Abstract
Previous reports suggested that humans and mice differ in their sensitivity to the genetic dosage of transcription factors that play a role in early testicular development. This difference implies that testis determination might be somewhat different in these two species. We report that the Fog2 and Gata4 transcription factors are haploinsufficient for testis determination in mice. Whether gonadal sex reversal occurs depends on genetic background (i.e., modifier genes). For example, C57BL/6J (B6) XY mice develop testes if they are heterozygous for a mutant Fog2 (Fog2−) or Gata4 (Gata4ki) allele. However, if the B6 Y chromosome (YB6) is replaced by the AKR Y chromosome (YAKR), B6 Fog2−/+ XYAKR mice develop ovaries, and B6 Gata4ki/+ XYAKR mice develop ovaries and ovotestes (gonads containing both ovarian and testicular tissue). Furthermore, DBA/2J (D2) Fog2−/+ XYAKR mice and (B6 × D2)F1 hybrid Gata4ki/+ XYAKR mice develop testes. Sry is expressed in the mutant XY gonads, indicating that the lack of Sry expression is not the cause of ovarian tissue development in B6 Fog2−/+ or Gata4ki/+ XYAKR mice. However, up-regulation of Sox9 expression, which is critical for normal testicular development, does not occur in mutant XY gonads that develop as ovaries. We conclude that under certain genetic conditions, Sox9 up-regulation depends on the proper dosage of Fog2 and Gata4. We propose that in humans the FOG2 and/or GATA4 genes might be haploinsufficient for normal testis determination and thus could be the cause of some previously unassigned cases of XY gonadal sex reversal.
Keywords: gonadal sex reversal, haploinsufficiency, sex determination, Sry, Sox9
During human fetal development, differentiation of the bipotential gonads into ovaries or testes depends on the proper dosage of transcription factors. For example, the presence of only a single functional copy of the autosomal transcription factor-encoding genes SF1, SOX9, or WT1 can result in the development of XY females, and duplication of a chromosomal region containing SOX9 can lead to the development of XX males (1–5). In contrast, the proper dosage of the transcription factors Sf1, Sox9, and Wt1 appears not to affect testicular development in mice because XY mice containing only a single copy of these genes develop testes (6–9). This apparent species difference suggested that the correct dosage of gonadal sex-determining transcription factors is necessary for normal testis development in humans, whereas mice are less sensitive to these gene dosage effects (10–12).
Studies in our laboratory indicate that genetic background plays an essential role in the process of gonadal sex determination in mice. Specifically, mice of the C57BL/6J (B6) inbred strain are exceptionally sensitive to disturbances in the early events of gonad development and thus provide a genetic “litmus test” for identifying genes that cause sex reversal. For example, the SryPOS gene carried on the Mus domesticus poschiavinus Y chromosome causes ovarian tissue development when present in C57BL/6J (B6) mice but not in DBA/2J (D2) or (B6 × D2)F1 mice (13, 14), and a mutant allele of the X-linked transcription factor Dax1 (Nr0b1) causes gonadal sex reversal in B6 but not D2 or (B6 × D2)F1 XY mice (15). This “B6 sensitivity” is even stronger if the AKR/J Y chromosome (YAKR) is present. For example, if B6 mice are heterozygous for the TOrl mutation, they develop testes. If the YAKR chromosome is present, however, they develop ovaries (16, 17). These findings suggest that genetic background, not species differences, could explain apparent differences in transcription factor dosage sensitivity for gonad development in XY humans and mice. The fact that not all XY humans are sex-reversed if only a single copy of a normal SF1 or WT1 allele is present (18–20) supports this possibility, as does the fact that XY males carrying a mutant SRY gene can pass this allele on to XY daughters (for review, see ref. 21).
To test the hypothesis that testis determination in XY mice is sensitive to levels of transcription factor gene dosage in a genetic background-dependent manner, we conducted experiments to determine whether the presence of only a single normal copy of the autosomal Fog2 (Zfpm2) or Gata4 gene causes XY sex reversal on specific genetic backgrounds. FOG2 is a multitype zinc finger cofactor that binds to and regulates the transcriptional activity of GATA4, a member of the GATA family of transcription factors (22–24). In mice, Fog2 and Gata4 are coexpressed in the somatic cells of XX and XY genital ridges as early as embryonic day (E) 10.5 (25–28). Previous work had shown that homozygosity for mutant alleles of Fog2 or Gata4 leads to abnormal fetal gonadal development in XY mice from an early developmental stage (29). But an abnormal gonadal phenotype was not observed in heterozygous mice on the mixed B6/129 genetic background, and the XX and XY heterozygotes appeared to have normal fertility.
We report that B6 XY mice develop testes if they are heterozygous for a Fog2 null (Fog2−) (30) or a Gata4 mutant (Gata4ki) (31) allele. Gata4ki encodes a mutant GATA4 protein that is unable to interact with FOG2. However, if a YAKR chromosome is substituted for the B6 Y chromosome, B6 Fog2−/+ XYAKR mice develop ovaries (i.e., are completely sex-reversed), and B6 Gata4ki/+ XYAKR mice develop ovaries and ovotestes. On the other hand, D2 Fog2−/+ XYAKR mice and (B6 × D2)F1 Gata4ki/+ XYAKR mice develop testes. These results indicate that, as is the case in humans, testis development in mice requires a sufficient gene dosage of autosomal transcription factors, and they suggest that genetic background (i.e., modifier genes) plays a role in testicular development in humans.
Results
Genetic Background Determines Whether the Presence of a Single Functional Copy of Fog2 and Gata4 Causes XY Gonadal Sex Reversal.
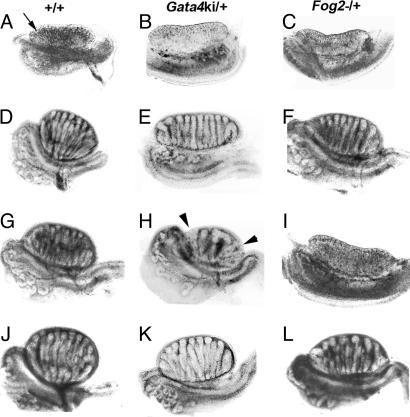
To explore the role of Fog2 and Gata4 transcription factor gene dosage in gonadal development and to determine whether genetic background affects their role in this process, we examined fetal gonad development in B6 and (B6 × D2)F1 Fog2−/+ and Gata4ki/+ mice. Testicular growth was abnormal in the gonads of B6 Fog2−/+ and Gata4ki/+ XY fetal mice (Table 1 and Fig. 1); the developing testicular cords in 12 B6 Fog2−/+ and 22 B6 Gata4ki/+ XY fetuses were shorter compared with those in control B6 XY normal (+/+) siblings, giving the testes a flat appearance (Fig. 1). This abnormal development, however, does not affect fertility in these mice. The fetal ovaries observed in B6 Fog2−/+ or Gata4ki/+ XX mice appeared normal (Fig. 1).
Table 1.
Depending on genetic background, the presence of a single functional allele of Fog2 or Gata4 causes XY gonadal sex reversal
| Fetal genotype | Gonadal phenotype |
|||||
|---|---|---|---|---|---|---|
| O-O | O-OT | OT-OT | OT-T | T-T | Total | |
| B6 Fog2−/+ XYB6 | 0 | 0 | 0 | 0 | 12* | 12 |
| B6 Fog2−/+ XYAKR | 16 | 0 | 0 | 0 | 0 | 16 |
| (B6 × D2)F1 Fog2−/+ XYAKR | 0 | 0 | 0 | 1† | 19‡ | 20 |
| D2 Fog2−/+ XYAKR | 0 | 0 | 0 | 0 | 14* | 14 |
| B6 Gata4ki/+ XYB6 | 0 | 0 | 0 | 0 | 22* | 22 |
| B6 Gata4ki/+ XYAKR | 8 | 5 | 2 | 0 | 0 | 15 |
| (B6 × D2)F1 Gata4ki/+ XYAKR | 0 | 0 | 0 | 0 | 18 | 18 |
Numbers indicate the no. of mutant XY gonad pairs examined at E14.5–16. O, ovary; T, testis; OT, ovotestis.
*Both testes contained shorter testicular cords.
†One abnormal testis and one ovotestis.
‡One fetus with two normal testes, and 18 fetuses with two testes showing attenuated cord growth.
Fig. 1.
Embryonic day (E) 14.5–15 gonad–mesonephros complexes from normal (+/+) (A, D, G, and J), and heterozygous mutant Gata4ki (Gata4ki/+) (B, E, H, and K), and Fog2− (Fog2−/+) (C, F, I, and L) fetuses. (A) B6+/+ XX ovary. (B) B6 Gata4ki/+ XX ovary. (C) B6 Fog2−/+ XX ovary. (D) B6+/+ XYB6 testis. (E) B6 Gata4ki/+ XYB6 testis. (F) B6 Fog2−/+ XYB6 testis. (G) B6+/+ XYAKR testis. (H) B6 Gata4ki/+ XYAKR ovotestis. (I) B6 Fog2−/+ XYAKR ovary. (J) (B6 × D2)F1+/+ XYAKR testis. (K) (B6 × D2)F1 Gata4ki/+ XYAKR testis. (L) (B6 × D2)F1 Fog2−/+ XYAKR testis. In each case, the gonad (arrow in A) lies above the mesonephros. (E) Testis with attenuated cord growth. (H) Ovotestis. The arrowheads in H point to ovarian tissue.
We also analyzed gonads from E14.5–16 B6 and (B6 × D2)F1 fetal mice carrying the YAKR chromosome. As shown in Fig. 1 and Table 1, the presence of only a single functional copy of Fog2 or Gata4 in B6 XYAKR mice had a major effect on testis development. The gonads of the 16 B6 Fog2−/+ XYAKR fetuses analyzed were ovaries (i.e., no testicular cords were observed), whereas the gonads of the 15 B6 Gata4ki/+ XYAKR fetuses were ovaries or ovotestes: eight fetuses contained bilateral ovaries, five contained one ovary and an ovotestis, and two contained ovotestes. The gonads of the 20 (B6 × D2)F1 Fog2−/+ XYAKR fetuses analyzed were testes or ovotestes: one fetus contained bilateral normal testes, 18 contained bilateral testes with short cords, and one contained an ovotestis accompanied by a testis with short cords (Table 1). The gonads of the 18 (B6 × D2)F1 Gata4ki/+ XYAKR fetuses analyzed were normal testes. Because one (B6 × D2)F1 Fog2−/+ XYAKR fetus contained an ovotestis and a testis, we also analyzed gonadal development in 14 D2 Fog2−/+ XYAKR mice. No ovarian tissue was observed, but the testicular cords were shorter than normal [see supporting information (SI) Fig. 5]. We conclude that the presence of only one functional copy of a normal Fog2 or Gata4 allele can disrupt gonadal development in XYAKR mice. Whether this disruption occurs, however, depends on genetic background.
B6 Fog2−/+ and Gata4ki/+ XYAKR Mice Develop Ovarian Tissue.
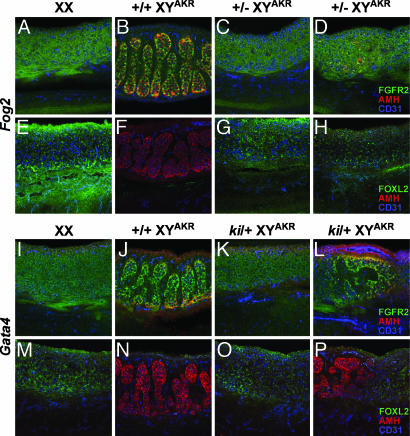
To conduct a more thorough morphological analysis of B6 Fog2−/+ and Gata4ki/+ XYAKR gonads, the spatial localization of selected protein markers was determined by using whole-mount immunohistochemical analysis of E13.5 gonad–mesonephros complexes (Fig. 2). The antibodies used specifically recognized anti-müllerian hormone (AMH) (Sertoli cell marker), CD31 (germ cell and vascular endothelial cell marker), fibroblast growth factor receptor 2 (FGFR2) (somatic cell marker, localized to the nucleus in Sertoli cells), and FOXL2 (female somatic cell marker). For 11 of the 13 B6 Fog2−/+ XYAKR and 9 of the 14 Gata4ki/+ XYAKR gonads analyzed, the expression and localization pattern of these four markers as well as the gonadal morphology resembled that observed in control XX ovaries. Because FOXL2 expression is ovary-specific and because it has been suggested to play a role in initiating the fetal ovarian pathway (32), these results indicate that the ovarian pathway is activated.
Fig. 2.
Whole-mount immunohistochemical analysis of marker gene expression in E13.5 B6 Fog2−/+ XYAKR (top two rows) and B6 Gata4ki/+ XYAKR (bottom two rows) gonads, and B6 XX and B6 XYAKR control gonads. First and third rows illustrate expression of FGFR2 (green), AMH (red), and CD31 (PECAM; blue); the second and fourth rows illustrate expression of FOXL2 (green), AMH (red), and CD31 (PECAM; blue). The gonads in the first column (A, E, I, and M) are normal XX ovaries, and gonads in the second column (B, F, J, and N) are normal B6 XYAKR testes. Gonads in the third column are B6 Fog2−/+ XYAKR (C and G) and B6 Gata4ki/+ XYAKR (K and O) ovaries that appear similar to the control B6 XX ovaries (first column). Gonads in the fourth column are B6 Fog2−/+ XYAKR ovaries (D and H) and B6 Gata4ki/+ XYAKR ovotestes (L and P). In each image, the gonad lies above the mesonephros.
Interestingly, 2 of the 13 B6 Fog2−/+ XYAKR and 3 of the 14 B6 Gata4ki/+ XYAKR gonads morphologically resembled ovaries but exhibited partial activation of the male pathway. In these gonads, a small number of centrally located cells expressed AMH and had nuclear localization of FGFR2, indicating that these cells had initiated differentiation as Sertoli cells (Fig. 2D). However, no testis cords were observed in these 5 gonads. On the other hand, 2 of the 14 B6 Gata4ki/+ XYAKR gonads were clearly ovotestes, with regions containing testis cords and expressing AMH and/or nuclear FGFR2. In these ovotestes, the ovarian and testicular regions were discrete, with the ovarian regions expressing FOXL2 or cell surface-localized FGFR2. None of the 13 B6 Fog2−/+ XYAKR gonads was ovotestes (i.e., contained testis cords).
Sry Is Expressed in B6 Fog2−/+ and Gata4ki/+ XYAKR Gonads.
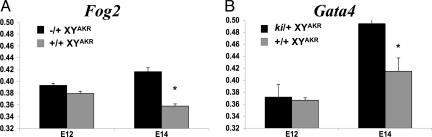
Real-time RT-PCR results indicated that absence of Sry expression is not responsible for the failure to induce normal testicular development in B6 Fog2−/+ and Gata4ki/+ XYAKR gonads. The level of Sry expression in E12 B6 Fog2−/+ XYAKR and B6 Gata4ki/+ XYAKR gonads was similar to that observed in control E12 B6 XYAKR gonads (Fig. 3). Contrary to what is observed in E14 B6 XYB6 gonads, Sry expression was still detected in E14 B6 XYAKR gonads. These results are consistent with a previous report that M. domesticus Sry alleles can exhibit prolonged expression in B6 gonads (33). However, at E14, Sry expression was significantly higher (P < 0.05) in B6 Fog2−/+ and Gata4ki/+ XYAKR ovaries compared with control B6 XYAKR testes (Fig. 3). These results are similar to the higher Sry expression observed in E14 B6 Dax1−/Y ovaries compared with control testes (15).
Fig. 3.
Relative Sry expression in E12 (A) and E14 (B) Fog2 and Gata4 B6 XYAKR homozygous normal gonads and heterozygous mutant gonads. At E14, only heterozygous mutant gonads classified as ovaries were used (i.e., no testicular cords observed). Expression levels are relative to 18S rRNA expression. The mean values represent the average values of a minimum of three cDNA samples (one cDNA sample represents both gonads from one fetus). *, Significant; P < 0.05 lower expression.
The Ovarian Genetic Pathway Is Activated in B6 Fog2−/+ and B6 Gata4ki/+ XYAKR Gonads.
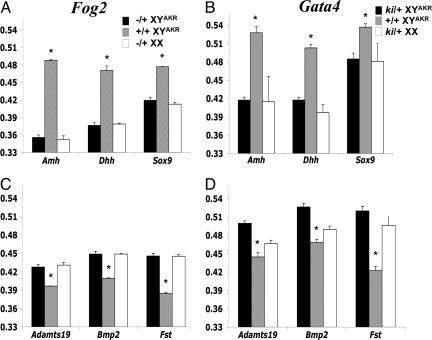
A multigene real-time RT-PCR assay was conducted to determine whether the ovarian, or testicular, or both developmental pathways were initiated in B6 Fog2−/+ XYAKR and B6 Gata4ki/+ XYAKR gonads. The gene expression results indicate that the ovarian genetic pathway is activated despite persistent Sry expression (Fig. 4 and SI Table 2). For example, the ovarian-specific genes (i.e., genes expressed at significantly higher levels in ovaries than testes) Fst, Wnt4, Bmp2, and Adamts19 were expressed at significantly (P < 0.05) higher levels, whereas the testicular-specific genes (i.e., genes expressed at significantly higher levels in testes than ovaries) Amh, Dhh, Sox9, and Fgf9, were expressed at significantly (P < 0.05) lower levels in E14 B6 Fog2−/+ XYAKR ovaries and B6 Gata4ki/+ XYAKR ovaries compared with control B6 XYAKR testes (Fig. 4 and SI Table 2). In addition, despite normal Sry expression at E12, Sox9 expression was significantly lower (P < 0.05) in E12 B6 Fog2−/+ XYAKR and B6 Gata4ki/+ XYAKR gonads compared with control gonads (2.0- and 2.3-fold, respectively) and Fst expression was significantly higher (P < 0.05) in E12 B6 Fog2−/+ XYAKR and Gata4ki/+ XYAKR gonads compared with control gonads (3.5- and 4.6-fold, respectively) (SI Table 2).
Fig. 4.
Expression pattern of fetal testicular-specific (A and B) and fetal ovarian-specific (C and D) genes in B6 Fog2 (A and C) and B6 Gata4 (B and D) homozygous normal and heterozygous mutant E14 gonads. Expression levels are relative to 18S rRNA expression. The mean values represent the average values of a minimum of three cDNA samples (one cDNA sample represents both gonads from one fetus). *, Significant (P < 0.05) higher (A and B) or lower (C and D) expression levels.
We conclude that Sox9 up-regulation fails even though the Sry gene is activated in B6 Fog2−/+ XYAKR and B6 Gata4ki/+ XYAKR gonads. The result is that the ovarian pathway is initiated, and ovarian development proceeds.
Discussion
We report that Fog2 and Gata4 are haploinsufficient for normal fetal testicular development in certain XY mice. The severity of the gonadal phenotype, i.e., the extent of ovarian tissue development, depends on genetic background: gonadal sex reversal is complete in B6 Fog2−/+ XYAKR mice, partial in B6 Gata4ki/+ XYAKR mice, and does not occur in D2 Fog2−/+ XYAKR and (B6 × D2)F1 Gata4ki/+ XYAKR mice. These results provide insight into the genetic control of mammalian gonadal development and have implications for human syndromes involving abnormal gonadal sex determination.
Haploinsufficiency for specific transcription factors resulting in gonadal sex reversal has previously been reported in humans but not mice. For example, human XY females have been identified who are heterozygous for a mutant SF1 or WT1 allele (1, 4), whereas XY mice that are heterozygous for a mutant Sf1 or Wt1 allele were reported to develop as normal males (6, 7). These observations led to the idea that humans and mice differ in their transcription factor dosage requirement for fetal gonad development (11). Results presented here demonstrate that mouse fetal gonadal development is sensitive to transcription factor dosage. Moreover, similar to Fog2 and Gata4, the presence of only a single functional copy of Sf1 or Wt1 also can cause gonadal sex reversal in XY mice (unpublished data), further underscoring the concept that testicular development in XY mice is sensitive to transcription factor dosage. When these findings are combined with the results involving B6 YPOS and B6 DAX1 sex reversal in mice (14, 15), it is clear that genetic background and transcription factor dosage are key players in mammalian gonad sex determination and point to the possibility that a single copy of a mutant FOG2 or GATA4 gene can cause XY sex reversal in humans and other mammals.
Similar to our observation in B6 Dax1−/Y gonads (15), the failure to initiate complete testicular cord formation in B6 Gata4ki/+ XYAKR and B6 Fog2−/+ XYAKR is not caused by a lack of Sry expression but is due to the failure to up-regulate Sox9 expression in the somatic support cells of the gonad, thereby preventing these cells from differentiating as Sertoli cells. Furthermore, it is likely that the extended Sry expression observed in E14 mutant XY ovaries is caused by the lack of Sox9 up-regulation. Results from Chaboissier et al. (9) indicate that Sox9 up-regulation is responsible for Sry down-regulation in pre-Sertoli cells.
Previously, Tevosian et al. (29) reported that at E11.5, homozygosity for the Fog2− allele lowered Sry expression and prevented testicular cord formation. It is possible that before E12, Sry expression is lower in B6 Fog2−/+ XYAKR gonads compared with B6 Fog2+/+ XYAKR gonads. If this possibility is correct, our data suggest that Sry levels increase rapidly such that at E12, Sry levels are comparable between B6 Fog2−/+ and control B6 XYAKR gonads. A single normal Fog2 allele then is sufficient to establish normal Sry levels at E12, a time when the SryAKR allele is normally expressed at peak levels in B6 XYAKR gonads [in B6 mice, the SryAKR allele peaks from 16 tail somites (E11.5) until 22 tail somites (E12) (34, 35)]. Future studies are needed to determine whether Sry expression is affected at earlier stages or whether FOG2 and GATA4 can physically interact with the Sry gene and regulate its expression.
All B6 Fog2−/+ XYAKR gonads were ovaries, whereas B6 Gata4ki/+ XYAKR gonads were ovaries or ovotestes. The more severe phenotype observed when only one functional copy of Fog2 is present compared with only one functional copy of Gata4 may reflect the nature of the mutant alleles. The Fog2− allele is a null allele, whereas the Gata4ki allele encodes a mutant GATA4 protein that fails to interact with FOG2. Our data indicate that the interaction between GATA4 and FOG2 is required for efficient Sox9 up-regulation. However, it is possible that the GATA4ki protein retains some function independent of its interaction with FOG2.
B6 Fog2−/+ and Gata4ki/+ XY mice develop testes and are fertile males. However, at E15, the testicular cords in these gonads are shorter compared with normal XY gonads, thus giving the testis a flat appearance (e.g., see Fig. 1 D and E). Fetal testicular cord development and growth are dependent on Sry-induced cell migration from the mesonephros into the gonad and cell proliferation (36–38). We propose that the presence of only a single functional Fog2 or Gata4 allele interferes with, but does not abolish, Sry-mediated signaling pathways involved in cell migration and proliferation, leading to attenuated testicular cord growth in B6 Fog2−/+ and Gata4ki/+ XYB6 gonads.
Sry is recognized as the mammalian testis-determining gene, although it alone is not sufficient to initiate testicular differentiation. Studies with mice have shown that in the absence of Sry, transgenic mice expressing Sox9 can initiate the testicular developmental pathway (39, 40). Sox9 up-regulation appears to be a complicated process involving interactions between Dax1 and Tda1 (15), and Fog2 and Gata4 (this work). The intricate regulation of Sox9 transcription in pre-Sertoli cells is not surprising considering the number of upstream and downstream sequences involved in Sox9 transcriptional regulation. Sequences ≈1 Mb upstream of Sox9 can regulate Sox9 expression in the gonad (41), and there are at least eight evolutionarily conserved regulatory elements located at the 5′ and 3′ of the Sox9 gene involved in tissue-specific Sox9 expression (42). How Dax1, Gata4, Fog2, and Tda1 control signaling pathways and which signaling pathways are involved in up-regulating Sox9 expression remain to be determined. Interestingly, Manuylov et al. (43) recently reported that the GATA4–FOG2 transcription complex regulates Sox9 expression in XX transgenic mice that are sex-reversed males, adding further support to the idea that GATA4–FOG2 is involved in regulating Sox9 expression.
In summary, the presence of only a single functional allele of the transcription factor Fog2 or Gata4 causes gonadal sex reversal in B6 XYAKR mice. Gonadal sex reversal is not observed in B6 Fog2−/+ and Gata4ki/+ XYB6, (B6 × D2)F1 Gata4ki/+ XYAKR, and D2 Fog2−/+ XYAKR mice. Ovarian development in B6 Gata4ki/+ and Fog2−/+ XYAKR mice results from a failure of Sox9 up-regulation. These results demonstrate that the process of gonadal sex determination in mice is sensitive to transcription factor gene dosage, and they strengthen the concept that genetic background plays an important role in mammalian gonadal development. We hypothesize that the presence of only a single functional FOG2 or GATA4 gene can cause gonadal sex reversal in some human XY individuals.
Materials and Methods
Mice.
The C57BL/6J (B6) and DBA/2J (D2) Fog2− and Gata4ki congenic strains were produced by transferring the Fog2− and Gata4ki alleles (30, 31) to the B6 and D2 mouse strains with successive backcrossing. The B6 YAKR (16, 17) and D2 YAKR consomic strains were generated by mating AKR/J males to B6 and D2 females, respectively, followed by successive backcrosses of XYAKR males to B6 or D2 females, respectively. All experiments were conducted by using mice that were at backcross generation N10 or greater. The Jackson Laboratory is American Association for Laboratory Animal Science-accredited, and The Jackson Laboratory Animal Care and Use Committee approved all animal procedures.
Embryo Staging and Genotyping.
Timed matings were performed, and noon of the day that a vaginal plug was observed was considered E0.5. Precise fetal age was assessed for fetuses younger than E13 by counting tail somites distal to the hind limbs (44). Fetuses at E13–14.5 were staged according to limb morphology, and at E15 development was confirmed by limb morphology and the extent of formation of the cranial blood vessel (45).
The presence of the Y chromosome was determined by using a multiplex genotyping PCR assay on tissue lysate as described previously (37). The presence of the Fog2− allele was identified by using a multiplex PCR assay with a primer pair that amplified a fragment of the myogenin gene as a positive control (5′-TTACGTCCATCGTGGACAGCAT-3′ and 5′-TGGGCTGGGTGTTAGTCTTAT-3′) and a primer pair that amplified a fragment of the neomycin cassette (5′-CTTGGGTGGAGAGGCTATTC-3′ and 5′-AGGTGAGATGACAGGAGATC-3′). The PCR cycle conditions were 95°C for 2 min followed by 40 cycles of 94°C (30 s), 60°C (30 s), 72°C (30 s), and 1 cycle at 72°C for 5 min. The presence of the Gata4ki and Gata4+ alleles was identified by using a primer pair that amplified both alleles (5′-TGCGGAAGGAGGGGATTCAAAC-3′ and 5′-TCTGAGAGAACTGAGGGGGTTAGC-3′) and the following PCR cycle conditions: 95°C for 3 min, followed by 39 cycles of 94°C (30 s), 55°C (30 s), 72°C (1 min), and 1 cycle at 72°C for 5 min. The amplified Gata4ki and Gata4+ products are ≈300 and 210 bp, respectively.
Whole-Mount Immunohistochemistry.
Whole-mount immunohistochemistry was performed on E13.5 gonad–mesonephros complexes and conducted as described previously (46) except the blocking buffer consisted of 10% donkey serum (Jackson ImmunoResearch, West Grove, PA)/3% BSA/0.01% Triton X-100/0.02% sodium azide in PBS. The following antibodies were used: CD31 (1:250, clone MEC13.3; BD PharMingen, San Diego, CA), FOXL2 (1:500; gift from Marc Fellous, Cochin Institute, Paris, France), FGFR2 (1:500, C17, sc-122; Santa Cruz Biotechnology, Santa Cruz, CA), and AMH (1:200, C20, sc-6886; Santa Cruz Biotechnology). All antibodies used in this work have been validated and used in previous published reports (15, 46–49). Images were obtained by using a Zeiss (Thornwood, NY) LSM 510 confocal microscope.
RNA Isolation and Multigene Real-Time RT-PCR Analysis.
Multigene real-time RT-PCR was conducted with gonadal tissue collected at E12 (the first time when several ovary- and testis-specific genes are differentially expressed; e.g., see ref. 15) and E14 (ovaries and testes can be distinguished morphologically). Only E14 XYAKR mutant gonads lacking any testicular cords (classified as ovaries) were used for the real-time RT-PCR analysis. E12 gonad–mesonephros complexes and E14 gonads were collected and placed in lysis buffer containing β-mercaptoethanol (RNAeasy kit; Qiagen, Valencia, CA), homogenized, and stored at −80°C until further use. Total RNA was isolated by using the RNeasy mini kit (Qiagen) as described previously (28), except tissue lysate was not applied to a QIAshredder spin column (Qiagen).
Multigene real-time RT-PCR was conducted by using the 56 gene-specific primer pairs described previously (28), and primers specific for Lhx1 (LIM homeobox protein 1 (5′-TCTCCCCCTTTTGATTTGCTAGT-3′ and 5′-GGAGCGACAGGGCAATTAGAG-3′) and the M. domesticus Sry gene (5′-TGCCTCAACAAAACTGTACAACCT-3′ and 5′-GGGATATCGACAGGCAGCA-3′). Significant changes in gene expression levels and fold changes were determined by using both Global Pattern Recognition (GPR version 2.0) (50) and Student's t test, as described previously (15, 28). At each developmental time point, a minimum of three cDNA samples (one cDNA sample is generated from gonads obtained from one fetus), except at E14, when only two cDNA samples were available from control B6 Gata4+/+ XYAKR gonads. Experiments were repeated at least once, and representative results are shown in Figs. 3 and 4. Because gene expression profiles did not differ between B6 XX heterozygous mutant and control ovaries (data not shown), gene expression profiles of B6 XX heterozygous mutant ovaries were used as controls.
Supplementary Material
Acknowledgments
We are grateful to Dr. Stuart Orkin (Harvard Stem Cell Institute, Boston, MA) for providing mice containing the Gata4ki and Fog2− alleles and Dr. Sergei Tevosian (Dartmouth College, Hanover, NH) for the Gata4ki and Gata4+ primer sequences used for genotyping. We thank Meredith Crane, Andrew Recknagel, and Lisa Somes for help with the maintenance of the mouse strains and conducting PCR assays for determining genotypes; Dr. Marc Fellous (Cochin Institute, Paris, France) for kindly providing the FOXL2 antibody; and Michelle Musson for help with the whole-mount immunohistochemistry. Finally, we thank Drs. Greg Cox, Mary Ann Handel, and George Seidel for helpful comments concerning an earlier version of this paper. This work was supported by National Institutes of Health Grants GM20919 (to E.M.E.), HD07065 (to G.J.B.), and HD042779 (to K.H.A.) and National Cancer Institute Core Grant CA34196 (to The Jackson Laboratory).
Abbreviations
- E
embryonic day
- AMH
anti-müllerian hormone
- FGFR2
fibroblast growth factor receptor 2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701677104/DC1.
References
- 1.Achermann JC, Ito M, Hindmarsh PC, Jameson JL. Nat Genet. 1999;22:125–126. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- 2.Foster JW, Dominguez-Steglich MA, Guioli S, Kowk G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 3.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 4.Hastie ND. Hum Mol Genet. 1992;1:293–295. doi: 10.1093/hmg/1.5.293. [DOI] [PubMed] [Google Scholar]
- 5.Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Am J Hum Genet. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Ikeda Y, Parker KL. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 7.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 8.Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Combrugghe B. Proc Natl Acad Sci USA. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, Van De Kant HJ, Wegner M, De Rooij DG, Behringer RR, Schedl A. Development (Cambridge, UK) 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 10.McElreavey K, Fellous M. Am J Med Genet. 1999;89:176–185. doi: 10.1002/(sici)1096-8628(19991229)89:4<176::aid-ajmg2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Veitia RA, Salas-Cortes L, Ottolenghi C, Pailhoux E, Cotinot C, Fellous M. Mol Cell Endocrinol. 2001;179:3–16. doi: 10.1016/s0303-7207(01)00460-9. [DOI] [PubMed] [Google Scholar]
- 12.Barrionuevo F, Bagheri-Fam S, Klattig J, Kist R, Taketo MM, Englert C, Scherer G. Biol Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- 13.Eicher EM, Washburn LL, Whitney JB, III, Morrow KE. Science. 1982;217:535–537. doi: 10.1126/science.7089579. [DOI] [PubMed] [Google Scholar]
- 14.Eicher EM, Washburn LL, Schork NJ, Lee BK, Shown EP, Xu X, Dredge RD, Pringle MJ, Page DC. Nat Genet. 1996;14:206–209. doi: 10.1038/ng1096-206. [DOI] [PubMed] [Google Scholar]
- 15.Bouma GJ, Albrecht KH, Washburn LL, Recknagel AK, Churchill GA, Eicher EM. Development (Cambridge, UK) 2005;132:3045–3054. doi: 10.1242/dev.01890. [DOI] [PubMed] [Google Scholar]
- 16.Washburn LL, Lee BK, Eicher EM. Genet Res. 1990;56:185–191. doi: 10.1017/s001667230003528x. [DOI] [PubMed] [Google Scholar]
- 17.Washburn LL, Albrecht KH, Eicher EM. Genetics. 2001;158:1675–1681. doi: 10.1093/genetics/158.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajima T, Sasaki S, Tanaka Y, Kusunoki H, Nagashima T, Nonomura K, Fujieda K. Horm Res. 2003;60:302–305. doi: 10.1159/000074249. [DOI] [PubMed] [Google Scholar]
- 19.Ruf RG, Schultheiss M, Lichtenberger A, Karle SM, Zalewski I, Mucha B, Everding AS, Neuhaus T, Patzer L, Plank C, et al. Kidney Int. 2004;66:564–570. doi: 10.1111/j.1523-1755.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Gu WX, Ozisik G, To WS, Owen CJ, Jameson JL, Achermann JC. J Clin Endocrinol Metab. 2006;91:3048–3054. doi: 10.1210/jc.2006-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron FJ, Sinclair AH. Hum Mutat. 1997;9:388–395. doi: 10.1002/(SICI)1098-1004(1997)9:5<388::AID-HUMU2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Patient RK, McGhee JD. Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 23.Viger RS, Taniguchi H, Robert NM, Tremblay JJ. J Androl. 2004;25:441–452. doi: 10.1002/j.1939-4640.2004.tb02813.x. [DOI] [PubMed] [Google Scholar]
- 24.Cantor AB, Orkin SH. Semin Cell Dev Biol. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Viger RS, Mertineit C, Trasler JM, Nemer M. Development (Cambridge, UK) 1998;125:2665–2675. doi: 10.1242/dev.125.14.2665. [DOI] [PubMed] [Google Scholar]
- 26.Ketola I, Anttonen M, Vaskivuo T, Tapanainen JS, Toppari J, Heikinheimo M. Eur J Endocrinol. 2002;147:397–406. doi: 10.1530/eje.0.1470397. [DOI] [PubMed] [Google Scholar]
- 27.Anttonen M, Ketola I, Parviainen H, Pusa AK, Heikinheimo M. Biol Reprod. 2003;68:1333–1340. doi: 10.1095/biolreprod.102.008599. [DOI] [PubMed] [Google Scholar]
- 28.Bouma GJ, Hart GT, Washburn LL, Recknagel AK, Eicher EM. Gene Expr Patterns. 2004;5:141–149. doi: 10.1016/j.modgep.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Development (Cambridge, UK) 2002;129:4627–4634. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- 30.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 31.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH. Genes Dev. 2001;15:839–844. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ottolenghi C, Uda M, Crisponi L, Omari S, Cao A, Forabosco A, Schlessinger D. BioEssays. 2006;29:15–25. doi: 10.1002/bies.20515. [DOI] [PubMed] [Google Scholar]
- 33.Lee CH, Taketo T. Dev Biol. 1994;165:442–452. doi: 10.1006/dbio.1994.1266. [DOI] [PubMed] [Google Scholar]
- 34.Nagamine CM, Morohashi K, Carlisle C, Chang DK. Dev Biol. 1999;216:182–194. doi: 10.1006/dbio.1999.9436. [DOI] [PubMed] [Google Scholar]
- 35.Bullejos M, Koopman P. Dev Biol. 2005;278:473–481. doi: 10.1016/j.ydbio.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Schmahl J, Capel B. Dev Biol. 2003;258:264–276. doi: 10.1016/s0012-1606(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 37.Capel B, Albrecht KH, Washburn LL, Eicher EM. Mech Dev. 1999;84:127–131. doi: 10.1016/s0925-4773(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 38.Tilmann C, Capel B. Development (Cambridge, UK) 1999;126:2883–2890. doi: 10.1242/dev.126.13.2883. [DOI] [PubMed] [Google Scholar]
- 39.Qin Y, Bishop CE. Hum Mol Genet. 2005;14:1221–1229. doi: 10.1093/hmg/ddi133. [DOI] [PubMed] [Google Scholar]
- 40.Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 41.Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 42.Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D. Genomics. 2001;78:73–82. doi: 10.1006/geno.2001.6648. [DOI] [PubMed] [Google Scholar]
- 43.Manuylov NL, Fujiwara Y, Adameyko II, Poulat F, Tevosian SG. Dev Biol. 2007;307:356–367. doi: 10.1016/j.ydbio.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Development (Cambridge, UK) 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- 45.Theiler K. The House Mouse. New York: Springer; 1989. [Google Scholar]
- 46.Albrecht KH, Eicher EM. Dev Biol. 2001;240:92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- 47.Schmahl J, Kim Y, Colvin JS, Ornitz DM, Capel B. Development (Cambridge, UK) 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- 48.Gao F, Maiti S, Alam N, Zhang Z, Deng M, Behringer R, Lecureuil C, Guillou F, Huff V. Proc Natl Acad Sci USA. 2006;103:11987–11992. doi: 10.1073/pnas.0600994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cocquet J, Pailhoux E, Jaubert F, Servel N, Xia X, Pannetier M, De Baere E, Messiaen L, Cotinot C, Fellous M, Veitia RA. J Med Genet. 2002;39:916–922. doi: 10.1136/jmg.39.12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akilesh S, Shaffer DJ, Roopenian D. Genome Res. 2003;13:1719–1727. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.