Abstract
The exosome is a 3′ to 5′ exoribonuclease central to many cellular processes, including mRNA decay. Liu et al. (2006) now present the biochemical reconstitution and crystal structure of the eukaryotic exosome. This remarkable achievement provides key insights into the composition and assembly of the human and yeast exosomes, revealing functions of individual subunits.
In eukaryotes, the exosome is an ordered conglomerate of 9–11 proteins that cleaves RNA molecules one base at a time in a 3′ to 5′ direction by removing the 3′-most phosphodiester bond. The exosome plays a central role in mRNA decay, mRNA surveillance, and in the processing of certain ribosomal and small nucleolar RNAs (reviewed in Baker and Parker [2004], Houseley et al. [2006], and Raijmakers et al. [2004]). More recently, the exosome has also been shown to degrade 5′ intermediate fragments produced in RNA interference (Orban and Izaurralde, 2005). Despite the importance and known physical composition of the exosome, it has been unclear how its enzymatic reaction is carried out and how the individual subunits contribute to its function.
In this issue, Liu et al. (2006) report the biochemical reconstitution of the yeast and human exosome complexes in vitro and provide the first structural view of the ninesubunit human exosome complex (hExo9) at 3.35 Å. Although previous reports suggested that several subunits of the yeast exosome possess phosphorolytic activity, the authors demonstrate that the individual and collective components of the reconstituted core of the yeast exosome (yExo9) lack intrinsic exonuclease activity. Instead, exonuclease activity is conferred by the hydrolytic activity of Rrp44p, either individually or as the tenth subunit of the yeast exosome complex (yExo10). In contrast to the yeast exosome, Liu et al. (2006) found that the hExo9 human exosome complex contains phosphorolytic activity, and this activity is due to the hRrp41/hRrp45 dimer. The remaining RNase phosphorolytic (PH) domain-containing subunits lack intrinsic exonuclease activity, both individually and within the context of the exosome. The structure also reveals the macromolecular composition of the human exosome and the proper architectural assembly mediated by protein-protein interactions.
The basic ring-shaped architecture of the RNA degradation machinery is structurally conserved throughout bacteria, archaea, yeast, and humans (Hernandez et al., 2006; Lorentzen et al., 2005). The archaeal exosome is a six-membered ring structure consisting of a trimer of Rrp41/Rrp42 capped on one side by a homotrimer of Rrp4 or Csl4 (Lorentzen et al., 2005). Its active sites are located near the opening of the ring structure on the opposite of the cap. In contrast, the human exosome uses six different polypeptides to form the ring structure, namely hRrp41/hRrp45, hRrp46/hRrp43, and Mtr3/hRrp42 (Figure 1, top). Curiously, the six-ring exosome does not form a stable structure in the absence of Rrp4, Csl4, and Rrp40, which form the top cap of the exosome to constitute the stable hExo9 unit. Despite the conservation in structure, biochemical analysis shows that the enzymatic properties among the exosomes are variable. For example, the7/25/2007 archaeal exosome contains three phosphorolytic active sites per complex, whereas the human exosome contains one active site. The yeast exosome also possesses one active site, although it is provided by the hydrolytic activity of the tenth subunit, Rrp44p, which is specific to yeast.
Figure 1. Structural Composition of the Exosome.
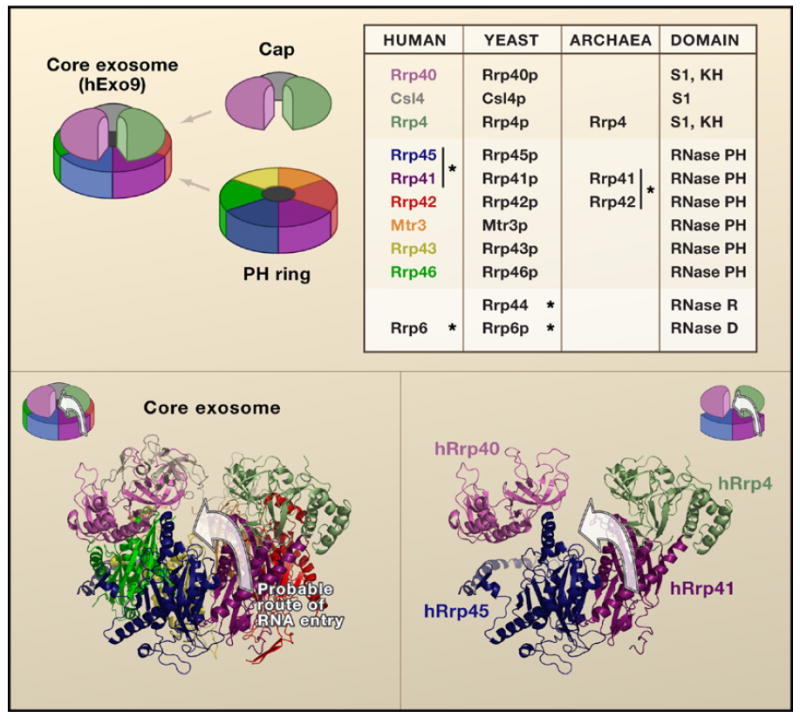
(Top) A schematic shows the nine subunit human exosome with the corresponding subunits in the table. The asterisk designates experimentally validated catalytic subunits. The human subunits are color coded. (Bottom) The structure of the exosome reveals the cleft between hRrp40 and hRrp4 within the context of the hExo9 complex crystal structure and schematic (left) and within the isolated subunits hRrp41, hRrp45, hRrp4, and hRrp40 (right). The arrow denotes the putative path of an RNA extending into the hRrp41/hRrp45 heterodimer through the cleft.
Previous reports have shown that nine nucleotides is the minimal length necessary for the 3′ end of the RNA substrate to enter from the cap side of the exosome and extrude through the central cavity to reach the active site of the archaeal exosome (Lorentzen and Conti, 2006). Liu et al. (2006) find that a minimum of ten nucleotides is required to detect human exosome activity. Curiously, the hRrp41/Rrp45 dimer in isolation also has the same length requirements. Therefore, the length of the central cavity does not necessarily determine the minimal size for a substrate for the human exosome. Instead, it is likely that an RNA needs to be at least ten nucleotides to form a stable association with the hRrp41/hRrp45 heterodimer.
Examination of the hExo9 structure reveals a cleft between the hRrp4 and hRrp40 cap proteins exposing the hRrp41/hRrp45 heterodimer (Figure 1). It is possible that an RNA enters the exosome through a direct association with the hRrp41/hRrp45 component rather than having to access the apex of the cap first (Figure 1, bottom). This raises interesting questions as to why the cap proteins contain RNA binding domains. How does the RNA binding potential of the exosome contribute to substrate specificity and regulation, and do the essential auxiliary proteins (Ski7 and Ski2/3/8 complex) regulate access of the RNA through the hRrp4-hRrp40 crevice? Mutational analysis of the protein subunits and cocrystal structures of the hExo9 with RNA substrate will begin addressing these questions.
It is also an open question as to why the hRrp41/hRrp45 heterodimer is within a larger complex when it can degrade the RNA by itself. In this regard the experiments by Liu et al. (2006) with the yeast counterpart are informative and suggest a regulatory mechanism. Although RNA is more efficiently hydrolyzed by the yeast Rrp44p catalytic subunit in isolation, it has reduced hydrolytic activity within the yExo10 complex, thus indicating that its catalytic activity is either less accessible or is allosterically regulated within the exosome complex. Interestingly, the distributive exonuclease activity of the nuclear yExo11 is broadly comparable to that of the nuclear restricted Rrp6p alone, suggesting that the active site of Rrp6p in the yExo11 complex is readily accessible as it is in the monomer.
The 3′ to 5′ model for mRNA decay predicts that the exosome degrades mRNAs that retain approximately ten adenosines following deadenylation. This size is consistent with the minimum size for an exosome substrate (Liu et al. 2006). It is somewhat surprising that the hExo9 and yExo10 weakly hydrolyzed RNA substrates with a 20 nucleotide polyadenosine tail, indicating that additional components facilitate exosome activity. These could be auxiliary proteins or might be cis elements (as is the case of AU-rich sequences that can be bound by hRrp45 [PM/SCL-75; Mukherjee et al. (2002)]). These results suggest that structural features of RNA, which can vary widely, might influence the way in which the exosome associates with its substrates. Recent studies indicate that an additional priming step promotes the recruitment of the exosome. The TRAMP complex (through its polyadenosine polymerase activity) is proposed to tag an RNA with a stretch of adenosines prior to exosome-mediated degradation in the nucleus (reviewed in Houseley et al. [2006]). The preferential hydrolysis of adenosine by Rrp6p and its nuclear localization are consistent with a role in the degradation of TRAMP complextagged nuclear RNAs. In contrast, the cytoplasmic hExo9 and yExo10, which lack Rrp6, function less efficiently on poly(A) RNA, thus ensuring the integrity and regulated deadenylation in the cytoplasm.
The structural elucidation and biochemical reconstitution of the human core exosome complex is a remarkable feat that reveals the protein machinery that is at the heart of multiple RNA processes. Likewise, the recent cocrystal structure of the Rrp44p family member, RNase II (Frazao et al., 2006), may also provide insights into the catalytic component of the yeast exosome. The reconstituted human exosome complex provides new opportunities to study the functional regulation of the exosome and may yield further insights into the role of noncatalytic subunits.
References
- Baker KE, Parker R. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Cell. 2006 doi: 10.1016/j.cell.2006.10.037. this issue. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. Cell. 2006;125:651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R, Schilders G, Pruijn GJ. Eur J Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]


