Abstract
Single rat ventricular myocytes and human ventricle tissue sections were labeled with antibodies against the ryanodine receptor (RyR) and α-actinin to examine the 3D distribution of RyRs with confocal microscopy. Image contrast was maximized by refractive index matching and deconvolution. The RyR label formed discrete puncta representing clusters of RyRs or “couplons” around the edges of the myofilaments with a nearest-neighbor spacing of 0.66 ± 0.06 μm in rat and 0.78 ± 0.07 μm in human. Each bundle of myofibrils was served by approximately six couplons, which supplied a cross-sectional area of ≈0.6 μm2 in rat and ≈0.8 μm2 in human. Although the couplons were in reasonable registration with z-lines, there were discontinuities in the longitudinal position of sarcomeres so that dislocations in the order of RyR clusters occurred. There was ≈53% longitudinal registration of RyR clusters, suggesting a nonrandom placement of couplons around the sarcomere. These data can explain the spherical propagation of Ca2+ waves and provide quantitative 3D data sets needed for accurate modeling of cardiac Ca2+-induced Ca2+ release. By quantifying labeling intensity in rat ventricular myocytes, a lower limit of 78 RyRs per cluster (on average) was obtained. By modeling the couplon as a disk wrapping around a t-tubule and fitting cluster images, 95% of couplons contained between 120 and 260 RyRs (assuming that RyRs are tight packed with a spacing of 29 nm). Assuming similar labeling efficiency in human, from the fluorescence intensity alone we estimate that human ventricular myocytes contain ≈30% fewer RyRs per couplon than rat.
Keywords: calcium-induced calcium release, excitation–contraction coupling, sarcoplasmic reticulum
In cardiac ventricular muscle, excitation–contraction (EC) coupling arises from Ca2+ release via clusters of ryanodine receptors (RyRs) in regions of close apposition between the sarcoplasmic reticulum (SR) and surface membranes in functional units called couplons (1, 2). Current work directed at understanding cardiac EC coupling is hindered by uncertainty in the size and 3D distribution of the couplons. Previous detailed analysis from electron micrographs has shown that typically 30–270 RyRs (depending on species) may be present in a couplon (1), but the thin sectioning associated with EM limits analysis of the spatial relationship between nearby and more distant couplons. Such knowledge is important, not only to make sense of the structures that underlie Ca2+ sparks (3, 4) but also for detailed mathematical modeling of cardiac Ca2+ metabolism.
In this study, we have used immunocytochemistry combined with 3D imaging and analysis to both reveal the 3D organization of RyR clusters and estimate the numbers of RyRs within the couplon. Our analyses generally support some detailed quantitative measurements from EM (1), but also provide insight into organization in 3D at spatial scales that would be extremely laborious (if not impossible) to achieve by using conventional thin sectioning. In addition, the antibody labeling reports the presence of the RyR epitope (5) and thereby avoids any uncertainty as to the possible identification of electron densities within EM thin sections. Although some previous studies (6, 7) have looked at the RyR distribution with fluorescence microscopy in the rat, no comparable data are available for human myocytes to our knowledge. Our data suggest that the reported fluorescence-derived intercouplon distances are generally too large and provide 3D data sets of RyR distribution in rat and human ventricular myocytes.
Results
RyR Cluster Distribution Within a Transverse Z-Disk.
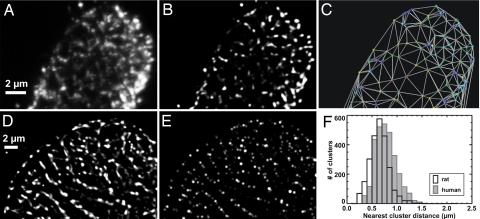
Transverse sections of intact rat ventricular myocytes labeled with an Ab against RyR2 (5) showed punctate labeling (see Fig. 1A). These data were further refined by digital deconvolution to provide high-contrast images at an in-plane diffraction limit of ≈250 nm. Some puncta were so close in 3D so as to give the impression of a more extended structure, but most puncta (>90%) were well separated and mostly diffraction-limited in size (≈250 nm). These observations and comparison with electron micrographs [see supporting information (SI) Fig. 5] suggest that these puncta represent individual RyR2 clusters. To analyze the cluster distribution, the labeling was detected by two independent algorithms (see Materials and Methods). The analysis is illustrated in Fig. 1C, where the distribution of detected puncta was subject to Delaunay triangulation (see Materials and Methods) to emphasize the geometric relationship between couplons. The nearest-neighbor distance between RyR2 clusters was 0.66 ± 0.18 μm (mean ± SD), a value significantly smaller than previously analyzed with antibody labeling (8). This finding can be explained by the limited confocal z-resolution that distorts measurements in longitudinal sections (see SI Fig. 6). The confidence in the precision of our measurement method is increased by noting that an edge-to-edge couplon spacing of ≈400 nm was reported by Franzini-Armstrong et al. (1), which for a cluster diameter of ≈250 nm would translate to a mean spacing of ≈650 nm, as reported here. In addition, our data show that the mean distance from a cluster to all surrounding nearest neighbors is ≈1.0 μm. Using the 3D data, we obtained a mean cluster density of ≈1.01 clusters per μm3 of cell volume. We also labeled transverse sections of human ventricular myocytes with the RyR2 Ab (Fig. 1 D and E). The labeling pattern in human cells was also punctate, but the couplon distribution was qualitatively different from that in rat (see also SI Fig. 7A). The histogram of cluster distances was right-shifted in the human (Fig. 1F), and quantitative analysis revealed that the nearest-neighbor distance between clusters was 0.78 ± 0.21 μm (mean ± SD), the mean distance to surrounding neighbors was 1.51 μm, and clusters were present at a significantly lower density of 0.52 per μm3. All mean values were significantly different from those in the rat (P < 0.001; see Table 1).
Fig. 1.
Detection of RyR clusters in transverse optical sections in rat and human ventricular myocytes. (A) The original data as a maximum projection of a shallow 1.5-μm stack containing a single z-disk showing punctate RyR labeling. (B) Before cluster detection, the fluorescence micrographs were enhanced by digital deconvolution, resulting in improved signal-to-noise ratio and resolution. (C) The geometrical arrangement of the couplons within a z-disk is clarified by calculating the Delaunay triangulation. Distances to the nearest-neighboring RyR clusters were calculated from the location data. (D) A deconvolved maximum projected stack containing couplons within a z-disk in a human ventricular myocyte. (E) The detected clusters where each circle represents a detected couplon. Note the similarity to the deconvolved data and that the calculated positions are limited to couplons within that z-disk [so that some of the weaker (out-of-focus) labeling seen in D is not reproduced in E]. (F) Using the procedure illustrated, a number of cells were analyzed, and the resulting nearest-neighbor distances between couplons are shown in the histograms that summarize pooled data from five rat and human cells, respectively. On average, the distance between a couplon and its nearest neighbor was 0.66 ± 0.18 μm in rat ventricular myocytes and 0.78 ± 0.21 μm in human ventricular myocytes (mean ± SD).
Table 1.
Couplon distribution in rat and human ventricular myocytes
| Myocyte type | Nearest-neighbor distance, μm* | Distance to surrounding neighbors, μm* | Local area served per couplon, μm2* | Couplons per cell volume, μm−3† | No. of RyRs per couplon‡ | Minimum myofibril width, μm‡ |
|---|---|---|---|---|---|---|
| Rat | 0.66 ± 0.06 | 1.02 ± 0.03 | 0.59 ± 0.02 | 1.01 ± 0.02 | 182 ± 6 | 0.57 ± 0.02 |
| Human | 0.78 ± 0.07§ | 1.51 ± 0.01§ | 0.81 ± 0.05§ | 0.52 ± 0.02§ | N/A | N/A |
Data are expressed as mean ± SEM. Student's t test was applied when appropriate. N/A, not available.
*Rat, n = 10 cells from four hearts; human, n = 10 cells.
†Rat, n = 8 cells from four hearts; human, n = 8 cells.
‡n = 6 cells from three hearts.
§Mean significantly different from rat (P < 0.001).
Longitudinal RyR Cluster Distribution and Relationship to Z-Disk Structure.
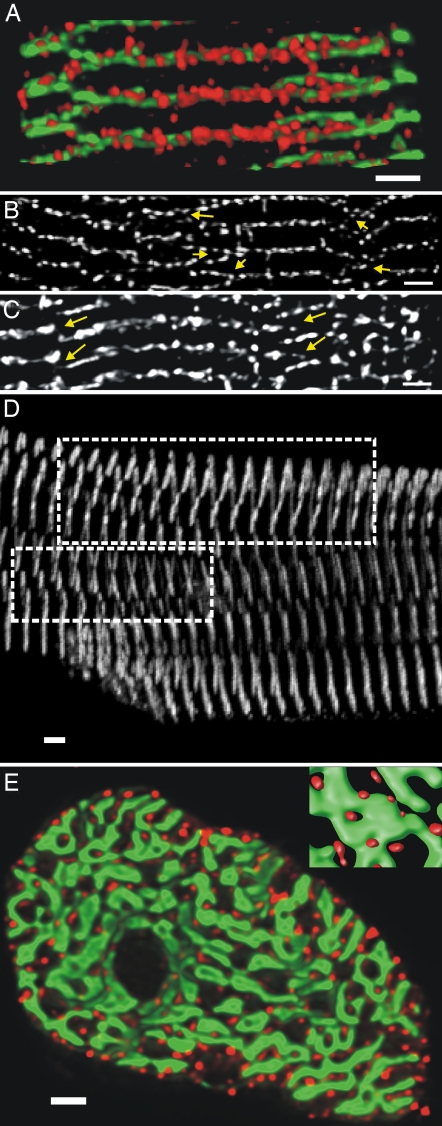
Across the z-disk, the RyR clusters were not in perfect longitudinal alignment, as illustrated in Fig. 2A, where double labeling with an Ab to α-actinin was used to simultaneously visualize the z-line structure in a rat myocyte. Most RyR clusters are located on or close to an adjacent region of α-actinin labeling. The mean longitudinal distance of the RyR cluster to the centroid of the z-disk was ≈160 nm, a value similar to the radius of t-tubules (9, 10). The distribution of α-actinin label in this rendered view also shows that z-disks in ventricular myocytes are not flat (as is assumed in most models of regenerative Ca2+ release; e.g., ref. 11) but curved with frequent bifurcations that allow connection to adjacent z-disks which are out of register (see also SI Movie 1). As shown in Fig. 2B, this complex z-disk architecture is also seen in the RyR cluster distribution where the z-disk bifurcations are evident as dislocations (8) in the otherwise regular appearance of RyR labeling and would provide a path for regenerative Ca2+ release to propagate between z-disks. Similar dislocations in the RyR pattern were also seen in human ventricular myocytes (Fig. 2C, arrows). Every cell inspected (>20 cells from four rat hearts and >10 human cells) had such z-disk bifurcations. Typically, regions with bifurcations extended over 30–40 μm spanning many z-disks, as illustrated in the projection of α-actinin labeling in a rat ventricular myocyte in Fig. 2D (see also SI Movie 2). Often several extended regions were observed across the length of a myocyte, suggesting that most z-disks exhibit bifurcations. Fig. 2E shows the general arrangement of RyR clusters across a transverse section of a rat myocyte, and the complex organization of myofibrils and their z-disks (green) is apparent. The myofibrils were not circular in cross-section but typically flattened with an aspect ratio of 4:1 and an average minimal width of ≈0.6 μm (see Table 1). These myofibril z-disks occupied ≈47% of the cell cross-sectional area in the rat, consistent with estimates from EM data of a myofilament volume fraction of ≈48% (12). The RyR clusters were arranged around the myofibril with approximately six clusters per fibril (depending on the size of the myofibril bundle) located <200 nm from the myofibril edge. The typical arrangement of RyR clusters around the myofibril is shown enlarged in Fig. 2E Inset.
Fig. 2.
RyR cluster distribution and relationship to z-disk structure. (A) Shown is the volume-rendered longitudinal distribution of RyR clusters (red) and its relationship to α-actinin labeling (green), a marker for the location of z-disks, in a rat ventricular myocyte. Note how z-disks bifurcate and the RyR cluster distribution follows this architecture. (B) In a projection of the longitudinal RyR distribution, the branching z-disk structure is apparent as dislocations in the otherwise quite regular transverse pattern in the rat data (see arrows). (C) Dislocations are also present in the RyR distribution in human ventricular myocytes (arrows). (D) These dislocations often extended across many z-disks as shown in a maximum projection of α-actinin staining in a 6-μm-thick stack through a rat ventricular myocyte (see boxed regions). (E) The transverse distribution of RyR clusters (red) in a rat myocyte in a complete z-disk around the myofibrils, which are labeled by anti-α-actinin Abs (green). (Inset) An enlarged rendering of the typical transverse arrangement of a ring of couplons around the contractile machinery. (Scale bars: 2 μm.)
Analysis of the Number of RyRs in a Cluster in the Rat.
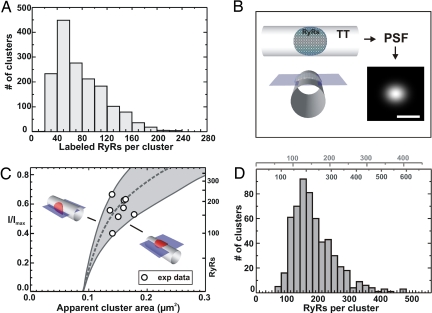
Although the punctate RyR labeling was similar in extent to the smallest feature produced by the microscope (≈250 nm; see above), the fluorescence data contain information that can be used to estimate the number of receptors present in a cluster. A lower limit can be obtained by comparing the intensity of the labeling to a known Ab concentration. Assuming binding of four primary Abs per RyR molecule (i.e., one Ab per subunit) and one secondary Ab per primary, we calculated that on average, the RyR cluster in the rat would contain 78.4 ± 39 RyRs (mean ± SD; see Fig. 3A). Although it is possible that up to two secondary Abs may bind to each primary, steric hindrance would probably prevent such a high degree of binding in the tight junctional space, and a 1:1 ratio of secondary-to-primary binding has recently been measured (13) and estimated (14). This estimate of RyRs in a cluster represents a lower limit because, although we took care to maximize labeling efficiency, it is unlikely that either the primary or secondary Ab labeling would be completely saturated.
Fig. 3.
Counting the number of RyRs in a couplon in rat ventricular myocytes. (A) A histogram of the number of labeled RyRs in a couplon obtained by calibrating the recorded cluster intensities with the signal from a solution containing a known concentration of secondary Abs. On average, ≈78 RyRs were labeled per cluster, which places a lower bound on the number of RyRs in a couplon. (B) Shown schematically is a 300-nm-diameter circular patch of RyRs wrapped around a t-tubule and a simulated micrograph of the resulting fluorescence distribution. (Scale bar: 500 nm.) (C) The relationship between the observed fluorescence intensity and the apparent extent of the couplon in confocal micrographs (measured as the area at half-maximal intensity) is shown. As a result of the asymmetry of the confocal PSF, the exact shape of the relationship depends on the orientation of the patch with respect to the image plane. The range of relationships spans the shaded area and is steepest when patches are orthogonal to that plane (left solid curve) and shallowest for parallel orientation (right solid curve); the dashed curve represents randomly oriented patches. The axis on the right shows the resulting calibration for the number of RyRs in the couplon. The nine largest-diameter well separated clusters observed in a z-disk are superimposed on the calculated relationship. (D) A histogram summarizing the number of RyRs per couplon for all clusters in a complete z-disk based on calibrations where the patches are oriented randomly. Alternative calibrations are also shown at the top for couplons oriented orthogonal or parallel to the image plane, respectively. Assuming randomly oriented couplons, the mean number of RyRs per couplon in this cell was 184 (see Table 1 for statistical data).
We developed a more powerful approach to quantify the number of RyRs in a cluster by examining RyR clusters that are above the resolution limit and calculating the maximum number of RyRs that could be packed into that volume. As shown in Fig. 3B, RyRs in a couplon can be envisioned as a near circular patch that is wrapped around a t-tubule, a geometry that is consistent with the appearance of couplons in electron micrographs (e.g., figure 23 in ref. 15 and ref. 16). When such a couplon is imaged by the confocal microscope, it is blurred by the known point-spread function (PSF). Using computer simulation of the blurring, and models of RyR clusters as shown in Fig. 3B, we were able to calculate the maximum number of RyRs that could be present in the imaged volumes. For close-packed RyRs with a unit packing as shown in Fig. 3B (a repeat spacing of 29 nm, originally from skeletal muscle data, e.g., ref. 17), we can derive the relationship between the apparent diameter of the couplon and the observed labeling intensity for any couplon orientation. The fluorescence intensities of the resolved (largest) clusters then provide a gauge for interpreting the observed fluorescence (F/Fmax) in terms of the number of RyRs present in any cluster, even those with projected areas that are below the resolution of the microscope (≤250 nm; see Fig. 3C). It should be noted that the asymmetric confocal PSF (with an axial extent of ≈700 nm) leads to couplons appearing brighter when oriented orthogonal to the image plane than when aligned parallel with the image plane. These two orientations therefore give upper and lower bounds for the relationship between the number of RyRs and fluorescence intensity. By picking the largest well separated clusters that were above the diffraction limit (circles in Fig. 3C), the fluorescence intensity for subresolution clusters could then be calibrated. Using this approach, we found that, on average, between 230 and 156 RyRs were present in each couplon. However, it is more likely that the couplon orientations are randomly distributed, and using the corresponding calibration, we calculate that the mean number of RyRs per couplon was 184 in this cell with a range between 100 and 400 (Fig. 3D). Analysis of data from six cells yielded a mean of 182 ± 6 RyRs per couplon (see Table 1). Although the accuracy of this estimate hinges on the accuracy of the close-packed RyR center-to-center distance of 29 nm, this value seems well supported by the extensive EM literature (e.g., ref. 1; for review, see ref. 18). It would be straightforward to correct our estimates for other close-packed distances if more accurate data became available. From comparison to the simple estimate based on Ab concentration (see above), these data suggest that the RyR Ab labeling efficiency may be ≈42%, a value that has never been determined before to our knowledge. As an independent test of our analysis, we also calculated the average junctional SR membrane area per unit cell volume by assuming that the RyR cluster area is synonymous with the junctional SR area. This calculation suggests that the junctional area should be ≈0.16 μm2/μm3, a value in good agreement with published values of 0.156 μm2/μm3 (calculated from total sarcotubular and dyadic membrane fractions in ref. 12).
Uniformity of RyR Cluster Distribution.
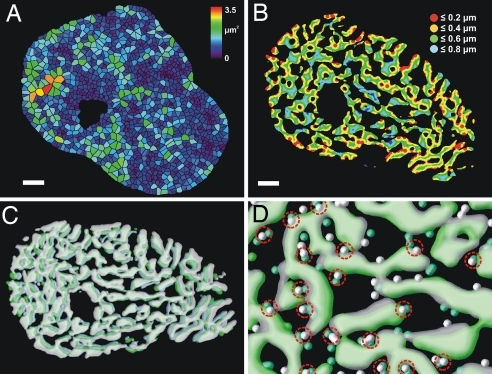
To achieve a uniform level of Ca2+ within the cell, the distance between RyR clusters and the volume they serve should not be too large. To examine this issue quantitatively, we calculated the local area served by each RyR cluster from the midpoints of the edges of the Delaunay triangles. Each of the resulting domains was color-coded for area and contains the points closest to an individual RyR cluster. In human ventricular myocytes (Fig. 4A), the average area was 0.81 μm2 with a range from 0.2 to 3.5 μm2. In rat cells, the average area was smaller at 0.59 μm2 per couplon with a range from 0.13 to 2.3 μm2 (as expected from the higher couplon density; see also Table 1 and SI Fig. 7B). Visual inspection of Fig. 4A suggests that most of the cross-section is reasonably uniform.
Fig. 4.
Uniformity of RyR cluster distribution. (A) A color-coded map of the areas served by each RyR cluster in a z-disk of a human ventricular myocyte. Each of the domains contains the points closest to an individual cluster with an average area of 0.81 μm2 per couplon (see also SI Fig. 7B for a similar map in the rat). (B) The effective diffusion distances from the RyR clusters across the myofibrillar space in a rat ventricular myocyte are shown as intersections between the myofibrillar space and circles of diffusion originating from surrounding RyR clusters. Most of the myofibrillar cross-sectional area (95%) is within a transverse diffusion distance ≤0.7 μm from the nearest couplon (n = 4). (C) A 3D-rendered view of three adjacent z-disks (front, white; middle, green; back, light blue) illustrating the subtle changes in myofibril outline from one sarcomere to the next in a rat myocyte. (D) The variation in RyR cluster position (spheres) between adjacent z-disks (front, white; back, light green). A larger proportion of clusters (53%) than expected (if randomly placed) are aligned longitudinally within ≤300 nm (dashed circles). Similar results were obtained in four other cells. (Scale bars: A, 5 μm; B, 2 μm.)
Using the α-actinin labeling to define the myofibrillar space through which Ca2+ diffuses, we calculated the effective diffusion distances from the RyR clusters. Fig. 4B shows a color-coded distance map for Ca2+ diffusion across the myofibrils in a rat ventricular myocyte. The net effect of this RyR cluster distribution was to reduce the transsarcomeric Ca2+ diffusion distance to ≈0.7 μm for 95% of myofibril cross-sectional area. In addition, no part of the myofilaments was >0.9 μm from a RyR cluster.
Because myofibrillar bundles had irregular outlines, it was of interest to examine the extent to which the outline was maintained from one z-line to the next. Fig. 4C shows three successive z-lines color-coded to emphasize the changes in cross-sectional shape. Visual inspection suggests that the irregular outline changed only slowly with longitudinal location (see also SI Movie 3). Although total transverse myofibril area did not change from one sarcomere to the next, the outline of the myofibrils exhibited subtle changes so that the projected z-disk cross-section occupied >80% of the area of the adjacent cross-section.
In contrast to the relative constancy of myofibrillar cross-section, the location of RyR clusters from one z-line to the next showed greater variability. Fig. 4D illustrates the longitudinal variation in RyR cluster position between adjacent sarcomeres. If couplons were randomly distributed, we would expect 23% to align within 300 nm (from the ≈1.3-μm mean distance between RyR clusters around the periphery of the myofibril). However, ≈53% of the RyR clusters were aligned longitudinally to this precision, suggesting that couplons are not simply randomly located around the myofibril.
Discussion
In this article, we have exploited high-resolution optical imaging of Ab labeling to clarify the 3D organization of RyR clusters and myofibrils in rat and human ventricular cardiomyocytes. Although some of our data confirm earlier measurements obtained from ultra-thin EM sections, our data add information about structural organization on a cell-wide scale. By using dual labeling, we were able to examine the relationship between RyR clusters and the myofilaments in the rat, which is important for understanding Ca2+ spark (4) genesis and Ca2+ wave propagation. By examining structure at high resolution in 3D, we found that the mean RyR cluster distance is significantly less than has been reported in previous confocal imaging studies. This finding is important because it removes the apparent discrepancy between optical (6–8) and electron microscopic (1) analysis of couplon spacing and also provides insight into the geometry that determines propagation of Ca2+ waves between RyR clusters. In addition, we have been able to develop a method for placing bounds on the number of RyRs in a typical rat cluster, and this information is important for understanding how concerted activation of RyRs within a couplon produces Ca2+ sparks.
Ca2+ Wave Propagation.
Less than half of the cross-section of the ventricular myocyte is occupied by contractile machinery (Fig. 2C), whereas the rest is occupied by other organelles such as mitochondria (12). Because diffusion depends on the gradient and the projected area through which diffusion occurs (Fick's first law), our data can explain the unexpected observation that, in the rat, the apparent diffusion coefficient for Ca2+ during a Ca2+ spark is twice as large in the longitudinal direction compared with the transverse direction (19). For saltatory wave propagation between RyR clusters, the velocity should be proportional to D/d, where D is the apparent diffusion coefficient and d is a mean distance between release sites (20). From our measurements, the longitudinal spacing of release sites in the rat is ≈1.8 μm, and the geometric mean transverse spacing is 0.8 μm. From Ca2+ spark properties, the apparent longitudinal and transverse diffusion coefficient are 7.9 and 17.1 μm2/s, respectively (19). Therefore the ratio of transverse-to-longitudinal wave propagation velocity should be 7.9/0.8 to 17.1/1.8 (respectively) or 1.04, i.e., close to unity. This idea may explain the paradoxical observation that Ca2+ waves are nearly spherical (21, 22), despite the underlying Ca2+ diffusion being asymmetric.
Sarcomeric Dislocations.
An unexpected finding was that, in both rat and human, sarcomeres remain in good longitudinal register only for short transverse distances before a dislocation occurs and a new registration develops. Although the cause of this architecture is uncertain, it is possible that it is the result of multiple embryonic myofibrils growing laterally within the cell. Additionally (or alternatively), the subsequent cell remodeling as the cell links to form cardiac fibers may cause realignment of internal contractile elements, which leads to dislocation in z-line alignment. It is also possible that dislocations arise from the need of a branching cell to make connections with adjacent cells that are not in sarcomeric registration. Regardless of the cause, RyR clusters span the regions of dislocation and would therefore limit nonuniformity in Ca2+ during activity and provide a facilitating path for regenerative release propagation along the cell.
Couplons and Ca2+ Sparks.
The close spacing between RyR clusters raises some uncertainty as to what functional unit underlies the Ca2+ spark. Although the mean spacing between couplons and the area they served was smaller in rat than in human, this difference need not translate to a less-uniform sarcomere activation. Because the human ventricular contraction (time to peak dP/dt) is ≈30% slower than the rat, there would be more time for diffusion of Ca2+ from the site of release. In connection with this point, we note that the nearest-neighbor distance between couplons in human is approximately times the rat couplon spacing so that at the time of peak contractility, sarcomeric Ca2+ nonuniformities should be no worse in human than in rat.
It has been suggested that Ca2+ sparks may exhibit quantal rates of rise that have been interpreted in terms of very low numbers of RyRs being involved in their genesis (23). Our data suggest that an alternative model should be considered in which Ca2+ sparks recorded at the periphery might recruit different numbers of clusters. In connection with this idea, we note that within the confocal measurement volume, we would expect 1–5 RyR clusters, which is the same as the number of quanta (assumed to be individual RyRs) thought to underlie a Ca2+ spark (23). On the other hand, if 1–5 RyRs are involved in the genesis of a spark, our finding of ≈180 RyRs per couplon suggests that the open probability of the RyR during a Ca2+ spark should be very low (<0.03). A similarly low open probability is obtained by considering a whole couplon quantal flux of ≈1 pA (23) with a maximum current of 180 RyRs/couplon × 0.5 pA/RyR (24), or 90 pA. Of course, it is also possible that the reported quantal current is not well measured in fluorescence recordings because of the limited kinetics of the indicator. These problems suggest that, with our data in mind, further experiments are needed to clarify the structural basis of the Ca2+ spark.
Couplon Size.
We have obtained hard upper and lower bounds for the numbers of RyRs in rat couplons. Ryanodine binding has been previously used to estimate the total numbers of RyRs within the cell. Estimates of RyR binding range from 680 to 833 fmol/mg protein in rat (25, 26), which can be used to calculate that there should be 2.1 to 2.6 × 106 RyRs in a 30-pl cell (cf. ref. 18). From our estimate of couplon density (1.01 μm−3), these binding data suggest that 70–90 RyRs per couplon is in good agreement with our lower estimate of ≈78 RyRs per couplon, but we suggest the former number may underestimate the true number because of problems in preservation of binding sites during protein isolation.
Our upper limit estimate (230 RyRs) is in good agreement with that of Franzini-Armstrong et al. (1) but based on a geometry where all couplons would be aligned orthogonal to the image plane. This scenario seems unlikely, and we suggest that the couplons should be oriented more randomly, giving a more likely mean value of 182 RyRs per couplon. If the brightest clusters are oriented orthogonally and the dimmest parallel to the image plane, inspection of Fig. 3D suggests that couplon size varies between 120 and 260 RyRs, or an approximate 2-fold variation. This value is interesting because it suggests that if Ca2+ release were simply proportional to the number of RyRs per couplon, there should be a 2-fold variation in Ca2+ spark amplitude between sites. Such variation in local Ca2+ (spark) amplitude should cause larger variations in force per half sarcomere, which would be undesirable. This argument suggests that additional local feedback mechanisms (which may reside in RyR gating and local SR load) are required to compensate for variations in couplon size.
From the human tissue labeling, we were unable to find sufficiently large and well separated couplons with which to gauge the labeling intensity. Nevertheless, the RyR labeling in human appeared ≈30% dimmer than in rat, suggesting that a similar reduction in the number of RyRs per couplon might be present. A previous study on mammalian couplons has suggested between 90, 128, and 267 RyRs per couplon (for dog, mouse, and rat, respectively) (1). Our data therefore suggest that human couplons are within this range.
Conclusion
Using a combination of confocal imaging and image processing, we have quantified, in rat and human ventricular myocytes in three dimensions, the distribution of RyRs, a key protein for cardiac EC coupling. This approach revealed structural information that would be difficult to obtain from EM thin sections. We found that RyR clusters are more closely spaced than expected from recent Ab labeling studies and retain partial registration from one sarcomere to the next. We have also developed a method for calibrating protein expression from antibody labeling using two different approaches. This methodology is analogous to Western blotting but retains the full 3D spatial distribution of the protein. Our data suggest that between 120 and 260 RyRs are present in a rat couplon, a figure that sets bounds on gating models for cardiac EC coupling. The methods reported here are applicable to other cell types and proteins and may give insight into a variety of cell signaling pathways.
Materials and Methods
For details, see SI Text. Enzymatically isolated rat cardiac myocytes were prepared as described (27), using protocols approved by the University of Auckland Animal Ethics Committee, and immediately fixed. Human ventricular tissue samples were obtained under protocols approved by the Northern Y Regional Ethics Committee, fixed, cryoprotected, cryosectioned to yield sections for immunolabeling, and mounted on glass slides. Fixed cells and tissue sections were indirectly immuno-labeled with an Ab against RyR2 by a sequence of permeabilization, blocking, incubation with primary Ab, and finally secondary fluorescent Ab. In some experiments, rat cells were double-labeled with Abs against RyR2 and α-actinin. Rat cells were mounted on glass slides for longitudinal imaging or embedded in agar for transverse imaging. The samples were imaged on a LSM410 laser scanning confocal microscope (Zeiss, Jena, Germany) using a high numerical aperture oil immersion objective. The raw image data were deconvolved, and couplons were detected by using a correlation algorithm. The couplon distribution was statistically analyzed with the software package IDL (ITT, Boulder, CO).
Supplementary Material
Acknowledgments
We thank Prof. Clara Franzini-Armstrong and Dr. Venkat Ramesh for permission to use unpublished electron micrographs of rat ventricular tissue sections, Isuru Dilshan for assistance with data acquisition, and Dr. Peter Ruygrok and the Auckland District Health Board for support in obtaining human tissue samples. This work was supported by grants from the Auckland Medical Research Foundation, the Health Research Council, and the Wellcome Trust (U.K.).
Abbreviations
- RyR
ryanodine receptor
- SR
sarcoplasmic reticulum
- EC
excitation–contraction
- PSF
point-spread function.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703016104/DC1.
References
- 1.Franzini-Armstrong C, Protasi F, Ramesh V. Biophys J. 1999;77:1528–1539. doi: 10.1016/S0006-3495(99)77000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern MD, Pizarro G, Rios E. J Gen Physiol. 1997;110:415–440. doi: 10.1085/jgp.110.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannell MB, Cheng H, Lederer WJ. Biophys J. 1994;67:1942–1956. doi: 10.1016/S0006-3495(94)80677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng H, Lederer WJ, Cannell MB. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 5.Seok JH, Xu L, Kramarcy NR, Sealock R, Meissner G. J Biol Chem. 1992;267:15893–15901. [PubMed] [Google Scholar]
- 6.Cleemann L, Wang W, Morad M. Proc Natl Acad Sci USA. 1998;95:10984–10989. doi: 10.1073/pnas.95.18.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kockskamper J, Sheehan KA, Bare DJ, Lipsius SL, Mignery GA, Blatter LA. Biophys J. 2001;81:2590–2605. doi: 10.1016/S0006-3495(01)75903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen-Izu Y, McCulle SL, Ward CW, Soeller C, Allen BM, Rabang C, Cannell MB, Balke CW, Izu LT. Biophys J. 2006;91:1–13. doi: 10.1529/biophysj.105.077180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soeller C, Cannell MB. Circ Res. 1999;84:266–275. doi: 10.1161/01.res.84.3.266. [DOI] [PubMed] [Google Scholar]
- 10.Stewart JM, Page E. J Ultrastruct Res. 1978;65:119–134. doi: 10.1016/s0022-5320(78)90050-3. [DOI] [PubMed] [Google Scholar]
- 11.Izu LT, Means SA, Shadid JN, Chen-Izu Y, Balke CW. Biophys J. 2006;91:95–112. doi: 10.1529/biophysj.105.077214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page E, McCallister LP, Power B. Proc Natl Acad Sci USA. 1971;68:1465–1466. doi: 10.1073/pnas.68.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCloskey KE, Comella K, Chalmers JJ, Margel S, Zborowski M. Biotechnol Bioeng. 2001;75:642–655. doi: 10.1002/bit.10040. [DOI] [PubMed] [Google Scholar]
- 14.Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP. Nat Chem Biol. 2005;1:196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]
- 15.Forbes MS, van Neil EE. Anat Rec. 1988;222:362–379. doi: 10.1002/ar.1092220409. [DOI] [PubMed] [Google Scholar]
- 16.Forbes MS, Hawkey LA, Jirge SK, Sperelakis N. J Ultrastruct Res. 1985;93:1–16. doi: 10.1016/0889-1605(85)90080-1. [DOI] [PubMed] [Google Scholar]
- 17.Dulhunty AF. J Membr Biol. 1989;109:73–83. doi: 10.1007/BF01870792. [DOI] [PubMed] [Google Scholar]
- 18.Bers DM. Excitation-Contraction Coupling and Cardiac Contractile Force. Dordrecht, The Netherlands: Kluwer; 2001. [Google Scholar]
- 19.Parker I, Zang WJ, Wier WG. J Physiol (London) 1996;497:31–38. doi: 10.1113/jphysiol.1996.sp021747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson SP, Keizer J, Pearson JE. Proc Natl Acad Sci USA. 1999;96:6060–6063. doi: 10.1073/pnas.96.11.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipp P, Niggli E. Biophys J. 1993;65:2272–2276. doi: 10.1016/S0006-3495(93)81316-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wussling MH, Salz H. Biophys J. 1996;70:1144–1153. doi: 10.1016/S0006-3495(96)79715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SQ, Stern MD, Rios E, Cheng H. Proc Natl Acad Sci USA. 2004;101:3979–3984. doi: 10.1073/pnas.0306157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mejia-Alvarez R, Kettlun C, Rios E, Stern M, Fill M. J Gen Physiol. 1999;113:177–186. doi: 10.1085/jgp.113.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bers DM, Stiffel VM. Am J Physiol. 1993;264:C1587–C1593. doi: 10.1152/ajpcell.1993.264.6.C1587. [DOI] [PubMed] [Google Scholar]
- 26.Thomas MJ, Hamman BN, Tibbits GF. J Exp Biol. 1996;199:1999–2009. doi: 10.1242/jeb.199.9.1999. [DOI] [PubMed] [Google Scholar]
- 27.Evans AM, Cannell MB. Cardiovasc Res. 1997;35:294–302. doi: 10.1016/s0008-6363(97)00117-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.