Abstract
Mucosal epithelial M cells provide an efficient portal of entry for microorganisms. Initially defined by their irregular microvilli and abundant transcytotic channels in the avian bursa of Fabricius, M cells also are found in the lymphoid follicle-associated epithelium of the mammalian appendix, Peyer's patches, and other mucosal surface-lymphoid interfaces. We describe here a previously unrecognized cathelicidin gene in chickens, chCATH-B1, that is expressed exclusively in the epithelium of the bursa of Fabricius. Like the mature peptides of previously identified cathelicidins, the carboxyl-terminal peptide of chCATH-B1 has broad antimicrobial activity against Gram-positive and Gram-negative bacteria. chCATH-B1 expression is restricted to the secretory epithelial cell neighbors of the M cells, whereas its mature peptide is transported to become concentrated on the fibrillar network surrounding basolateral surfaces of the M cells that overlie the bursal lymphoid follicles. We conclude that chCATH-B1 is well placed to serve a protective antimicrobial role at the M cell gateway.
Keywords: antimicrobial peptides, follicle-associated epithelium, innate immunity, bursa of Fabricius
The survival of all multicellular organisms depends on an effective immune response to microbial pathogens. The first hurdle to microbial entry is provided by our epithelial surfaces. Microbial pathogens that mount this barrier encounter innate immunity elements as a first line of defense. Innate immune responses also facilitate the ensuing adaptive immune responses that vertebrates use to clear infectious agents. The mucosal M cells are an important microbial portal of entry because of their highly efficient pinocytotic channels. Initially identified as specialized epithelial cells overlying the lymphoid follicles of the avian bursa of Fabricius that have irregular microvilli and efficient transcytotic capability (1), M cells were also found to be conserved in the lymphoid follicle-associated epithelium of mammalian appendix and intestinal Peyer's patches (1, 2). The M cells have since been found in other mucosal lymphoid tissues, including those of the upper and lower airways, oropharynx, salivary glands, stomach, colon, and eye (3, 4), where they provide an efficient conduit for transporting microorganisms and other antigenic substances into the underlying lymphoid structures to initiate immune responses (5, 6). Despite the physiological importance of this entry portal, there is limited information about the differentiation of the M cells, their transport mechanism(s), and how the microbes that constantly enter the body via the M cells are rendered noninvasive.
Antimicrobial peptides are well known as front-line participants in microbial defense (7–10). Two evolutionary groups of antimicrobial peptides, the cathelicidins and the defensins, provide endogenous peptide-based defense against microbial invasion (11–13). Cathelicidins and defensins are produced by many cell types and have broad spectrum antimicrobial activity against bacteria, fungi, and viruses. As one example, the human cathelicidin LL-37 (also called hCAP-18, FALL-39, and CAMP) is produced by neutrophils, B cells, γδ T cells, natural killer cells, monocytes, and macrophages (14–16); it is also found in the squamous epithelium of the mouth, tongue, and esophagus, as well as in the colonic and bronchial mucosal epithelium (17). LL-37 expression is negligible in normal skin, but epidermal cells are induced to express high levels of LL-37 in inflammatory conditions, such as psoriasis and contact dermatitis (18). Conversely, deficiencies in the LL-37 cathelicidin and the HBD-2 defensin may underlie the Staphylococcus aureus skin infections that plague patients with atopic dermatitis (19). Recent studies have also indicated the importance of epithelial cathelicidin in the maintenance of the sterility of the human urinary tract (20). Remarkably, neither cathelicidins nor defensins have been identified at the M cell interface, where one might anticipate their need. We report here an avian cathelicidin that appears to fulfill this expectation. This peptide was identified during a search for genes expressed preferentially in the bursa of Fabricius.
Results
Identification of a Bursa-Specific Cathelicidin, chCATH-B1.
Our search for bursa-specific genes began with the cloning of bursal cDNA subtracted by splenic cDNA and yielded cDNA clones, some of which have been reported (21). Among these, BFG7 is expressed exclusively in the bursa of Fabricius as shown by Northern blot analysis (Fig. 1). Sequence analysis of a full-length BFG7 cDNA did not yield a match with then-reported genes. However, a BLAST search of the National Center for Biotechnology Information protein database revealed a BFG7 cathelin domain sequence, which is a conserved hallmark of the cathelicidin gene family (Fig. 2). Although conserved cathelin regions of mammalian cathelicidins share 50% or greater amino acid identity (12), the BFG7 cathelin region has only 20–30% homology with mammalian cathelicidins. This is in the “twilight” zone of sequence similarity but is within the range of identity shared by many avian and mammalian orthologs (22). Provisionally, we have named this cathelicidin relative “chicken cathelicidin-B1,” or chCATH-B1, in view of its selective expression in the bursa of Fabricius.
Fig. 1.
Survey of chicken tissues for BFG7 expression by Northern blot analysis. RNA from different tissues was placed on the membrane before hybridization with an 800-bp fragment of a BFG7 cDNA as the probe.
Fig. 2.
Sequence of chCATH-B1 cDNA and the deduced protein. The deduced amino acid sequence of exons is shown below the nucleotide sequence. Nine octamer repeats (PGLDGSXS) are underlined. The cathelin domain is boxed. The potential polyadenylation signal (AATAAA) is double underlined. The nucleotide sequence of the chicken cathelicidin B1 cDNA has been deposited in the DNA Data Bank of Japan/GenBank/European Molecular Biology Laboratory database (AB307733).
Proteolytic cleavage of mammalian cathelicidin proproteins yields mature C-terminal peptides with antimicrobial activity. In this context, a comparative alignment of chCATH-B1 with mammalian cathelin region sequences predicted a cationic peptide of 40 C-terminal amino acid residues with a high pI value (pI = 12.2) (Fig. 2). This peptide sequence appears to belong to the α-helical group of antimicrobial peptides. A Shiffer–Edmundson wheel projection analysis of the first 20 residues of the chCATH-B1 peptide predicts an amphipathic structure that is characteristic for α-helical cathelicidin peptides (23) [supporting information (SI) Fig. 7]. In addition to the signal peptide of 22 aa, a cathelin domain and predicted mature peptide, a region of 105 aa flanking the N terminus of the chCATH-B1 cathelin domain, includes nine octamer repeats (Fig. 2).
chCATH-B1 Differs from Other Chicken Cathelicidin Orthologs.
A single hybridization band was observed when DNA from several chicken strains was digested with Bgl II or HindIII and analyzed by Southern blotting with an 800-bp BFG7 fragment as the probe (data not shown). This finding suggested that chickens lack close relatives of chCATH-B1 in their genome. In a preliminary search for chCATH-B1 relatives in other species, hybridization of a “Zoo blot” under low-stringency conditions yielded a discrete band for bovine DNA, but no specific bands of hybridization were seen with DNA from human, monkey, mouse, rat, or rabbit (SI Fig. 8). Given that the sequence homology between chCATH-B1 and the currently recognized bovine cathelicidins is not higher than that found between chCATH-B1 and other mammalian cathelicidins (Fig. 3 and data not shown), the hybridizing band in bovine DNA may represent an unidentified cathelicidin gene.
Fig. 3.
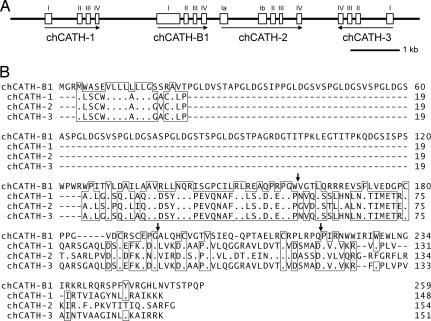
Gene organization and amino acid sequences of chicken cathelicidins. (A) Schematic representation of the gene organization of chicken cathelicidin locus. Exons are indicated by numbered boxes, and introns are indicated by intervening lines. Arrows indicate transcriptional orientation. (Scale bar, 1 kb.) (B) Comparison of the deduced amino acid sequence of chicken cathelicidins. Bars indicate gaps introduced to optimize the alignment, and dots denote identical amino acids with the top sequence. The amino acid residues identical with at least three of the sequences are boxed. Sites of intron interruptions are indicated above the aligned sequences by arrows. Numbers on the right indicate positions in the amino acid sequence. The nucleotide sequence for the locus has been deposited in the DNA Data Bank of Japan/GenBank/European Molecular Biology Laboratory database (AB308318).
In view of its unique sequence and discrete tissue localization, we sought to identify additional chicken cathelicidins with structural and tissue distribution characteristics more similar to known mammalian cathelicidins. A BLAST search of the chicken EST database using the chCATH-B1 cathelin sequence failed to yield candidate cathelicidin clones with high homology (E < 0.4), but, when we queried the then-available chicken EST database with a mammalian cathelin sequence (May 30, 2003), four cathelin domain-containing sequences were identified (BQ484540, BU106516, CB018183, and AJ393748). Notably, this search failed to identify the chCATH-B1 EST clones. Genomic cloning and sequencing indicated that the chCATH-B1 gene is flanked by three additional chicken cathelicidin-like genes, which we provisionally designated chCATH-1 (corresponding to BU106516), chCATH-2 (partly identical with AJ393748 and CB018183), and chCATH-3 (BQ484540) (Fig. 3). These closely linked genes are aligned in the order of chCATH-1, chCATH-B1, chCATH-2, and chCATH-3; chCATH-3 potentially is transcribed in the inverted orientation when compared with the orientation of the others. All of these cathelicidin genes have four exons, except for chCATH-2, which has an additional exon, Ib, that is predicted to be incapable of encoding functional protein products (data not shown). The three chicken cathelicidin relatives of mammalian cathelicidins have been identified recently by other investigators on the basis of sequence similarity and antimicrobial activity (24–26) and have been given the names fowlicidin 1, 2, and 3 (26).
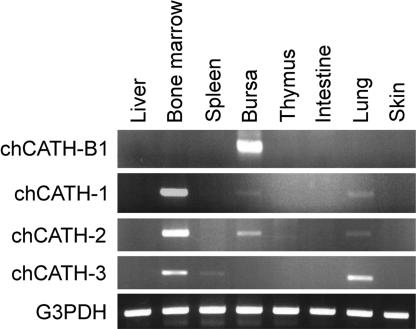
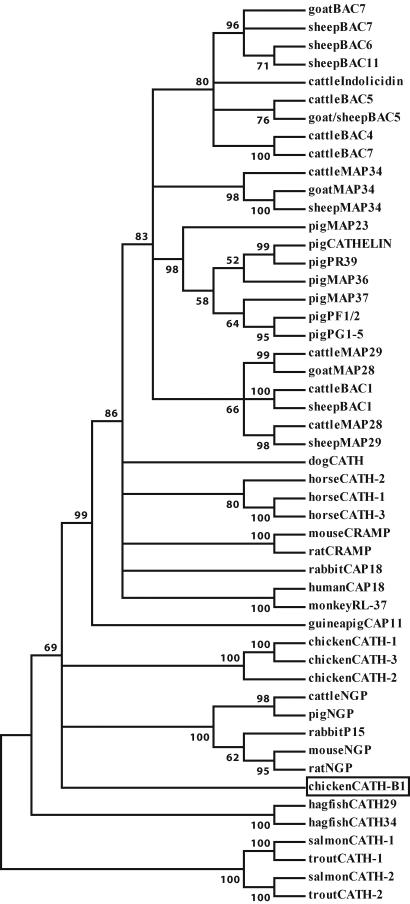
In contrast to the selective bursal expression of chCATH-B1, mRNA analysis indicates a broader tissue distribution for the other three chCATH genes (Fig. 4). chCATH-1 (fowlicidin1) and chCATH-2 (fowlicidin2) are more highly expressed in bone marrow than in the bursa and are demonstrable at relatively low levels in the lung. chCATH-3 (fowlicidin3) transcripts are not found in the bursa but are present in the bone marrow, lung, and spleen. chCATH-1, -2, and -3 encode highly homologous proteins, each of which has the typical structural organization of mammalian cathelicidins, a 17-aa signal peptide, a cathelin region, and a C-terminal peptide (Fig. 3A). The overall sequence identity for the predicted proteins is 98% between chCATH-1 and chCATH-3, 80% between chCATH-1 and chCATH-2, and only 40% between chCATH-B1 and chCATH-1. chCATH-1, -2, and -3 have predicted C-terminal basic mature peptides of 26 aa (pI = 11.7), 32 aa (pI = 12.9), and 29 aa (pI = 12.1), respectively. Both chCATH-1 and -3 have a potential cleavage site (V-R) for elastase (27). A phylogenetic analysis of the relationship between cathelin sequences of chicken cathelicidin with their mammalian, bony fish (28), and hagfish (29) counterparts indicates that chicken cathelicidins, chCATH-1, -2, and -3, fall between the two major clusters of mammalian cathelin regions: classical cathelicidins and neutrophilic granule proteins. chCATH-B1 appears to be an outlier, and the cathelin sequences of fish proteins belong to another branch of the phylogenetic tree (Fig. 5).
Fig. 4.
RT-PCR analysis of cathelicidin gene expression pattern in various chicken tissues using gene-specific primers with G3PDH as a control.
Fig. 5.
Phylogenetic comparison of cathelicidins. The tree was constructed with the protein sequences of the cathelin regions of mammalian, chicken, and hagfish cathelicidins and is shown in the “condensed” form in which nodes with bootstrap values <50 are removed. Chicken CATH-B1 is boxed.
Antimicrobial Activity of chCATH-B1 C-Terminal Peptide.
Because the chCATH-B1 cleavage site is presently unknown, we used a peptide containing the maximal number of C-terminal residues distal to the cathelin region to test for antimicrobial potential. This synthetic chCATH-B1 peptide proved to have bactericidal activity against Escherichia coli and S. aureus at micromolar concentrations (Table 1). The level of antimicrobial activity for the chCATH-B1 peptide was only slightly less than that seen for the PG-1gly19 acid peptide, a broad-spectrum antimicrobial peptide in pigs (12). Comparable antimicrobial activity for these two peptides also was demonstrable for Pseudomonas aeruginosa and Bacillus anthracis (data not shown). The C-terminal chCATH-B1 peptide thus appears to have potent antimicrobial potential.
Table 1.
Antimicrobial activity of cathelicidin peptides
| Bacteria, 2 × 103 cfu | Minimal inhibitory concentration, μM |
||
|---|---|---|---|
| chCATH-B1 | PG-1 | Control | |
| E. coli | 2.5 | 0.63 | >10 |
| S. aureus | 1.25 | 0.63 | >10 |
| P. aeruginosa | 0.63 | 0.63 | >10 |
Minimal inhibitory concentration was determined as the peptide concentration that gave no visible bacterial growth after 24 h of incubation. The PG-1 Gly19 acid peptide was used in this assay.
Cellular Distribution of chCATH-B1 Transcripts and chCATH-B1 Peptide.
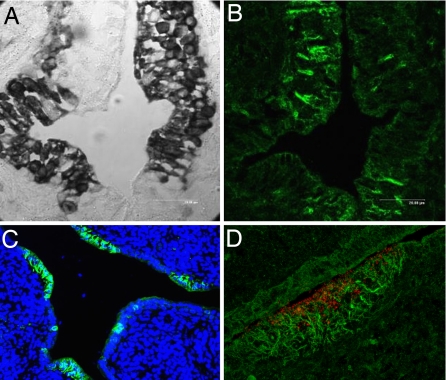
To identify the type(s) of cells that might produce chCATH-B1, we performed in situ hybridization studies using cryosections of different tissues. In these experiments, chCATH-B1 transcripts were detected only in the bursa and not in other chicken tissues. More surprisingly, the chCATH-B1 transcripts were confined to the secretory enterocytes that form the interfollicular bursal epithelium and were not detectable in the neighboring M cells (Fig. 6A). To determine the localization pattern for the antimicrobial peptide, we generated a peptide-specific mAb by fusing a nonproductive plasmacytoma cell line and lymph node cells from mice hyperimmunized with a synthetic C-terminal peptide mimic (see Materials and Methods). When the mAb recognizing the C-terminal chCATH-B1 peptide was used for immunofluorescence analysis of bursal sections, peptide staining was evident in the M cell regions overlying lymphoid follicles and not in the adjacent secretory epithelial cells (Fig. 6 B and C). To refine the chCATH-B1 peptide localization pattern, we introduced fluorescent microbeads into the bursal lumen and later examined bursal cryosections by immunofluorescence microscopy to trace the transcytotic passage of the beads through M cells. This analysis revealed the endocytosed red microbeads in addition to the green anti-BFG7 antibody staining in the M cell region. However, juxtaposition of two fluorochromes could not be verified by this analysis (Fig. 6D). Instead, the anti-chCATH-B1 peptide staining was found to be most heavily concentrated on the filamentous structures that form a basket-like network surrounding the basal and lateral surfaces of bursal M cells (30). These results indicate that, although chCATH-B1 is synthesized by the interfollicular secretory epithelial cells, the mature peptide must be transported to become deposited onto a fibrillar network lining the M cell basolateral surfaces; a likely route of transport is via M cell transcytosis before or after peptide cleavage from the preproprotein.
Fig. 6.
Expression of chCATH-B1 peptide and chCATH-B1 mRNA in the bursa. (A) The bursal section was subjected to in situ hybridization with a cathelin domain anti-sense RNA probe (dark stain). (B) A consecutive bursal section was stained with a FITC-labeled mAb specific for a chCATH-B1 C-terminal peptide. (C) The bursal section was stained with FITC anti-BFG7 and counterstained with the DNA-binding dye DAP1. (D) Red FluoSphere microbeads introduced into bursa duct were taken up by bursal M cells, whereas anti-BFG (FITC) stained a fibrillar network surrounding the base of the M cells.
Discussion
These studies identify a previously unrecognized cathelicidin gene in the chicken, chCATH-B1, that is expressed exclusively by a distinct subpopulation of bursal epithelial cells. In addition to this unique feature, chCATH-B1 has other unusual characteristics in comparison with cathelicidins that have been identified in other species and with the other three cathelicidins in the chicken. Members of the cathelicidin family typically have an N-terminal signal peptide, a highly conserved prosequence known as the cathelin region, and a structurally variable mature peptide at the C terminus that has broad antimicrobial activity. The cathelin region of chCATH-B1 is more distantly related to the mammalian cathelin regions than are the cathelin regions of chCATH-1, -2, and -3, also known as fowlicidins 1, 2, and 3 (26). Although the chCATH-B1 C-terminal peptide differs from the corresponding region in other cathelicidin family members, it has bactericidal activity for Gram-positive and Gram-negative bacteria comparable to that of the representative mature peptide of PG-1gly19 acid pig cathelicidin. A notably atypical feature of chCATH-B1 is that it has nine octamer repeats (PGLDGSXS) located between the N-terminal signal peptide and the cathelin region. This region potentially could affect intracellular preproprotein compartmentalization or stability. A more interesting possibility is raised by a BLAST search performed using the residues in this region; it reveals closest sequence homology with a protein kinase-like protein, 1G5. However, kinase catalytic function has not been demonstrable for 1G5 (31) and remains to be tested for chCATH-B1.
The bursal epithelial cells in which the chCATH-B1 gene is expressed are interfollicular secretory enterocytes. The neighboring M cells that overlie bursal lymphoid follicles, wherein clonally diverse B lymphocytes are generated through gene conversion (32), are devoid of chCATH-B1 transcripts. In contrast, immunofluorescence analysis with a C-terminal peptide-specific antibody indicates that the chCATH-B1 antimicrobial peptide is located in the M cell region, most prominently decorating the fibrillar network surrounding the M cell basolateral surfaces. This remarkable distribution pattern suggests the following hypothetical scenario. The enterocytes around the follicle-associated M cells secrete their chCATH-B1 proprotein product into the bursal lumen, where it becomes available for pinocytosis by neighboring M cells. During its subsequent passage through the transcytotic channels of M cells, the secreted proprotein is cleaved to yield the mature peptide epitope that is detected by our mAb. Microbes entering the body via M cells encounter the antimicrobial peptide within the transcytotic channel or after reaching the fibrillary network surrounding the basolateral surfaces of the M cells or possibly in both locations depending on where the proprotein cleavage occurs. An understanding of the mechanism for processing the antimicrobial peptide is needed to confirm or refute this hypothesis. In this regard, cleavage sites are predictable for chCATH-1, -2, and -3, (V-R for chCATH-1 and -3 and V-L for chCATH-2), whereas a candidate cleavage site is not yet obvious for chCATH-B1. Identification of the cleavage enzyme for chCATH-B1 proprotein therefore is clearly needed to resolve this issue.
In conclusion, chCATH-B1 is an antimicrobial defense element with the functional potential and appropriate cellular localization to guard against invasion by viable microbes via the mucosal M cell gateway. The suggestion of a bovine chCATH-B1 relative may be a helpful clue for addressing the interesting question of whether a similar antimicrobial defense factor is present at the mammalian M cell gateway.
Materials and Methods
Isolation of chCATH-B1 cDNA and Genomic Clones.
The cDNA from chicken bursa was subtracted by cDNA from spleen by using the PCR-select cDNA subtraction kit (BD Clontech, Palo Alto, CA), and the subtracted PCR products were cloned into pCR2.1 plasmid vector (Invitrogen, San Diego, CA). Each subtracted cDNA fragment was screened for bursa-specific expression by Northern blot analysis by using chicken tissues. One of the fragments that specifically hybridized to the bursa RNA was designated BFG7 and was used to screen an oligo(dT)-primed bursa cDNA library constructed in pME18S plasmid vector to isolate a full-length cDNA. The 1.0-kb positive clone was sequenced by the dideoxy-chain termination method with an automatic DNA sequencer (Applied Biosystems, Foster City, CA). Two overlapping genomic clones containing the chCATH-B1 gene were isolated by screening a chicken genomic library (Stratagene, La Jolla, CA) with a full-length chCATH-B1 cDNA as the probe. After restriction enzyme mapping of the clones, subfragments were sequenced.
Sequence and Phylogenetic Analysis.
Sequences were used for BLAST searches against the chicken EST or nonredundant DNA/protein database at the National Center for Biotechnology Information. Protein sequences of the cathelin regions for the presently known cathelicidins were aligned using ClustalX, version 1.8 (33). Phylogenetic and molecular evolutionary analyses were conducted using MEGA, version 3.0 (34) by employing the neighbor joining algorithm with pairwise deletion of gaps, P distance, and 1,000 bootstraps to test the inferred phylogenetic tree.
Northern Blot, Southern Blot, and RT-PCR Analysis.
Total RNA samples (10 μg) extracted from chicken tissue samples and cell lines using the acid guanidium-phenol method (35) were electrophoresed in 1.2 M formaldehyde/1.2% agarose gel before transfer to nylon membranes. These blots were hybridized with a 800-bp fragment of BFG7 cDNA obtained from the subtraction screening. Filters were stripped and rehybridized with a chicken G3PDH. Southern blot hybridization using a full-length or KpnI–NotI fragment of chCATH-B1 cDNA as probes was conducted as described in ref. 35 except that a low stringency washing condition (2× SSC at 42°C) was used for the Zooblot analysis. Expression of chCATH mRNA was also analyzed by RT-PCR by using the following specific primer pairs: 5′-CAGAGGAGGAGGTTGCAGGTGGCC-3′ and 5′-TCATGGCTGAGGAGTGCTGGTGAC-3′ for chCATH-B1; 5′-GGAGCACGGGGTGGGCACGGGGTG-3′ and 5′-CACTTCTTCTTGATCGCCCGGTAG-3′ for chCATH-1; 5′-GCACGGGGTGGGAACAGGGCAAGG-3′ and 5′-CAGCCAAAGCGTGCGCTGCCCTGG-3′ for chCATH-2; 5′-GGGTGGGCACGGGGTGAGGATGCTGAGCTG-3′ and 5′-GGACAGCGGTGACGGTGGCTCTGGGCATGG-3′ for chCATH-3. G3PDH mRNA was also amplified by PCR as a control by using primers 5′-ATTTGGCCGTATTGGCCGCC-3′ and 5′-CATAAGACCCTCCACAATGCC-3′.
Antimicrobial Activity.
Bacterial strains used for antimicrobial testing included E. coli (ATCC25927), S. aureus (ATCC29213), P. aeruginosa (ATCC27853), and B. anthracis (USAMRIID at Fort Detrick). A peptide corresponding to the C-terminal, 40-aa residues of chCATH-B1 was synthesized and used to examine antimicrobial activity (Alpha Diagnostic International, San Antonio, TX). The pig cathelicidin protegrin-1 peptide (PG-1gly19 acid, 19 aa, pI = 10.7) and a terminal deoxynucleotidyl transferase peptide (TdT, 20 aa, pI = 10.0) were synthesized for use as positive and negative controls, respectively. Antimicrobial activity of the test peptides was examined by using a microtiter broth dilution modification of a method recommended by the National Committee of Laboratory Safety and Standards (36). Briefly, a serial dilution of the test peptide was added to the same number of log-phase bacteria in a 96-well polystyrene microtiter plate. Inhibition of growth was assessed after 18 to 24 h of incubation. Minimal inhibitory concentration was determined as the peptide concentration that gave no visible bacterial growth.
Generation of a Monoclonal Anti-chCATH-B1 Antibody.
The predicted mature peptide of the chCATH-B1 C terminus (40 aa) and a conjugate of keyhole limpet hemocyanin with a hydrophilic 15-aa stretch (NGIRKRLRQRSPFYV) of the chCATH-B1 C terminus predicted to be antigenic were used to hyperimmunize mice, from which lymph node cells were fused with the Ag8.653 plasmacytoma cell line (37). Hybridoma culture supernatants were screened by ELISA for reactivity with the individual peptides. A positive hybridoma clone, whose supernatant reacted with the peptide immunogens by ELISA and Western blot analysis, was subcloned, and the mAb product (anti-BFG7, IgM kappa isotype) was purified by using an anti-mouse IgM affinity column.
In Situ Hybridization and Immunohistochemistry.
In situ hybridization was performed according to a method described in ref. 38. Briefly, a chCATH-B1 anti-sense RNA probe was generated by using the cathelin region as the template and was labeled with digoxigenin (RNA Labeling kit; Roche Molecular Biochemicals, Indianapolis, IN). Tissue section slides were hybridized and developed with alkaline phosphatase-conjugated anti-digoxigenin followed by exposure to the nitroblue tetrazolium substrate (Roche Molecular Biochemicals). Immunohistochemical analysis of BFG7 antibody staining was also performed on serial cryosections as described in ref. 39. In some experiments, red FluoSphere microbeads [0.2 μm, 2% solid, 1:4 dilution (Molecular Probes, Eugene, OR)] were introduced into the bursa duct through a small polyethylene tube. The chicks were killed 1 h later, and bursal cryosections were prepared for staining by immunofluorescence (39) with anti-BFG7 antibody.
Supplementary Material
Acknowledgments
We thank Yolanda Hartman for technical assistance, Drs. Pat Bucy and Dale Bockman for very helpful advice, and Matthew Alder for help with the phylogenetic analysis. This work was supported in part by National Institutes of Health Grants AI139561 and AI039816 and U.S. Department of Agriculture Grant 95-37204-2143.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan database, www.ddbj.nig.ac.jp (accession nos. AB307733 and AB308318 for chCATH-B1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707037104/DC1.
References
- 1.Bockman DE, Cooper MD. Am J Anat. 1973;136:455–477. doi: 10.1002/aja.1001360406. [DOI] [PubMed] [Google Scholar]
- 2.Owen RL. Semin Immunol. 1999;11:157–163. doi: 10.1006/smim.1999.0171. [DOI] [PubMed] [Google Scholar]
- 3.Gebert A, Pabst R. Semin Immunol. 1999;11:165–170. doi: 10.1006/smim.1999.0172. [DOI] [PubMed] [Google Scholar]
- 4.Kraehenbuhl JP, Neutra MR. Annu Rev Cell Dev Biol. 2000;16:301–332. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 5.Neutra MR. Am J Physiol. 1998;274:G785–G791. doi: 10.1152/ajpgi.1998.274.5.G785. [DOI] [PubMed] [Google Scholar]
- 6.Sansonetti PJ, Phalipon A. Semin Immunol. 1999;11:193–203. doi: 10.1006/smim.1999.0175. [DOI] [PubMed] [Google Scholar]
- 7.Lehrer RI, Ganz T. Curr Opin Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 8.Hancock RE, Scott MG. Proc Natl Acad Sci USA. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 10.Gallo RL, Murakami M, Ohtake T, Zaiou M. J Allergy Clin Immunol. 2002;110:823–831. doi: 10.1067/mai.2002.129801. [DOI] [PubMed] [Google Scholar]
- 11.Lehrer RI, Lichtenstein AK, Ganz T. Annu Rev Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 12.Gennaro R, Zanetti M. Biopolymers. 2000;55:31–49. doi: 10.1002/1097-0282(2000)55:1<31::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Ganz T. Science. 2002;298:977–979. doi: 10.1126/science.1078708. [DOI] [PubMed] [Google Scholar]
- 14.Cowland JB, Johnsen AH, Borregaard N. FEBS Lett. 1995;368:173–176. doi: 10.1016/0014-5793(95)00634-l. [DOI] [PubMed] [Google Scholar]
- 15.Agerberth B, Gunne H, Odeberg J, Kogner P, Boman HG, Gudmundsson GH. Proc Natl Acad Sci USA. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, Kiessling R, Jornvall H, Wigzell H, Gudmundsson GH. Blood. 2000;96:3086–3093. [PubMed] [Google Scholar]
- 17.Bals R, Wang X, Zasloff M, Wilson JM. Proc Natl Acad Sci USA. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. J Invest Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 19.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DY. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 20.Zasloff M. Nat Med. 2006;12:607–608. doi: 10.1038/nm0606-607. [DOI] [PubMed] [Google Scholar]
- 21.Goitsuka R, Fujimura Y, Mamada H, Umeda A, Morimura T, Uetsuka K, Doi K, Tsuji S, Kitamura D. J Immunol. 1998;161:5804–5808. [PubMed] [Google Scholar]
- 22.Chen CH, Six A, Kubota T, Tsuji S, Kong FK, Gobel TW, Cooper MD. Curr Top Microbiol Immunol. 1996;212:37–53. doi: 10.1007/978-3-642-80057-3_5. [DOI] [PubMed] [Google Scholar]
- 23.Tossi A, Sandri L, Giangaspero A. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 24.Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT, Fares MA, Mulcahy G, O'Farrelly C. Immunogenetics. 2004;56:170–177. doi: 10.1007/s00251-004-0675-0. [DOI] [PubMed] [Google Scholar]
- 25.van Dijk A, Veldhuizen EJ, van Asten AJ, Haagsman HP. Vet Immunol Immunopathol. 2005;106:321–327. doi: 10.1016/j.vetimm.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Cai Y, Bommineni YR, Fernando SC, Prakash O, Gilliland SE, Zhang G. J Biol Chem. 2006;281:2858–2867. doi: 10.1074/jbc.M507180200. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez JF, Hoh F, Strub MP, Aumelas A, Dumas C. Structure (London) 2002;10:1363–1370. doi: 10.1016/s0969-2126(02)00859-6. [DOI] [PubMed] [Google Scholar]
- 28.Chang CI, Zhang YA, Zou J, Nie P, Secombes CJ. Antimicrob Agents Chemother. 2006;50:185–195. doi: 10.1128/AAC.50.1.185-195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzzell T, Stolzenberg ED, Shinnar AE, Zasloff M. Peptides. 2003;24:1655–1667. doi: 10.1016/j.peptides.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Davenport WD, Jr, Allen ER. Anat Record. 1995;241:155–162. doi: 10.1002/ar.1092410203. [DOI] [PubMed] [Google Scholar]
- 31.Godbout M, Erlander MG, Hasel KW, Danielson PE, Wong KK, Battenburg ELF, Foye PD, Bloom FE, Sutcliffe JG. J Neurosci. 1993;14:1–13. doi: 10.1523/JNEUROSCI.14-01-00001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynaud CA, Bertocci B, Dahan A, Weill JC. Adv Immunol. 1994;57:353–378. doi: 10.1016/s0065-2776(08)60676-8. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 35.Goitsuka R, Chen CH, Cooper MD. J Immunol. 1997;159:3126–3132. [PubMed] [Google Scholar]
- 36.Wu M, Hancock RE. J Biol Chem. 1999;274:29–35. doi: 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- 37.Chen CL, Ager LL, Gartland GL, Cooper MD. J Exp Med. 1986;164:375–380. doi: 10.1084/jem.164.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panoskaltsis-Mortari A, Bucy RP. BioTechniques. 1995;18:300–307. [PubMed] [Google Scholar]
- 39.Morimura T, Miyatani S, Kitamura D, Goitsuka R. J Immunol. 2001;166:3277–3283. doi: 10.4049/jimmunol.166.5.3277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.