1016/0027-5107(95)00149-d. [DOI] [PubMed] [Google Scholar]
- 33.Perlow-Poehnelt RA, Likhterov I, Scicchitano DA, Geacintov NE, Broyde S. J Biol Chem. 2004;279:36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 34.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 35.Yang W. FEBS Lett. 2005;579:868–872. doi: 10.1016/j.febslet.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Kolbanovskiy A, Wu X, Zhang Y, Wang Z, Zhuang P, Amin S, Geacintov NE. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 37.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Streisinger G, Owen J. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Diaz M, Kunkel TA. Trends Biochem Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Kroth H, Yagi H, Sayer JM, Kumar S, Jerina DM. Chem Res Toxicol. 2001;14:708–719. doi: 10.1021/tx0002637. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 44.Jones TA, Zou JY, Cowan SW. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. Pymol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.