Abstract
As a result of large-scale sequencing projects and recent splicing-microarray studies, estimates of mammalian genes expressing multiple transcripts continue to increase. This expansion of transcript information makes it possible to better characterize alternative splicing events and gain insights into splicing mechanisms and regulation. Here, we describe a class of splice sites that we call dual-specificity splice sites, which we identified through genome-wide, high-quality alignment of mRNA/EST and genome sequences and experimentally verified by RT-PCR. These splice sites can be alternatively recognized as either 5′ or 3′ splice sites, and the dual splicing is conceptually similar to a pair of mutually exclusive exons separated by a zero-length intron. The dual-splice-site sequences are essentially a composite of canonical 5′ and 3′ splice-site consensus sequences, with a CAG|GURAG core. The relative use of a dual site as a 5′ or 3′ splice site can be accurately predicted by assuming competition for specific binding between spliceosomal components involved in recognition of 5′ and 3′ splice sites, respectively. Dual-specificity splice sites exist in human and mouse, and possibly in other vertebrate species, although most sites are not conserved, suggesting that their origin is recent. We discuss the implications of this unusual splicing pattern for the diverse mechanisms of exon recognition and for gene evolution.
Keywords: alternative splicing, competition, mRNA/EST
Eukaryotic genes are split into exons and introns, which in the vast majority of cases are marked by a GU dinucleotide (5′ splice site) at the exon/intron boundary and an AG dinucleotide (3′ splice site) at the intron/exon boundary. To produce a mature transcript from a pre-mRNA, the introns are spliced out and the exons are ligated by a large protein/small nuclear RNA complex, the spliceosome (1, 2). The accuracy and efficiency of exon and intron recognition and splicing are dictated by: (i) primary splicing signals, including the splice sites, a polypyrimidine tract, and a branch site (2); (ii) nearby exonic or intronic regulatory sequences acting as splicing enhancers or silencers (3–5); (iii) spatial and structural constraints, such as exon and intron size (6, 7) and RNA secondary structure (8); and (iv) interactions of these cis-acting elements with splicing factors (9). Any compromise or disruption of these splicing elements or changes in the levels or properties of the factors may result in regulated alternative splicing (AS) or aberrant splicing events (10).
With the availability of genome sequences and a large amount of mRNA/EST data, especially in human and mouse, genome-wide bioinformatic analysis has revealed that a majority (>60%) of mammalian genes are alternatively spliced in various patterns (11, 12). Typical types of AS events include exon skipping/inclusion (cassette exons), alternative 5′ or 3′ splice sites, mutually exclusive exon use, intron retention, and various combinations thereof (10). Despite the complexity of splicing patterns and regulation, in all of these cases, 5′ and 3′ splice sites are defined unambiguously and recognized by distinct sets of spliceosomal components, usually at the earliest stages of spliceosome assembly (Fig. 1A) (1). The splice sites have degenerate consensus sequences, although GU and AG are nearly invariant at the 5′ and 3′ intronic borders, respectively. Interestingly, CAG|GU defines the consensus sequence of both 5′ and 3′ splice sites, although with a different extent of degeneracy (Fig. 1C). This observation raises interesting questions concerning how the splicing machinery distinguishes 5′ and 3′ splice sites, and whether the same site can be used as both a 3′ and a 5′ splice site.
Fig. 1.
Illustrative representation of dual splicing. (A and B) Schematic diagram of canonical splicing (A) and dual splicing (B). Boxes represent exons, and lines are introns. The dual splice site is labeled in B. (C and D) The motifs of canonical (constitutive) 5′ and 3′ splice sites (C) and of dual splice sites (D). Dotted arrows and boxes indicate the similarity of dual sites with constitutive splice sites. Uridine is shown as thymine in the logos.
In this study, we investigate unusual AS events associated with splice sites that can be used as either 5′ or 3′ splice sites. We refer to these sites as dual-specificity splice sites (or dual splice sites). We detected these dual-specificity sites with high-quality mRNA/EST and genome sequence alignment evidence. In these cases, a particular splice site is used as a 3′ splice site in some transcripts, and in other transcripts, the same site is used as a 5′ splice site. When the dual splice site is recognized as a 3′ splice site, the sequences upstream of the site are removed as an intron, whereas the sequences downstream are retained as an exon. However, this situation is reversed in alternative isoforms, in which the dual site is used as a 5′ splice site and the sequences downstream of the site are removed as an intron (Fig. 1B). Thus, the resulting exon/intron flip-over in different isoforms affects the nature of the protein products. We validated the occurrence of dual-specificity splicing in vivo by RT-PCR and direct sequencing and found that the use of the site as a 5′ or 3′ splice site can vary in a tissue-specific manner. Bioinformatic analysis revealed unique features that are consistent with the dual-specificity character and predictive of the splicing outcome. The implications for protein coding and gene-structure evolution are also discussed. We conclude that the use of dual-specificity splice sites as either a 5′ or 3′ splice site represents an additional class of AS.
Results
Identification and Classification of Dual-Specificity Splice Sites.
We built a database of classified AS events (dbCASE), using high-quality transcripts (mRNA/EST) and genome alignment for multiple species. A data structure called splicing graph (13) was applied and extended to efficiently detect various alternative and constitutive splicing events and to track supporting transcripts (see Materials and Methods). During this process, we found that previous data structure could not represent the transcript data in some cases because of the presence of dual splice sites. In total, we found 594 human (and 195 mouse) putative dual splice sites with supporting transcript (mRNA/EST) evidence. We also extracted strictly constitutive exons and introns (in the sense that no violating transcripts were detected) as a comparative dataset to further analyze the nature of these dual splice sites.
Because most canonical introns have GU and AG dinucleotides at their 5′ and 3′ termini, respectively (14), we first examined whether the dual splice sites conform to this AG|GU rule. Overall, 155 dual splice sites (26%) conform to the AG|GU rule. This percentage is lower than that expected compared with constitutive splice sites (Table 1). There are several explanations that may account for this difference. First, sites with few supporting transcripts may be unreliable because they could reflect aberrant splicing or RT-PCR errors. Second, repetitive elements, sequencing errors in the transcripts (especially ESTs) or in the genome, polymorphisms, and transcripts from paralogous or pseudo genes may result in spurious alignments. The third point, which is not mutually exclusive with the two preceding explanations, is that we observed 64 human (and 9 mouse) genes with clusters of dual splice sites. These genes seem to be highly conserved across vertebrate species but are enriched in exonic SNPs (data not shown). They account for approximately half of the total number of putative dual splice sites. Most of these sites (≈85%) do not match the AG|GU pattern, and it is unclear whether they are authentic examples of dual splice sites or whether they represent artifacts.
Table 1.
Percentage of dual splice sites conforming to the AG|GU rule.
| Site | All | AG|GU sites |
|---|---|---|
| 5′ splice site | 27,556 | 15,455 (56.1%) |
| 3′ splice site | 27,556 | 5,022 (18.2%) |
| dual site | 594 | 155 (26.1%) |
| dual site.2 | 85 | 46 (54.1%) |
| dual site.3 | 40 | 28 (70.0%) |
| dual site.singleton | 319 | 119 (37.3%) |
| dual site.singleton.2 | 39 | 31 (79.5%) |
| dual site.singleton.3 | 26 | 23 (88.5%) |
dual site.2 (dual site.3), dual splice sites with two (three) or more supporting transcripts for each isoform; dual site.singleton, singleton dual splice sites (no other dual splice sites from the same gene). dual site.singleton.2 and dual site.singleton.3 are similarly defined.
To increase the level of confidence in dual-splice-site prediction, we explored ways to increase the stringency of our criteria for dual-splice-site classification. The percentage of AG|GU sites increased greatly when two or more supporting transcripts were required for each isoform (Table 1). We also considered gene transcripts with only one dual splice site (singletons) by removing all genes with two or more sites, to eliminate potential noise from other classes of transcripts, as described above. This filtering step further increased the proportion of AG|GU sites. For example, 23 of 26 (88.5%) singleton sites with three or more supporting transcripts for each isoform conformed to the AG|GU pattern; this percentage is significantly higher compared with constitutive splice sites (P = 0.0006 for 5′ splice sites, P = 10−14 for 3′ splice sites, Fisher's exact test). Thus, we surmise that most authentic dual splice sites follow the AG|GU rule, which is likely an important feature to specify dual splicing, probably by the U2-type spliceosome (2).
To characterize the features of dual splice sites, we derived a high-confidence dataset by limiting dual splice sites to AG|GU sites with two or more supporting transcripts for each isoform. We further removed nine sites from the UBC gene (because this gene contains multiple repetitive coding units (15), which are prone to alignment uncertainties) and also three other sites that lacked perfectly matching alignments in sequences flanking the sites. The final high-confidence dataset has 36 dual-specificity splice sites [supporting information (SI) Table 2], which were used for the analyses below. Among these splice sites, 11 (31%) have RefSeq or mRNA supporting evidence for both isoforms, whereas the remaining 25 (69%) have only ESTs as supporting evidence for one or both isoforms.
The dual splicing pattern can be classified according to the nature of the resulting alternative transcripts. The most prevalent class of dual splice sites is associated with the first exon (12 of 36 cases) (class I, SI Fig. 5). This is unlikely to be attributable to sequence-alignment artifacts, because all spurious terminal exons <25 nt were removed, so that each intron is flanked by two reliable exons. Instead, this first-exon preference suggests a possible link between alternative promoters and dual-splice-site choice (SI Fig. 5). Other dual sites create an upstream or downstream alternative exon (class II, SI Fig. 6 and class III, SI Fig. 7) or result in intron retention (class IV, SI Fig. 8) or exon truncation (class V, SI Fig. 9).
Dual Splice Sites Resemble the 5′ and 3′ Splice Site Consensus Sequences.
To study the specificity of recognition as 5′ splice sites and 3′ splice sites more quantitatively, we derived the position weight matrices (PWMs) of dual splice sites, and canonical 5′ and 3′ splice sites from constitutive exons (16) (Fig. 1C). Compared with the constitutive splice sites, it is readily discernible that the PWM of dual splice sites (Fig. 1D) is approximately the juxtaposition of the intronic portions of the constitutive 5′ and 3′ splice site matrices, with CAG|GURAG (R represents A or G) as a core in the consensus. The GC content around dual splice sites is higher than that of the corresponding portions of constitutive splice sites (SI Table 3). This finding could reflect either that exonic sequences generally have a higher GC content than intronic sequences (11) or perhaps unknown mechanistic reasons related to the recognition and splicing of dual splice sites.
One could argue that the resemblance of the dual splice site matrix to both canonical 5′ and 3′ splice site matrices of constitutive splice sites may be an artifact of contamination with both types of splice sites, which are erroneously classified as dual splice sites. To exclude this possibility, we scored each individual dual splice site with both canonical 5′ and 3′ splice site matrices, using methods described in ref. 16 (see Materials and Methods for details).
As shown in Fig. 2, the canonical 5′ and 3′ splice sites of constitutive exons fall into two distinct but overlapping populations in the space of 5′ and 3′ splice site matrix scores. Most 5′ splice sites have low scores, using the PWM for 3′ splice sites, and vice versa. In contrast, dual splice sites have relatively high scores, using both matrices (Fig. 2 A and C). For example, only 2–4% of constitutive splice sites have both scores for a single site greater than the first quantile (0.25 in the abscissa in Fig. 2C), whereas ≈50% (19 of 36) of dual splice sites have both matrix scores greater than the same threshold.
Fig. 2.
Motif scores of dual and constitutive splice sites. (A) Graphical representation of motif scores of constitutive 5′ (blue), 3′ (cyan), and dual (red) splice sites. (B) Similar to analysis in A, except that only constitutive splice sites with the AG|GU pattern are shown. (C) Resemblance of dual splice sites to the canonical 5′ and 3′ splice-site consensus motifs. Matrix scores were ranked and converted into quantiles according to constitutive splice sites, and different thresholds (quantile 0 to 1, in steps of 0.01) were applied to count the number of sites whose scores exceed both thresholds.
To ensure that this difference between constitutive and dual splice sites is not an artifact reflecting our choice of dual splice sites with the AG|GU pattern, which conforms to the consensus of both 3′ and 5′ splice sites, we performed a stringent comparison of dual splice sites to the subset of constitutive splice sites with the AG|GU pattern (Fig. 2 B and C). This increased the percentage of constitutive splice sites with high scores by both PWMs, which nevertheless was still significantly lower than that of dual splice sites. For example, only 8–13% of AG|GU constitutive splice sites have both scores greater than the first quantile, compared with ≈50% for dual splice sites (P < 10−7 in both comparisons with 5′ and 3′ splice sites, Fisher's exact test).
Thus, the resemblance of dual splice sites to both 5′ and 3′ splice site consensus motifs strongly suggests that they are authentic splice sites with dual specificity as both 5′ and 3′ splice sites. It is also worth noting that relatively few dual splice sites have top scores (e.g., greater than the third quantile, 0.75 in the abscissa) for both matrices (Fig. 2C), most likely reflecting the difficulty in simultaneously satisfying the constraints of both matrices in a perfect manner.
Competitive Recognition Predicts Splicing Outcome.
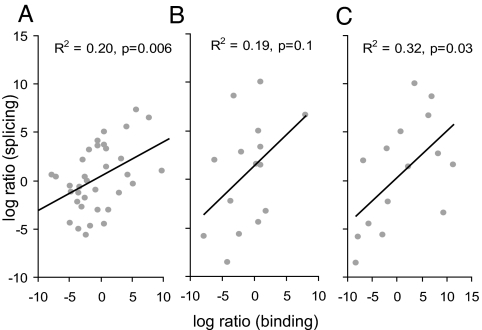
We further reasoned that if the dual splice sites are authentic, the sequence should dictate the competition between 5′ splice-site-associated and 3′ splice-site-associated spliceosomal components, which would be reflected in the relative use of each site and hence the number-of-transcripts evidence for each isoform. The difference between 5′ and 3′ splice-site matrix scores of each dual splice site, Δb, reflects the log-likelihood ratio of the site being recognized as a 5′ splice site to it being recognized as a 3′ splice site (Eq. 2 in Materials and Methods). We assumed a linear relationship between the binding-likelihood ratios to splicing ratios and examined their Pearson correlation. Indeed, we observed a significant correlation (R2 = 0.2, P = 0.006), which means that 20% of the variation in splicing outcome can be explained by the binding affinity to the splice sites (Fig. 3A). A simple classifier according to Δb at the threshold of zero gives 26 of 36 (72%) correct predictions. This correlation and accuracy of prediction is surprising, given that the number of sequenced transcripts pooled from all sources is only an approximation of the real splicing outcome (17) and that other sequences around the splice sites are also likely to be important determinants of splice-site selection.
Fig. 3.
Predicting splicing outcome, using binding specificity. Each point represents a dual splice site. The x axes show the log-likelihood ratio of the binding probabilities, and the y axes show the log ratio of supporting-transcript number for the 5′ splice-site isoform relative to that of the 3′ splice-site isoform. (A) Competition at the dual splice site. The log-likelihood ratio of binding is calculated as Δb by using the high-confidence dataset. (B) Similar to A, except by using the extended set. (C) Competition by exon definition or alternative 5′/3′ splice sites. The log-likelihood ratio of binding is calculated as Δb2 by using the extended set.
To test the latter hypothesis, we evaluated the importance of upstream and downstream splice sites in determining splicing outcome. We reasoned that the strength of the splice site that pairs with the dual 5′ or 3′ splice site across the exon [as per exon definition (6)] may influence the splicing outcome. For simplicity, we limited our analysis to dual sites that give rise to alternative exons, i.e., class II and class III, as defined above (see also SI Figs. 6 and 7). In the high-confidence dataset, 6 of 36 cases belong to this category. To expand the sample size, we examined 109 AG|GU dual sites with a single supporting transcript for either isoform, and with perfect local alignment, and included nine additional cases in the category. For each of the 15 cases in total, we calculated the scores of the upstream 3′ splice site and downstream 5′ splice site, together with the scores of the dual site, and derived a measure of competition Δb2 (see Eq. 3 in Materials and Methods for details). This measure can have two alternative interpretations: the difference in the strength of exon definition of the two isoforms or the difference in the strength of alternative 5′ and 3′ splice-site competition. As shown in Fig. 3B, the competitive recognition of dual splice sites alone measured by Δb explains 19% of the variation, consistent with the results in Fig. 3A, although the significance level drops because of the limited sample size (P = 0.1). A classifier according to Δb at a threshold of zero gives 10 of 15 (67%) correct predictions. Importantly, including upstream and downstream splice sites in the competition model explains 32% of the variation (P = 0.03) (Fig. 3C). A classifier according to Δb2 at a threshold of zero gives 12 of 15 (80%) correct predictions. Therefore, we conclude that the strength of the upstream and downstream splice sites, and probably other regulatory sequences, also contributes to the dual splicing pattern.
Splice Sites Are Used as Both 5′ and 3′ Splice Sites in Cells.
To confirm that dual splice sites are used as both 5′ splice sites and 3′ splice sites, we analyzed splicing of the endogenous Smac/Diablo (DIABLO), UBE2C, POLR2G, and UROD transcripts in two human cell lines. In each case, RT-PCR analysis verified the presence of the two isoforms with the expected sizes (Fig. 4 and SI Fig. 10; see primer sequences in SI Table 4). Each pair of isoforms was identified in HeLa cells and the neuronally derived Weri-Rb1 cell line. The use of the predicted 5′ and 3′ splice sites of the Smac/Diablo and UBE2C dual splice sites was further confirmed by sequence analysis of the RT-PCR products (data not shown).
Fig. 4.
Validation of dual-splice-site recognition in human and mouse. RT-PCR analysis of RNA from human brain (b), testis (te), tonsil (to), and thymus (th) tissues (Clontech) and HeLa (H), Weri-Rb1(W), NIH 3T3 (3), and NSC-34 (N) cells. Diagram (A–C) of the general splicing pattern and location of primers (arrowheads) used to detect isoforms that result from the use of the 5′ splice site or 3′ splice site of Smac/Diablo (3′ss, 234 nt; 5′ss, 205 nt) (B) and UBE2C (3′ss, 287 nt; 5′ss, 233 nt) (C). In the case of UBE2C, the exon containing the dual splice site is also skipped to give an additional isoform (skip, 82 nt). (D) Quantitative analysis of RT-PCR results. The histogram represents the percentage of the products that are generated by use of the 5′ splice site.
Dual splice sites are essentially alternative splice sites and are potentially subject to regulation. We tested the relative use of the Smac/Diablo and UBE2C dual 5′ and 3′ splice sites in a number of tissues and cell lines. We observed variations in the use of the splice sites (Fig. 4), suggesting that use of the dual splice sites is regulated. If a specific trans-acting factor determines whether the site is used as a 5′ or 3′ splice site, then a specific cell type or tissue might be expected to show a general preference for the 5′ or 3′ splice site of all dual splice sites. However, there did not appear to be a consistent bias for the 5′ or 3′ splice site in any of the tissues or cell types we tested for the two pre-mRNAs we examined.
To explore the functional implications of dual splicing, we examined the splicing patterns that can potentially generate functional protein products (SI Table 2). In six cases (17%), we found protein products for both isoforms, and the alternatively spliced region of each isoform was at least partially coding. In 21 cases, protein products for one or both isoforms were not found, most likely because of the incompleteness of the protein sequence database in GenBank (18) and/or because of the presence of premature termination codons in some isoforms that are presumably subject to nonsense-mediated mRNA decay (19). In the remaining cases, the dual splice sites were in the untranslated regions, making it difficult to link transcripts to protein sequences directly, although protein sequences that are compatible with the transcripts were found.
Dual Splice Sites in Mouse.
Dual splice sites are not limited to human: We also found evidence for 195 putative dual splice sites in mouse. Using the same filtering criteria as applied in human, the mouse high-confidence dataset contains 18 sites (SI Table 5). The difference in number is likely because EST coverage in human is significantly higher than that in mouse (7 million compared with 4 million). Dual splice sites were also detected in rat, zebrafish, and fly, although infrequently and with less supporting evidence (data not shown). We performed a detailed analysis of mouse dual splice sites in the same way as we did for human. The properties of mouse dual splice sites, such as the motif itself, were generally very similar to those of human sites (data not shown). We performed a human–mouse comparison by examining the conservation of dual-splice-site sequences and the splicing patterns. Although the splice sites are often conserved at the sequence level, it appears that the conservation of flanking exonic and/or intronic sequences is low (SI Figs. 5–9 and data not shown). Most of the sites lack supporting evidence for conservation of the dual splicing pattern, except in two cases: MYL6 and PHC (SI Figs. 11 and 12). Both of these sites follow the AG|GU rule in both species. However, neither of these two sites was included in our high-confidence set because of an insufficient number of supporting transcripts. Therefore, the conservation rate in dual splicing appears to be very low, with an upper bound of 5% [2 of 38 (36 + 2)], which is much lower than that of cassette-type splicing events (10–20%) (20–22). Indeed, we could only detect a single isoform of Smac/Diablo and UBE2C in the mouse neuronal cell line NSC-34 and in mouse NIH 3T3 fibroblasts (Fig. 4).
Discussion
Large-scale sequencing projects in the past decade and recent applications of splicing microarrays have made clear the extent and complexity of AS in mammalian genes (23). In this study, we identify a class of splice site and associated AS pattern. A dual splice site is a composite of canonical 5′ and 3′ splice sites, which makes it possible for a single site to be recognized as either a 5′ splice site or a 3′ splice site and results in an exon becoming an intron and vice versa (exon/intron flipping). There was only one previously documented example of a dual-specificity splice site in the IRF3 gene (24). In this case, dual splicing generates isoforms that can potentially code for proteins with different functions. We show that this form of AS is more prevalent than previously appreciated. We identified hundreds of potential dual splice sites in human and mouse, among which at least 36 in human and 18 in mouse were identified with high confidence. The greatly expanded list of dual sites allowed us to uncover unique features of these sites.
Several lines of evidence, including multiple supporting transcripts, the resemblance of the sites to both 5′ and 3′ splice site consensus motifs, the correlation between binding specificity and splicing outcome, and the presence in different species, strongly suggest that AS via dual sites is an authentic pattern. For several cases, the presence of these sites and the splicing pattern were further validated by RT-PCR and sequencing. It is possible that many of the dual splice sites not included in our high-confidence dataset are also authentic but currently have a limited number of supporting ESTs or mRNA transcripts for reasons implicit in the splicing event. For example, the AS events associated with the use of the 5′ or 3′ splice site may be rare in certain tissues, or one of the splicing events may generate a premature termination codon, resulting in a transcript that is subject to nonsense-mediated mRNA decay.
The capacity of a dual splice site to switch its identity between a 5′ splice site and a 3′ splice site has implications for many aspects of pre-mRNA processing and raises important questions regarding the mechanisms of splice-site recognition, regulation, and competition. First, to our knowledge, dual splice sites are the first type of splice site to lack unambiguous identity as either a 5′ or a 3′ splice site. The use of a dual splice site likely involves competition between the 5′ and 3′ splice sites through a stochastic process, as the two isoforms can coexist in the same tissue type. It is of interest to know at which step of spliceosome assembly the choice of 5′ or 3′ splice site is made. Second, what are the determinants of dual-splice-site use? Except for two cases, the dual AS pattern does not appear to be conserved between human and mouse. However, in many cases, including two that we tested (Fig. 4), the sequence of the dual splice site is conserved. Despite this sequence conservation, the sites do not appear to be used as dual splice sites in mouse. In fact, thousands of splice sites have dual character comparable with the observed dual splice sites, but they do not appear to have dual splicing. Our preliminary analysis suggests that the flanking splice sites also contribute to the splicing outcome, together with the dual sites. Other splicing signals, such as the strength of the polyrimidine tract and the distribution of splicing enhancers and silencers, are also likely important determinants of dual-splice-site use.
At the present time, the functional significance of this unusual AS pattern is not clear. Our results suggest that most dual splice sites have a recent evolutionary history, appearing independently in each species. Recently, introns with significant sequence similarities at their 5′ and 3′ splice sites were described in ref. 25. It was proposed that sequences bearing cryptic splice sites can be duplicated to serve as the termini of a new intron. Such a mechanism could be one possible evolutionary origin of dual splice sites before the duplicated cryptic splice sites had a chance to evolve into unambiguous 5′ or 3′ splice sites.
As with other AS patterns, many of the new isoforms might have arisen as splicing errors or may represent evolutionary precursors (26). However, by inserting an exon and simultaneously deleting another exon, dual splicing may in some cases generate a transcript with adaptive value and thus serve as a mechanism for genomic diversification and expansion of coding capacity. In some cases, both isoforms appear to be abundant. For instance, there are 344 transcripts aligned to the WDR73 locus, among which 43 (13%) and 99 (29%) directly support one of the two isoforms resulting from dual splicing, respectively. In addition to cases that are predicted to yield unproductive transcripts by inducing nonsense-mediated mRNA decay, we found several cases, including Smac/Diablo and UBE2C, in which both isoforms code for distinct protein products with potentially altered biochemical properties (SI Table 2). Our RT-PCR analysis reveals that both isoforms of Smac/Diablo and UBE2C are abundant at the mRNA level. Furthermore, the levels of each isoform vary among tissues and cell types, suggesting regulation of the use of the dual splice site as a functional 5′ or 3′ splice site. This regulation may have important functional consequences for protein activity. In the case of Smac/Diablo, which codes for a proapoptotic protein, the alternative isoforms have different abilities to bind to effector molecules and differential cellular localization, although the dual splicing was not previously noted (27).
There are interesting similarities and differences between dual splice sites and the recursive splice sites reported in refs. 28 and 29, which are thought to be used as intermediate steps in the splicing of long introns. The consensus motifs for both types of splice sites look like a composite of the canonical 5′ and 3′ splice-site consensus motifs. However, in reported examples of recursive splicing, a splice site first functions as a 3′ splice site, and then, after ligation to the upstream exon, a 5′ splice site is regenerated. This new 5′ splice site is subsequently spliced to a downstream 3′ splice site. Thus, recursive splice sites are generated, in part, as a result of the splicing reaction, in contrast to dual splice sites, for which both the 5′ and 3′ splice sites are present in the pre-mRNA and are functional.
Another difference between these two classes of splice sites is that recursive splicing at intronic sites does not directly affect the final mRNA product, whereas AS of dual splice sites does. In addition, although in principle the two sequential steps of recursive splicing might be reversed, with a 5′ splice site used first and regenerating a functional 3′ splice site, a recent study argued against this reversibility (29). Therefore, competition is not involved in recognition of the recursive splice site as a 5′ or 3′ splice site in the first splicing reaction because only one functional splice site is initially present and used. In contrast, for dual splice sites, both the 5′ splice site and the 3′ splice site are present simultaneously and probably compete for binding of their respective splicing factors. Steric hindrance presumably forces the use of a dual site in a given pre-mRNA molecule as either a 5′ or 3′ splice site because once 3′ splice-site factors bind to the site, 5′ splice-site factors are effectively excluded, and vice versa, a consideration that also applies to microexons (30).
Despite the above differences, it is possible that some dual splice sites could function as sites of recursive splicing as well. For 10 of 36 high-confidence dual splice sites, there is transcript evidence that the exon in which the dual splice site resides can be skipped (e.g., Fig. 4 A and C). Recursive splicing at a dual splice site would result in an mRNA isoform lacking an exon, which would be indistinguishable from a mature mRNA arising from a conventional exon-skipping event. Examples of recursive splicing resulting in exon skipping are described in refs. 28 and 29. More direct experimental evidence will be required to determine whether exon skipping is actually generated by recursive splicing at the dual splice sites we found.
In summary, by using transcripts and genome alignment in human and mouse and experimental validation, we have identified and characterized a class of splice sites with dual specificity as 5′ and 3′ splice sites. The functional significance of these sites and of the AS events they specify is underscored by their direct effects on the corresponding protein products, in some cases in a tissue-specific manner. Importantly, this class of AS via dual splice sites suggests even greater versatility of the splicing machinery than was previously recognized.
Materials and Methods
Detection of Splicing Patterns with Splicing Graphs.
We built a database of classified AS events (dbCASE; http://rulai.cshl.edu/dbCASE), using high-quality transcripts (mRNA/EST) and genome alignment for human and mouse (coverage >85%, identity >95%). Briefly, transcripts from UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene, build 196 for human, build 158 for mouse) and RefSeq (ftp://ftp.ncbi.nih.gov/refseq/release, release 20) (31) were aligned to genomic sequences (hg18 and mm8) by using sim4 (32). The alignment of all transcripts to the same gene locus was then converted into a splicing graph, in which each splice site is represented by a node and each exon/intron is represented by an edge (13). In contrast to Sugnet et al. (13), we allowed the same position to be both 5′ and 3′ splice site, and the transcript evidence was recorded for each form separately, which was critical for this study. AS patterns (in particular dual-specificity splice sites) were detected by analyzing subnetwork topologies. In addition, strictly constitutive exon and introns (in the sense of no violation of transcript evidence) can be detected efficiently by graphic analysis.
Construction of PWMs for Canonical and Dual Splice Sites.
To measure the presumptive binding specificity of the spliceosome, we first constructed PWMs for canonical 5′ and 3′ splice sites from constitutively spliced exons (27,556 in human and 36,262 in mouse, with four or more supporting transcripts). Thirty nucleotides surrounding the splice junction (15 nt on each side) were extracted, and PWMs were built from these sequences (16). Each dual splice site, as well as constitutive splice site, was scored by both matrices as follows:
and
where s5ss (s3ss) is the score of the 5′ (or 3′) splice site matrix, i is the position in the matrix, and bi is the base of the site at position i. fi,bi5ss, fi,bi3ss, and fbi0 represent the frequency of base bi in 5′ splice sites, 3′ splice sites, and background sequences, respectively. A matrix of dual splice sites was built in a similar manner.
Competition at Dual Splice Sites or by Exon Definition.
We considered two models of competition to determine the splicing outcome at a dual splice site. In the first model, the recognition of the dual splice site as a 5′ or 3′ splice site results from the competition of 5′ splice-site-associated and 3′ splice-site-associated spliceosomal components at the dual splice site. Therefore, the difference between 5′ and 3′ splice site matrix scores of each dual splice site reflects the log-likelihood ratio of the site being recognized as a 5′ splice site to it being recognized as a 3′ splice site.
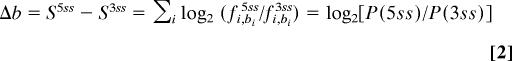 |
In the second model, the splicing outcome results from competition between exon definition by pairing the dual splice site with the upstream 3′ splice site or with the downstream 5′ splice site (Fig. 1B). The competition is represented by
 |
where the scores of the dual sites are shown by the subscripts, Sup3ss is the matrix score of the upstream 3′ splice site, and sdown5ss is the matrix score of the downstream 5′ splice site. An alternative interpretation of this model is the difference in the strength of alternative 5′ (3′) splice-site competition, as shown on the bottom part of Eq. 3.
Identification of Protein Products.
For each dual splice site, representative supporting transcripts were retrieved from dbCASE and searched against the nonredundant protein database of GenBank (18). All protein sequences with significant matches (>10 aa) were retrieved and BLATed against the genomic sequence in the UCSC genome browser (http://genome.ucsc.edu) (33). The protein sequences that aligned properly with the desired pattern were subsequently identified.
Statistical Analysis.
Fisher's exact test in R was used to evaluate the significance of 2 × 2 contingency tables (34).
RT-PCR.
RNA collected from cells by using TRIzol Reagent (Invitrogen, Carlsbad, CA) or RNA from tissue samples (Clontech, Mountain View, CA) was reverse transcribed by using SuperScript II reverse transcriptase (Invitrogen) with oligo(dT) primers. PCR with AmpliTaq Gold (Roche, Indianapolis, IN) was carried out for 40 amplification cycles (95°C for 30 s, 60°C for 60 s, and 72°C for 60 s) in reactions containing [α-32P]dCTP. Primer sequences are provided in SI Table 4. Products were separated on 6% native polyacrylamide gels. Quantitation was based on phosphorimage analysis (FLA-5100; Fujifilm, Valhalla, NY).
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grants GM42699 (to A.R.K.) and GM74688 (to M.Q.Z.) and the Dart Neurogenomics Alliance Funds (M.Q.Z).
Abbreviations
- AS
alternative splicing
- PWM
position weight matrix.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703773104/DC1.
References
- 1.Moore JM, Query CC, Sharp PA. In: The RNA World. Gesteland RF, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. pp. 303–357. [Google Scholar]
- 2.Black DL. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Ladd A, Cooper T. Genome Biol. 2002;3:reviews0008. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Z-M. J Biomed Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastings ML, Krainer AR. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 6.Berget SM. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 7.Fox-Walsh KL, Dou Y, Lam BJ, Hung S-P, Baldi PF, Hertel KJ. Proc Natl Acad Sci USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratti E, Baralle FE. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CWJ, Valcárcel J. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 10.Cartegni L, Chew SL, Krainer AR. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 11.Lander E, Linton L, Birren B, Nusbaum C, Zody M, Baldwin J, Devon K. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y, Gaasterland T RIKEN GER Group; GSL Members. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugnet C, Kent W, Ares MJ, Haussler D. Pac Symp Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 14.Burset M, Seledtsov IA, Solovyev VV. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker R, Board P. Am J Hum Genet. 1989;44:534–542. [PMC free article] [PubMed] [Google Scholar]
- 16.Stormo GD. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 17.Modrek B, Lee CJ. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 18.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y-F, Imam JS, Wilkinson MF. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 20.Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Sorek R, Shamir R, Ast G. Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blencowe BJ. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Karpova AY, Howley PM, Ronco LV. Genes Dev. 2000;14:2813–2818. doi: 10.1101/gad.813800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo D, Madden R, Elela SA, Chabot B. Proc Natl Acad Sci USA. 2007;104:882–886. doi: 10.1073/pnas.0604777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Krainer AR, Zhang MQ. Trends Genet. 2007 doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Roberts DL, Merrison W, MacFarlane M, Cohen GM. J Cell Biol. 2001;153:221–228. doi: 10.1083/jcb.153.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatton AR, Subramaniam V, Lopez AJ. Mol Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 29.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlo T, Sierra R, Berget SM. Mol Cell Biol. 2000;30:3988–3995. doi: 10.1128/mcb.20.11.3988-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ihaka R, Gentleman R. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.