Abstract
Many cancers harbor homozygous DNA deletions (HDs). In contrast to other attributes of cancer cells, their HDs are immutable features that cannot change during tumor progression or therapy. I describe an approach, termed deletion-specific targeting (DST), that employs HDs (not their effects on RNA/protein circuits, but deletions themselves) as the targets of cancer therapy. The DST strategy brings together both existing and new methodologies, including the ubiquitin fusion technique, the split-ubiquitin assay, zinc-finger DNA-recognizing proteins and split restriction nucleases. The DST strategy also employs a feedback mechanism that receives input from a circuit operating as a Boolean OR gate and involves the activation of split nucleases, which destroy DST vector in normal (nontarget) cells. The logic of DST makes possible an incremental and essentially unlimited increase in the selectivity of therapy. If DST strategy can be implemented in a clinical setting, it may prove to be curative and substantially free of side effects.
Keywords: split nucleases, split ubiquitin, zinc fingers
A major obstacle to drug-based therapies of human diseases that are both efficacious and substantially free of side effects is the massive interconnectedness and redundancy of molecular circuits in living cells. In the case of cancer, the problem is exacerbated by genomic instability of many, possibly most, cancers. This property increases heterogeneity of malignant cells in the course of tumor progression or anticancer treatment and is one reason for the failure of most drug-based cancer therapies (1, 2). A few relatively rare cancers, such as testicular carcinoma, Wilm's kidney tumor, and some leukemias in children, can often be cured through chemotherapy but require cytotoxic treatments of a kind that cause severe side effects and are themselves carcinogenic (3, 4). Several recent advances, including the use of antiangiogenic compounds and inhibitors of specific kinases, hold the promise of efficacious, curative therapies (5–7). Nevertheless, major human cancers are still incurable once they have metastasized.
In the present work, I suggest an approach to cancer therapy that involves homozygous deletions (HDs). Recent studies have demonstrated that many human cancers, including major ones, contain a significant number of scattered homozygous deletions (8–21). A salient property of an HD that involves DNA sequences not present elsewhere in the genome is that HD cannot revert. For this and other reasons, HDs may prove to be a particularly appropriate target for therapy. The difficulty here is that HD is an “absence,” and therefore it cannot be a conventional molecular target. Nevertheless, an HD-specific anticancer regimen is feasible through a strategy described below (Figs. 1–3).
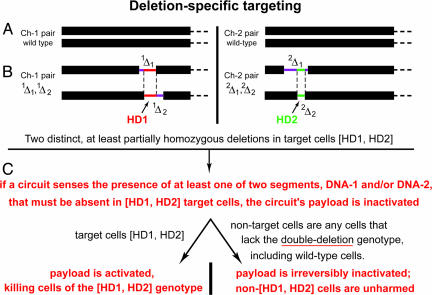
Fig. 1.
DST. (A) Chromosome pairs 1 and 2 (arbitrarily numbered) in a diploid cell. (B) The same chromosomes in a cell that contains two HDs, termed HD1 and HD2. Each of two HDs results from overlapping hemizygous deletions 1Δ1 and 1Δ2 in chromosome 1 and 2Δ1 and 2Δ2 in chromosome 2. The nonoverlapping parts of hemizygous deletions are in purple. Their overlapping parts, which comprise, respectively, HD1 and HD2, are in red on chromosome 1 and in green on chromosome 2. In Figs. 2 and 3 and in the main text, the segments of DNA that had been removed from wild-type cells as a result of homozygous deletions HD1 and HD2 are called, respectively, DNA-1 and DNA-2. (C) Outline of DST.
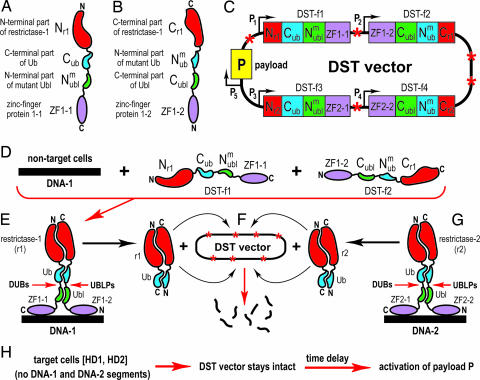
Fig. 2.
DST devices and their implementation. See DST Circuit and Its Operation for descriptions of specific components and circuits shown. (A) DST-fusion-1 (DST-f1). (B) DST-f2 fusion. (C) The ORFs of DST vector, which contains at least five ORFs. The ORF(s) encoding “payload” P (e.g., a conditionally toxic protein) is in yellow. See item 11 in the main text for a description of payload's design and induction. (D) DST vector enters normal (nontarget) cells, which contain either one or both of DNA segments, DNA-1 and DNA-2, that are absent in target cells, owing to the homozygous deletions HD1 and HD2. Only DNA-1 and its ligands DST-f1 and DST-f2 are shown in D. The other pair of DST fusions, which recognize DNA-2, is shown in G. (E) The operation of DST circuit in a normal (nontarget) cell, which is depicted, without loss of generality, to contain both DNA-1 and DNA-2. It may also contain just one of two DNA segments. DUB, deubiquitylating enzyme; UBLP, Ubl-specific protease. (F) Conditional destruction of DST vector. The released r1 nuclease becomes free to target its cleavage sites in DST vector, digesting it to small fragments and thereby halting the expression of its ORFs, including the (possibility of) expression of vector's payload P. Note that payload's expression had not been induced as yet at this stage. (G) If both DNA-1 and DNA-2 are present in a non-target cell, as shown here, an otherwise identical liberation of the reconstituted nuclease r2 takes place as well. (H) Events in a target cell, i.e., the one that contains both HD-1 and HD-2, and had received DST vector. Because neither DNA-1 nor DNA-2 are present in such cells, the reconstitution of r1 or r2 (which require the binding of ZF domains to DNA-1 or DNA-2) does not take place, and the DST vector stays intact. After an empirically optimized time delay, to allow DST circuits to search for DNA-1 and/or DNA-2, the vector's payload P (e.g., a cytotoxic protein) can be activated.
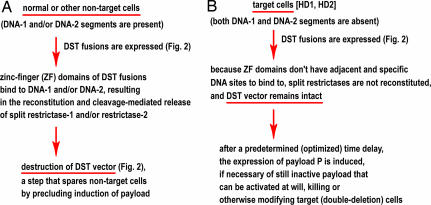
Fig. 3.
Summary of DST strategy and its logic. (A) DST in normal (nontarget) cells, which contain either both DNA-1 and DNA-2 segments or at least one of them. All of such cells are spared, given the logic of DST circuits described here, in the legend to Fig. 2, and in the main text. (B) DST in target cells, lacking both DNA-1 and DNA-2 segments.
This strategy, termed deletion-specific targeting (DST), employs homozygous deletions as “negative” targets of cancer therapy. The DST strategy is implemented through molecular circuits that combine both existing and new methodologies. One of the methods is the ubiquitin (Ub) fusion technique (22, 23). In addition, an essential part of DST strategy is based on “helper-dependent” split-protein devices, introduced by Johnsson and Varshavsky (24) in 1994 with the split-Ub assay and thereafter extended to other split-protein constructs, including dihydrofolate reductase (DHFR) (25), GFP (26–28), and β-lactamase (29, 30). Split-protein domains coupled to DNA-recognizing proteins (31) are also a component of DST strategy. Yet another part of DST is a conditional destruction of DST vectors by (reconstituted) split-restriction nucleases and a time delay in activating a vector's payload (Figs. 1–3). A major advantage of DST is its essentially unlimited selectivity. As described below, the logic and scope of DST is relevant to applications beyond HDs, as well as outside of cancer.
Homozygous DNA Deletions in Many, Possibly Most, Cancers.
Germline DNA of phenotypically normal humans has been shown to contain, on average, ≈30 hemizygous deletions larger than 5 kb, with regions of hemizygocity owing to deletions encompassing ≈550 kb altogether (<0.02% of the genome) (32, 33). Given the rarity of these hemizygous deletions, the bulk of homozygous deletions observed in cancer cells are de novo ones, acquired during tumor initiation and/or progression. Cancer-associated deletions that are relevant to the DST strategy are exclusively those that are at least partially homozygous and involve unique nucleotide sequences (Fig. 1). Although a hemizygously deleted DNA can be phenotypically similar to a homozygous deletion, owing, for example, to methylation of the region's remaining copy (34), only HDs are relevant to the DST strategy. Besides their advantage of zero reversion frequency, an HD is also a “digital” entity, in that the absence versus presence of a DNA sequence enables more robust designs that use deletions as targets.
Recent methods for detecting copy number changes in large genomes added a number of cancer-associated homozygous deletions to relatively few HDs that were unambiguously identified in earlier studies. Many cancers harbor HDs at fragile sites, defined (outside the context of cancer) as chromosomal regions that stain weakly in mitotic chromosome spreads of cells subjected to stress during DNA replication. For example, human cell lines derived from carcinomas of the stomach, lung, breast, ovary and colon often contain HDs, of a few to 200 kb in size at one or both of the two fragile sites called FRA3B and FRA16D (ref. 8 and references therein). Many other homozygous deletions were also identified in various cancers and in cell lines derived from them. These HDs encompass specific tumor suppressor genes such as, for example, SMARCB1/INI1 or PTEN, and other regions that are either known or suspected to contain tumor suppressors (9–20). One example is a study of multiple myeloma, a cancer originating from B-lymphocytes. Myeloma cells were shown to exhibit a broad range of copy-number changes in specific DNA regions, including multiple HDs whose sizes varied from 20 kb to 11 Mb (21). Whereas some deletions were observed in several myeloma patients, other HDs were apparently patient-specific (21). This pattern, which recurs in other cancers as well, suggests that some HDs are under positive selection in an evolving tumor, whereas other HDs of the same cancer, in the same patient, may be quasineutral, randomly retained deletions.
DST and Its Implementation.
Employing homozygous deletions, not their effects on tumor suppression and RNA/protein circuits but deletions themselves, as a target of therapy is, to my knowledge, a previously undescribed idea. Although the number of known cancer-associated HDs is already large, their current set is still the tip of the iceberg. In other words, future mapping studies would be likely to find that specific cancers in individual patients can be (nearly) always relied on to contain at least two homozygous deletions that satisfy the constraints of DST (Fig. 1). The DST strategy is independent, to a striking extent, of considerations that underlie other approaches to cancer therapy. For example, DST does not involve a function of deleted DNA, or its levels of expression in normal cells, or tumorigenic alterations of RNA/protein circuits in cancer cells, or cell-surface differences between them and normal cells.
Two HDs that are chosen for therapy are the sole selectivity determinants of DST strategy. The operation of the DST circuit, including the final stage at which the “decision” is made to either destroy a DST vector (if it entered a normal cell) or to allow the activation of a vector's payload (e.g., a cytotoxic protein) is described, step by step, in DST Circuit and Its Operation. Given the logic of DST (Figs. 1–3), a third and even fourth HD can be added as concurrent targets. Because the negative “spare this cell” output of a DST circuit is a part of its operation as a Boolean OR gate, an incremental addition of HDs as targets would increase the selectivity of treatment exponentially rather than linearly. The challenge, then, is to design a circuit that can sense, in effect, a DNA-dosage difference simultaneously for two (or more) loci at an error rate that can be made arbitrarily small. In a “complete” DST drug, a Boolean output of such a circuit is used to either irreversibly destroy a DST vector, thereby precluding activation of its payload, or to activate the payload if the circuit reports the presence of both HDs in a cell (Figs. 1–3). In a clinically relevant DST strategy, a DNA-based DST vector would be (nonspecifically) delivered into cells of a patient by using, for example, nonreplicating viruses or liposome-encapsulated DNA.
At this stage, the nearest aim is not a clinically realistic DST design but rather the circuit's ability to function as a DST device. Given the focus of DST on targets that cannot be altered in the course of tumor progression, a relevant design must be sensitive to HDs themselves, i.e., to the absence of specific DNA sequences. In other words, a DST device should not target, for example, a junctional DNA sequence at a deletion's breakpoint, because the latter, although unique to target cells, is not immune to change, in contrast to HD itself. Another desirable specification of a DST circuit is modularity, including the feasibility of increasing the circuit's selectivity by adding modules similar to those already present. These and other considerations led to a specific DST design whose components and operation are described in Fig. 2 and summarized in Fig. 3.
DST Circuit and Its Operation.
Fig. 1B depicts a pair of overlapping hemizygous deletions, 1Δ1 and 1Δ2. A segment of removed DNA in common between these deletions is termed DNA-1, and the corresponding homozygous deletion is termed HD1. 2Δ1 and 2Δ2 are another pair of hemizygous deletions. In Fig. 1B, they are located on a different pair of chromosomes, but they can also be located on the same pair of chromosomes as the first set of hemizygous deletions. DNA-2 is the segment of removed DNA in common between 2Δ1 and 2Δ2, and the corresponding (second) homozygous deletion is termed HD2 (Fig. 1B). What follows is a brief description of DST constructs and the circuit they comprise (Figs. 2 and 3).
As discussed above (Fig. 1), one identifies at least two homozygous deletions, termed HD1 and HD2, in a population of target cells. These deletions removed DNA segments termed DNA-1 and DNA-2. Although HD1 and HD2 should encompass unique DNA sequences, their minimally acceptable sizes can be as small as ≈100 bp (see item 6).
One “half” of DST circuit is implemented by two “complementary” protein fusions termed DST-f1 and DST-f2 (Fig. 2 A and B). The other, mechanistically identical “half” of the circuit is implemented by fusions termed DST-f3 and DST-f4 (Fig. 2G; see items 6 and 10). The fusion DST-f1 (Fig. 2A) consists of the following domains, beginning at its N terminus.
The first domain of DST-f1 (Fig. 2A) is an N-terminal fragment of a restriction endonuclease (restrictase-1, or r1) whose specific DNA cleavage site is absent from human DNA but is present, at multiple locations, in the DNA of the DST vector (Fig. 2C). Similarly to the previously characterized “helper-dependent” split proteins (see Introduction), the split restrictase r1 is constructed in such a way that a moderate-level coexpression of its N-terminal and C-terminal fragments (Fig. 2 A and B) cannot reconstitute the enzymatically active r1 restrictase, but it can be reconstituted if these fragments are brought into spatial proximity in vivo, as shown in Fig. 2E. One example of a site-specific endonuclease with requisite cleavage specificity is yeast SceI, which cuts DNA at an 18-bp-long recognition site (35). SceI is a member of the large and extensively characterized class of “homing” endonucleases, which mediate, in particular, the activity of selfish genetic elements (reviewed in ref. 36). Another class of site-specific endonucleases that can also be used to design a split restrictase includes artificial (engineered) ZF nucleases (ZFNs) (37–41). Although no split versions of restriction nucleases were described so far, extensive studies with other split-protein designs over the last decade (refs. 24–30 and references therein) suggest that a split restriction nuclease with required properties is feasible.
The next domain of DST-f1 (Fig. 2A) is Cub, a C-terminal half of the 76-residue Ub moiety and a part of the previously characterized split-Ub sensor (24).
The third domain of DST-f1 is a mutated N-terminal half of a Ub-like (Ubl) protein, for example NEDD8 or SUMO (42), that is a part of the additional (Ubl-based) split-protein sensor in the current DST design. Linking split-Ub and split-Ubl in the same pair of fusions (Fig. 2 A and B) makes possible a conditional cleavage of two polypeptide chains at once (after the last residue of Ub in DST-f1 and after the last residue of Ubl in DST-f2) and the resulting release of (reconstituted) restrictase r1, as shown in Fig. 2 E and F. A properly placed and double (as distinguished from single) proteolytic cleavage may be essential: a single cleavage would not release the reconstituted (previously split) restrictase r1 moiety, because the Ub halves and the Ubl halves of DST-f1/DST-f2 remain associated at this stage. No split versions of Ubl proteins were described so far. However, because all Ub-like proteins share the central feature of the Ub fold (a short α-helix over a β-fold), and because Ub fold-destabilizing mutations (in the N-terminal half of Ub) were previously characterized with the split-Ub sensor (24), it is nearly certain that a split Ubl protein with required properties is feasible. Technical note: it is possible (but remains to be verified) that a simpler design is feasible as well. A split Ubl moiety (instead of the second split-Ub moiety) was envisioned in DST-f1/DST-f2 fusions (Fig. 2 A and B) to bypass the problem of intramolecular (as distinguished from intermolecular) reconstitution of the Ub moiety. In other words, if the Cub half of Ub is followed, in DST-f1, by the Nub half of Ub, the two halves may be able to associate intramolecularly, an event to avoid: hence the use of split Ubl, which would be designed to be incapable of cross-associating with split Ub. However, if the linker sequences involved are made sufficiently short, steric constraints may prevent intramolecular folding and the (undesirable) reconstitution of the Ub moiety. If so, it may be possible to construct DST-f1/DST-f2 fusions with two otherwise identical split-Ub moieties oriented in opposite directions, instead of split-Ub and split-Ubl. Whether this simpler design is feasible remains to be determined.
The fourth and last domain of DST-f1 is ZF1–1, a ZF protein domain, similar to the previously described and extensively characterized ZF proteins that recognize specific DNA sequences (43–46). The ZF domains ZF1–1 and ZF1–2, of the fusions DST-f1 and DST-f2, respectively (Fig. 2 A and B), are designed to bind to two adjacent ≈9 bp (if necessary, longer) sequences of DNA-1 (Fig. 2D). The two ZF-binding sequences of DNA-1 are spaced apart to position their binding surfaces on the same side of the DNA double helix. Coexpression of DST-f1 and DST-f2, and the binding of ZF1–1 and ZF1–2 to the above two sequences of DNA-1 would bring the rest of DST-f1 and DST-f2 fusions into close proximity, thereby inducing reconstitution of the split restrictase r1, of Ub, and of Ubl, as shown in Fig. 2 D–F. This reassembly leads to cleavages by constitutively present deubiquitylating (DUB) enzymes after the last residue of Cub (24) and by (also constitutively present) Ubl-specific proteases (UBlPs) (42) after the last residue of Cubl, as shown in Fig. 2 E and F. These cleavages of both DST-f1 and DST-f2 release the now-active (reconstituted) restrictase r1 from its association with human DNA and lead to r1-mediated destruction of the DST vector (Fig. 2F). This vector encodes, in particular, the DST-f1 and DST-f2 fusions and, in addition, contains multiple cleavage sites, indicated by red asterisks in Fig. 2C, for restrictases r1 and r2, whose cleavage specificities are identical. The overall result of r1 activation (Fig. 2 E and F) is the destruction of DST vector, and thus the prevention of induction of its payload P under conditions in which the above DST circuit had detected the presence of DNA-1. Fig. 2G describes an identically designed pair of fusions, termed DST-f3 and DST-f4, that differ from the pair DST-f1/DST-f2 (Fig. 2 A, B, and D) in containing a distinct pair of zinc fingers (ZF2–1 and ZF2–2) that recognize DNA-2, a segment of DNA whose sequence differs from that of DNA-1 (Fig. 2 E–G). The involvement of DNA-2 converts a DST circuit into a Boolean OR gate in that a DST vector would be destroyed if just one of two DNA segments, DNA-1 or DNA-2, is engaged by corresponding DST fusions (Fig. 2 D–G). The DNA sequence-enabled assembly of a split protein, a part of the above circuit (Fig. 2 E and F), was described previously in a different (unrelated to DST) context of reconstituting, through the binding of ZF proteins to DNA, a split GFP protein (31, 47).
Fig. 2B depicts DST-f2, a fusion “complementary” to DST-f1. The two fusions can interact as a result of specific binding of the fusions' ZF1–1 and ZF1–2 moieties to adjacent DNA sequences of DNA-1, as described above and in Fig. 2 E and F.
Fig. 2C describes the ORFs of the DST vector, which contains at least five ORFs. The ORF(s) encoding “payload” P (e.g., a conditionally toxic protein) is in yellow. See item 11 for a description of the payload's design and induction. The other ORFs of the DST vector encode DST-f1 (Fig. 2A), DST-f2 (Fig. 2B), DST-f3, and DST-f4 (Fig. 2G). The latter pair of ORFs is identical to the one encoding DST-f1 and DST-f2, except for the following differences: ZF2–1 and ZF2–2 ZF domains of DST-f3 and DST-f4 recognize ≈9-bp DNA sequences distinct from those recognized by ZF1–1 and ZF1–2 (Fig. 2 A and B); and the split restrictase r2 of DST-f3/DST-f4, although of the same cleavage specificity as the restrictase r1, cannot “cross-reconstitute” with it. Red asterisks denote the cleavage site recognized by r1 and r2 that is present in multiple copies in the DST vector's DNA but is absent from human DNA. The DST vector is delivered into cells as nonspecifically as possible, either as a part of DNA viruses that are being developed for use in gene therapy, including therapy of cancer, or by any other route (e.g., a liposome–DNA complex) that involves a nonreplicating delivery vector, minimizes immunogenicity of the procedure and maximizes its efficiency (48–52).
Fig. 2 E–G describes the operation of the DST circuit in a normal (nontarget) cell. The latter is shown, without loss of generality, to contain both DNA-1 and DNA-2. (It can also contain just one of two DNA segments.) Upon expression by DST vector of DST-f1 and DST-f2, their DNA-recognizing domains ZF1–1 and ZF1–2 locate and bind to their adjacent recognition sites on human DNA-1. This double binding by ZF domains of the two fusions brings their polypeptide chains together and triggers reconstitution of both the restrictase r1, the Ub moiety, and the Ubl moiety of DST-f1/DST-f2. Reconstitution of the Ub and Ubl moieties results in cleavages of peptide bonds after the last residue of Cub in DST-f1 and after the last residue of Cubl in DST-f2 (see item 6). Because of the way in which Cub and Cubl are placed in DST-f1/DST-f2, these cleavages liberate the active (reconstituted) restrictase r1 moiety. This nuclease becomes free to target its cleavage sites in the DST vector, digesting it to small fragments (Fig. 2F) and thereby irreversibly stopping the expression of its ORFs, including, crucially, the (possibility of) expression of the vector's payload. Note that the payload's expression had not been induced as yet at this stage (see item 11).
Fig. 2G depicts the DNA sequence-directed association of the other two fusions, DST-f3 and DST-f4, followed by reconstitution of the split Ub moiety, the split Ubl moiety, and the split restrictase r2. The latter has the same cleavage specificity as r1 but differs from r1 in being unable to cross-associate with the halves of r1. Mechanistically, the association of DST-f3 and DST-f4 is similar to the association of DST-f1 and DST-f2, but the f3/f4 fusions recognize the presence of DNA-2, whereas f1/f2 recognize the presence of DNA-1. Because just one of these two association events (let alone both of them) would suffice for the activation of a restrictase(s) and destruction of DST vector (Fig. 2 E–G), any nontarget cell, i.e., a cell containing at least one of two DNA segments, DNA-1 and/or DNA-2, would be spared.
Fig. 2H summarizes events in a target cell (the one containing both HD1 and HD2 deletions) that received DST vector. Neither DNA-1 nor DNA-2 are present in such a cell, owing to the deletions HD1 and HD2. As a result, the reconstitution of r1 and/or r2 restrictases, which require the binding of ZF domains ZF1–1/ZF1–2 to (nonexistent) DNA-1, and/or of ZF2–1/ZF2–2 to (nonexistent) DNA-2, does not take place, and the DST vector stays intact. After an (empirically optimized) time delay, to allow DST circuits to search for DNA-1 and/or DNA-2, the expression of the vector's payload P (Fig. 2C) can be activated. Although later versions of DST may “automate” the induction of payload P, by making that induction a part of additional (time-delay) circuit encoded by DST vector, the delayed induction of payload P is “manual” in the current design, i.e., it is controlled by experimenter. It is possible that the P-induction step will stay manual in later elaborations of DST as well, because it may be beneficial to control the all-important step of payload's activation from “outside,” e.g., through the administration, to a patient, of an inducer of payload's toxicity after the state of DST vector (intact or destroyed) is verified by independent tests in at least some cells of a patient.
If the aim, in the end, is to kill target cells, rather than, for example, to induce their terminal differentiation, the range of possible toxic proteins to serve as a payload of the DST vector is quite broad and includes either bacterial or plant toxins, for example, diphtheria toxin or ricin. A gene that encodes payload P can be controlled, for example, by a nonleaky inducible promoter that can be activated by a small-compound inducer, such as, for instance, doxycycline or ecdysone (53, 54). If one wishes to interpose yet another level of temporal control, so that payload P (Fig. 2C) is expressed as a conditionally toxic protein, the range of current choices includes, for example, the herpes simplex virus thymidine kinase (HSV-tk) in the presence of its substrate acyclovir (55). Other options include, for instance, the use of small-compound dimerizers (56, 57), in which case the payload P (Fig. 2C) can be expressed as a split (conditional) toxin, with domains that bind to a cell-penetrating dimerizer, bringing the two halves of a toxic protein together in the presence of dimerizer and thereby making it possible to uncouple the induction of the payload's expression from the step of actually killing a target cell. Given several realistic choices of both the DST payload and its mode of activation, specific details of this last step (Fig. 2 C and H) are much less important at present than the nature of strict control over the possibility of payload expression that stems from the ability of the DST vector to self-destruct in response to entering a nontarget cell (Fig. 2F).
As mentioned in foregoing descriptions, the delivery of DST vector into cells can be nonspecific, because the selectivity of DST is an intracellular effect. If an actual, working DST circuit can be made as selective for target cells as the current designs suggest it may be, it would be best to employ a nonspecific, high-efficiency delivery, so that a sizable fraction of patient's cells, without regard to their nature, locations, or cell-cycle positions, receive a (transient) visit by DST vector in a given round of DST therapy. This would maximize the probability of not missing any target cells, irrespective of their heterogeneity in surface properties and other traits. For a more compact summary of DST and its mode of operation, see Fig. 3. Although fairly elaborate, the overall design (Fig. 2) is within reach of modern construction and expression routines.
Concluding Remarks
The DST strategy brings together both existing and new methodologies, such as the Ub fusion technique (22, 23); the split-Ub assay (24); ZF DNA-recognizing proteins (43–45); restriction nucleases that are derived either from engineered ZF proteins (37–41) or from homing endonucleases (35, 36) and that are configured (in DST designs) as split nucleases; DNA sequence-enabled assembly of a split protein (31); a new arrangement of split Ub-type domains in a polypeptide chain that enables a double proteolytic cleavage once two chains associate in vivo; and a new feedback mechanism that receives input from a circuit operating as a Boolean OR gate and involves the activation of split nucleases, which destroy the DST vector in normal (nontarget) cells (Figs. 1–3). A certainty that an HD in a cancer cell will be present in that cell and its progeny for as long as those cells endure may lead to a changed perspective on the nature of curative therapies. In what follows, I shall assume, for the sake of argument and without proof, that the DST strategy will advance, one day, from a diagram in the present work to an efficacious cancer therapy.
The above description of DST circuit emphasized its fundamentals rather than its technical details. Therefore it may be helpful to mention that reconstituted nucleases, which mediate the destruction of DST vector in nontarget cells (Fig. 2), can also be, for example, split recombinases such as Cre of the phage P1. A corresponding Cre-sensitive DST vector would contain multiple copies of loxP, the target of Cre, or (for example) a silent, loxP-containing, Cre-activated transcriptional promoter upstream of a gene encoding an intact (nonsplit) homing endonuclease. In the latter case, a DST vector would contain multiple cleavage sites for the endonuclease. In addition, the junction between a ZF domain and a split-Ub (or a split Ub-like) module in a DST fusion (Fig. 2) can be designed to activate a ZF-linked cryptic degron upon the fusion's cleavage, resulting in a short-lived ZF and multiple cycles of split-nuclease reconstitution on a single-copy DNA segment.
A DST system verifies the physical absence of a (homozygously) deleted DNA. In doing so, this circuit operates as a Boolean OR gate, erring on the side of caution: If the DST system (Figs. 2 and 3) appears to detect even one (of two or more) segment of DNA that is supposed to be absent from a target cell, the DST vector's genome is designed to irreversibly self-destruct, and the cell is spared. This happens before the cytotoxic step (the activation of the vector's payload) is even “considered” by a circuit. Depending on the specifics of a DST regimen, such an extent of double-checking may result, stochastically, in letting go of some target cells that should have been destroyed. Note, however, that if the therapy's selectivity for target cells versus nontarget (normal) cells is high enough, the resulting disappearance of side effects brings forth an opportunity that other, less-selective therapies are less able to afford: the option of repeated treatments. The nonreversion property of homozygous deletions is synergistic with the possibility of making the frequency of DST's error (i.e., the error of killing nontarget cells) arbitrarily low. As a result, a DST treatment can be administered repeatedly, with the usual concerns about side effects or alterations of targets either diminished or nonexistent.
Although HDs themselves would never be a reason for increased resistance to treatment, other sources of “acquired” resistance would be there, of course. However, they can be dealt with, because repeated treatments would now be feasible. The current DST strategy (Figs. 1–3) is based on macromolecular structures, and requires that a vector encoding them enters cells. Thus, a resistance to DST treatment can build up in ways that are similar to the routes that increase resistance to other cancer therapies as well. For example, if a DST vector is delivered into cells using a virus or a liposome-encapsulated DNA, the changing genetic landscape of a patient's cancer (in part because previous rounds of DST therapy eliminated a subset of cancer cells) may result in the remaining cancer cells being more resistant to entry of DST's carrier. A remedy, given the possibility of repeated DST treatments, would be to retain the DST vector but to deliver it through a different virus or modified liposomes. Strategies of this kind would also be expected to deal with the problem of a patient's immune responses to a viral vector. A potentially more difficult problem is an immune response to intracellular DST-specific proteins (Fig. 2). I mention such problems but do not discuss them here, because they recur in other protein-based anticancer therapies as well. One hopes that continuing advances in modulating immunological circuits selectively enough to reduce an undesirable response without shutting off the immune system will eventually address this difficulty.
The flexibility of DST in regard to repeated treatments has yet another advantage: it is unnecessary to focus on the same set of homozygous deletions throughout a DST therapy. Although Fig. 1 depicts the HDs HD1 and HD2 as being on separate chromosomes, this is not an actual constraint, because a set of relevant HDs can be present, a priori, anywhere in the genome. Cancer-associated HDs are presumed to form early in the process that led to a specific cancer, in part because some HDs eliminate genes for tumor suppressors. However, the known cancer-associated HDs (8–21) were identified either in cell lines established from tumors or by examining advanced cancers. Therefore it is still unclear whether a given cancer-associated HD is present in all cancer cells of a patient or in a large subset of them. Because the choice of HD targets remains flexible throughout DST therapy, the above (potential) complication can be dealt with by altering, if necessary, a set of targeted HDs. Apart from its therapeutic usefulness, such an alteration may also illuminate the temporal position of specific HDs in the history of a cancer. For example, if a choice of specific HDs as DST targets leads to eradication of cancer cells in a patient, this result would suggest that the chosen HDs were present in the earliest population of tumorigenic cells that gave rise to that cancer.
Although this work described DST ideas in the context of homozygous deletions (Figs. 1–3), a DST-type strategy is also relevant to any setting in which one wishes to distinguish amongst sets of cells that contain or lack specific DNA sequences and to target one or the other such set. Thus, cell populations with specific missense mutations, chromosomal translocations, hemizygous deletions (as distinguished from HDs), and copy-number increases in specific DNA regions (gene amplification) can also be a part of DST-type strategies. A major difference between DST-relevant homozygous deletions and other genetic alterations are both the permanence of HDs and their “digital” (all-or-none) quality, accentuated by relatively large sizes of cancer-associated HDs: hence the focus on homozygous deletions.
It is commonly assumed that if a universally applicable, curative, low-collateral-damage therapy of cancer is feasible at all, it would be attained through the understanding of tumor suppressors, oncoproteins, immune surveillance and their perturbations by cancer-causing mutations. The understanding of cancer's biology is already quite advanced (1). It is unknown, at present, whether the DST strategy will make the transition from a set of concepts to an efficacious cancer therapy. If it ever does, it would be striking that a curative treatment is unrelated to biological understanding of cancer, apart from its DNA-deletion specifications.
Acknowledgments
I thank Bert Vogelstein, Christopher Brower, Michael Grunstein, and Cheol-Sang Hwang for their helpful comments on the paper. This study was supported by National Institutes of Health Grants GM31530 and DK39520.
Abbreviations
- DST
deletion-specific targeting
- HD
homozygous deletion
- Ub
ubiquitin
- ZF
zinc finger
- ZFN
ZF nuclease.
Footnotes
The author declares no conflict of interest.
References
- 1.Weinberg RA. The Biology of Cancer. New York: Garland; 2006. [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Einhorn LH. Proc Natl Acad Sci USA. 2002;99:4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardman JG, Limbird LE, Gilman AG. The Pharmacological Basis of Therapeutics. New York: McGraw–Hill; 2001. [Google Scholar]
- 5.Folkman J. Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 6.O'Hare T, Corbin AS, Druker BJ. Curr Opin Genet Dev. 2006;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Sawyers C. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 8.Finnis M, Dayan S, Hobson L, Chevenix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Hum Mol Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 9.Kost-Alimova M, Imreh S. Sem Cancer Biol. 2007;17:19–30. doi: 10.1016/j.semcancer.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Modena P, Lualdi E, Facchinetti F, Galli L, Teixeira MR, Pilotti S, Sozzi G. Cancer Res. 2005;65:4012–4019. doi: 10.1158/0008-5472.CAN-04-3050. [DOI] [PubMed] [Google Scholar]
- 11.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 12.Jönsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringnér M, Höglund M, Borg Å. Genes Chromosomes Cancer. 2007;46:543–558. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, Xu J. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 14.Nakaya K, Yamagata HD, Arita N, Nakashiro K, Nose M, Miki T, Hamakawa H. Oncogene. 2007;26:1–9. doi: 10.1038/sj.onc.1210330. [DOI] [PubMed] [Google Scholar]
- 15.Cox C, Bignell G, Greenman C, Stabenau A, Warren W, Stephens P, Davies H, Watt S, Teague J, Edkins S, et al. Proc Natl Acad Sci USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struski S, Helias C, Gervais C, Audhuy B, Zamfir A, Herbrecht R, Lessard M. Cancer Genet Cytogenet. 2007;174:151–160. doi: 10.1016/j.cancergencyto.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh TJ, Welsch P, King M-C, Wigler MH. Proc Natl Acad Sci USA. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hustinx SR, Hruban RH, Leoni LM, Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Brown PN, Argani P, Ashfaq R, Fukushima N, et al. Cancer Biol Ther. 2005;4:83–86. doi: 10.4161/cbt.4.1.1380. [DOI] [PubMed] [Google Scholar]
- 19.Kasahara T, Bilim V, Hara N, Takahashi K, Tomita Y. Anticancer Res. 2006;26:4299–4306. [PubMed] [Google Scholar]
- 20.Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins VP. Genes Chromosomes Cancer. 2005;43:181–193. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 21.Largo C, Saéz B, Alvarez S, Suela J, Ferreira B, Blesa D, Prosper F, Calasanz MJ, Cigudosa JC. Haematologica. 2007;92:795–802. doi: 10.3324/haematol.11052. [DOI] [PubMed] [Google Scholar]
- 22.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 23.Varshavsky A. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 24.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Ma C, Chalfie M. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 28.Kerppola TK. Nat Rev Mol Cell Biol. 2006;7:449–556. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 30.Ooi AT, Stains CI, Ghosh I, Segal DJ. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. J Am Chem Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 32.Conrad DF, Andrews TD, Carter NP, Hurles MF, Pritchard JK. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 33.Sebat J, Lakshmi B, Troge J, Alexander J, Young JM, Lundin P, Måner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 34.Liu TX, Becker MW, Jelinek J, Wu WS, Deng M, Mikhalkevich N, Hsu K, Bloomfield CD, Stone RM, DeAngelo DJ, et al. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 35.Jasin M. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 36.Stoddard BL. Quart Rev Biophys. 2006;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 37.Porteus MH, Baltimore D. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 38.Porteus MH, Carroll D. Nat Biotech. 2006;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 39.Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 40.Szepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 41.Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L, Rupniewsky I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. Nat Biotech. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 42.Kerscher O, Felberbaum R, Hochstrasser M. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 43.Choo Y, Sanchez-Garcia I, Klug A. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 44.Jamieson AC, Miller CJ, Pabo CO. Nat Rev Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 45.Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, Brachat A, Brakenhoff RH, Quigley JP, Erdmann D, Barbas CFI. Proc Natl Acad Sci USA. 2005;102:11716–11721. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papworth M, Kolasinska P, Minczuk M. Gene. 2006;366:27–38. doi: 10.1016/j.gene.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh I, Stains CI, Ooi AT, Segal DJ. Mol BioSyst. 2006;2:551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 48.Verma IM, Weitzman MD. Annu Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 49.McCormick F. Oncogene. 2005;24:7817–7819. doi: 10.1038/sj.onc.1209064. [DOI] [PubMed] [Google Scholar]
- 50.Vasileva A, Jessberger R. Nat Rev Microbiol. 2005;3:837–847. doi: 10.1038/nrmicro1266. [DOI] [PubMed] [Google Scholar]
- 51.Limberis MP, Wilson JM. Proc Natl Acad Sci USA. 2006;103:12993–12998. doi: 10.1073/pnas.0601433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schillinger KJ, Tsai SY, Taffet GE, Reddy AK, Marian AJ, Entman ML, Oka K, Chan L, O'Malley BW. Proc Natl Acad Sci USA. 2005;102:13789–13794. doi: 10.1073/pnas.0506807102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schönig K, Schwenck F, Rajewsky K, Bujard H. Nucleic Acids Res. 2002;30:e134. doi: 10.1093/nar/gnf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saez E, Nelson MC, Eschelman B, Banayo E, Koder A, Gho GJ, Evans RM. Proc Natl Acad Sci USA. 2000;97:14512–14517. doi: 10.1073/pnas.260499497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 2006. [Google Scholar]
- 56.Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. Chem Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 57.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]