Abstract
Oligodendrocyte progenitor (OP) cell differentiation is a critical process of developmental myelination, tumor formation, and remyelination in the CNS. Activation of the fibroblast growth factor 2 (FGF2) or notch pathway can inhibit differentiation of OP cells. The current study examines the interaction of FGF2 and notch signaling components in regulating OP differentiation. Cultured neonatal rat brain OP cells were used for transfection-based promoter assays and for infection with retroviruses expressing a GFP reporter to monitor OP differentiation into oligodendrocytes or astrocytes. FGF2 treatment resulted in a 4-fold increase of transcriptional activity from the promoter region of Hes5, a notch pathway target gene. FGF2 inhibition of OP differentiation into oligodendrocytes was perturbed by retroviral expression of a dominant negative construct for mastermind-like 1, which is an important co-activator of transcription for notch target genes. OP differentiation into oligodendrocytes was reduced by co-culture with fibroblasts expressing Jagged1, a ligand for notch receptors. This Jagged1 inhibition of OP differentiation was not altered by retroviral expression of a dominant negative FGF receptor construct. Constitutive activation of notch signaling, by retroviral expression of the Notch1 intracellular domain, greatly reduced OP differentiation into either oligodendrocytes or astrocytes and did not require FGF2 signaling. These findings indicate that inhibition of OP differentiation through the Notch1 pathway was not influenced by FGF2 signaling. However, FGF2 signaling may interact with down stream components of the notch signaling pathway, including mastermind-like1 and Hes5, to inhibit OP differentiation into oligodendrocytes.
Keywords: oligodendrocyte, astrocyte, Notch1, FGF2, mastermind, jagged
Introduction
Differentiation of neural stem cells and progenitor cells must be regulated precisely to generate appropriate numbers of distinct cell populations during CNS development and repair, while aberrant differentiation can result in dysplasia or tumor formation. Differentiation of oligodendrocyte progenitor (OP) cells into myelinating oligodendrocytes is regulated by a balance of positive and negative signals that coordinate myelination relative to axonal maturation and integrity. Inhibition of OP differentiation is necessary for spatial and temporal regulation of oligodendrogenesis during development but may also be detrimental for efficient remyelination and so has emerged as an area of interest for modulating endogenous cell repair of demyelinating diseases. To better understand how inhibition of OP differentiation is regulated, the current study examines the interaction of signaling components of two inhibitors of OP differentiation, fibroblast growth factor 2 (FGF2) and Notch1.
Our work in FGF2 null mice demonstrated that FGF2 inhibits differentiation of oligodendrocyte progenitor (OP) cells into myelinating oligodendrocytes during both development and remyelination [3, 14, 15]. Importantly, FGF2 limits effective remyelination following chronic demyelination [2]. FGF2 inhibition of OP differentiation in vivo occurs through direct activation of fibroblast growth factor receptor (FGFR) signaling in oligodendrocyte lineage cells and may predominantly occur through FGFR1 [25].
Notch1 potently inhibits OP differentiation and myelination during CNS development [21]. Mice with Notch1 inactivation in oligodendrocyte lineage cells have abnormalities of the spatial and temporal regulation of OP differentiation resulting in accelerated and/or ectopic myelination [6, 7]. Pharmacological disruption of notch signaling promoted remyelination in mice with demyelination from experimental allergic encephalomyelitis [9]. Notch1 receptors on OP cells may be activated by cell-cell interaction with astrocytes expressing Jagged1 ligand as indicated from the pattern of expression of these notch signaling components during development and in non-remyelinating areas of multiple sclerosis lesions [8, 21].
FGF2 and Notch1 signaling may interact in regulating neural cell development. FGF2 can induce Notch1 expression in neuroepithelial precursor cells and in a cell line with an immature oligodendrocyte phenotype [4, 5]. Conversely, Notch1 signaling can increase responsiveness to FGF2 in telecephalic progenitors [24]. To more specifically examine the interaction of FGF2 signaling with the notch pathway in OP cells, we used the well characterized in vitro system of neonatal rat OP cell differentiation to modulate ligand and receptor activity of both pathways as well as down steam components of the notch signaling cascade. Jagged1 activation of classical Notch1 signaling is transmitted by means of cleavage of the Notch1 intracellular domain (Notch1IC) which then translocates to the nucleus to bind CSL (CBF-1/RBP-jk, Su(H), Lag-1), an interaction that is stabilized by binding of mastermind-like 1 (Maml1) co-activator of transcription. Targets of notch transcriptional regulation include the basic helix-loop-helix transcription factors, Hes1 and Hes5. Hes5 is an important inhibitor of myelin gene expression that is progressively down regulated during OP differentiation [11, 12, 21] yet elevated in demyelination associated with failed remyelination [8]. Our analysis of FGF2 and notch pathway interactions in regulating OP differentiation shows that the Notch1 pathway was not influenced by FGF2 signaling. In contrast, FGF2 signaling may interact with down stream components of the notch signaling pathway, including Maml1 and Hes5, to inhibit OP differentiation into oligodendrocytes.
Materials and Methods
OP cultures, retroviral vectors, infections, and immunocytochemistry
Rats were handled as approved by the USUHS Institutional Animal Care and Use Committee to promote health and minimize pain or discomfort. OP cultures were prepared from neonatal rat brains [1] and grown in defined medium [DMEM containing high insulin (5 μg/ml), T3 (Triiodothyronine; 30 nM), and 0.5% fetal bovine serum (FBS) (all from Sigma, St. Louis, MO)] to induce differentiation into mature oligodendrocytes. The defined medium was supplemented with mitogens, platelet-derived growth factor AA (PDGF; 10 ng/ml) and FGF2 (10 ng/ml)(both from R&D Systems, Minneapolis, MN) to promote OP proliferation and prevent differentiation. To induce OP differentiation into astrocytes, cultures were grown in DMEM with 10% FBS.
Retrovirus generation and infections were performed as previously described [16, 25]. Dr. Kitamura (Tokyo University) provided the PMXs replication-incompetent retroviral vector containing a multiple cloning site followed by an internal ribosome entry site (IRES) sequence and enhanced GFP [10]. In this vector, the inserted gene is transcribed first followed by GFP in a single transcript so that GFP expression can estimate viral transduction of both genes. FGFRdn plasmid, (PMXs with a dominant negative form of FGFR2 that heterodimerizes with all FGFR isoforms) has been characterized previously [25]. Notch1IC, from Dr. Nye (Pharmacia; Kalamazoo, MI) [17], was inserted into PMX to create Notch1IC-PMXs. MSCV-Maml1dn plasmid, containing a dominant negative form of Maml1, was a gift from Dr. Aster (Brigham and Women's Hospital; Boston, MA) [22]. Maml1dn relevance in OP cells was demonstrated by immunostaining for endogenous Maml1 in non-infected OP cultures using anti-Maml1 polyclonal antibody (Chemicon). OP cultures were incubated with retrovirus for 6 hrs (approximately 3000 CFU/well which typically resulted in GFP expression among the majority of cells). The cells were maintained for 2 days in defined medium with PDGF and FGF2, and refed medium to promote differentiation (see above) over 3 days prior to fixation and immunostaining [16, 25].
For co-culture experiments, OP cells were infected with PMXs (control virus) or FGFRdn retrovirus, as above. The following day, control fibroblasts (parental L cell line) or Jagged1-expressing fibroblasts were added at a ratio of 1.6:1 (fibroblasts:OP) [19]. Co-cultures were then grown for 72 hrs in defined medium without mitogens or in DMEM with 10% FBS.
Cultures were immunostained for O1 or glial fibrillary acidic protein (GFAP) to monitor differentiation of infected OP cells into oligodendrocytes or astrocytes, respectively [16]. To estimate cell proliferation, bromodeoxyuridine (BrdU; 2 μM; Sigma) was added for 20 hrs of culture prior to fixing and immunostaining cells to detect BrdU incorporation [16, 25]. The Chi-square test with the Mantel-Haenzel subgroup analysis was used to compare immunostaining results among retrovirally labeled cell populations.
Transcription activity assays
Plasmids with the firefly luciferase gene under control of the promoter for Hes1 (pHes1; −467 to +46) and Hes5 (pHes5; −800 to +73) were gifts from Dr. Kageyama (Kyoto University) [18]. OP cells were co-transfected with pHes1 or pHes5 along with pRL-CMV (SV40 promoter drives expression of Renilla luciferase; Promega) to estimate transfection efficiency [16]. After incubation overnight (16 hrs), the cells were then transferred to defined medium with or without FGF2 (10 ng/ml) and grown for 10 hrs. Transcriptional activity was evaluated using the Dual-Luciferase Reporter System (Promega). The relative luciferase units (RLU) were calculated from firefly luciferase values divided by the Renilla luciferase values to normalize for transfection efficiency [16]. The Student's t-test was used to compare relative luciferase values.
Results
Infection of OP cells with replication-incompetent retroviral vectors was used to directly monitor OP differentiation into oligodendrocytes (Figure 1). This approach targets integration of genes-of-interest, including a GFP reporter, to cycling OP cells within the cultures and those genes then continue to be expressed by the initial cell and its progeny. Immunostaining for O1, an oligodendrocyte marker, in combination with GFP detection then demonstrated differentiation of infected OP cells into mature oligodendrocytes (Figure 1 A, C). To examine the potential interaction between FGF2 and notch signaling components in OP differentiation, truncated gene constructs were inserted into the retroviral vectors to modulate activity of relevant receptors or downstream signaling components. Each vector included a GFP reporter to monitor viral transduction. FGFR signaling was attenuated by expression of a dominant negative form of FGFR (FGFRdn; Figure 1 A) [25]. Activity of the notch pathway was potentiated by expression of the Notch1 intracellular domain (Notch1IC; Figure 1B) which is constitutively active [17], or alternatively, was perturbed by expression of a dominant negative form of Maml1 (Maml1dn; Figure 1 C) [22]. In addition, each pathway was tested in the presence or absence of relevant ligands, either by addition of exogenous FGF2 to the defined medium and/or by co-culture of OP cells with fibroblasts expressing the Jagged1 ligand for Notch1.
Figure 1. OP differentiation into oligodendrocytes.
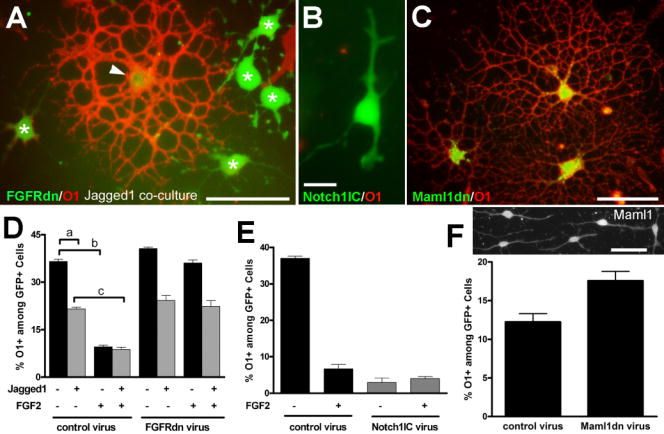
OP cells were infected with retroviral constructs to modulate receptor and ligand elements of the FGF2 and notch signaling pathways. OP differentiation into oligodendrocytes was monitored by O1 immunostaining. A: OP cells infected with retrovirus expressing a dominant negative form of fibroblast growth factor receptor (FGFRdn) and green fluorescent protein reporter (GFP; green). After 3 days of co-culture with Jagged 1 expressing fibroblast cells (not fluorescently labeled) many of the retrovirally labeled cells have immature characteristics (green; asterisks) in contrast to the differentiated oligodendrocyte (arrowhead) with highly branched processes and immunolabeling for O1 (red). Scale bar = 50 μm. B: OP cell infected with retrovirus expressing the intracellular domain of Notch 1 (Notch1IC), a constitutively active form, and GFP (B, green) has an immature morphology and lacks O1 immunoreactivity (red absent). Scale bar = 15 μm. C: OP cells infected with a retrovirus expressing a dominant negative form of mastermind-like1 (Maml1dn), a co-activator of notch target genes, and GFP (green) exhibit elaborate processes and O1 immunoreactivity (red), indicative of differentiation into mature oligodendrocytes. Scale bar = 50 μm. D, E, F: Quantitation of the proportion of O1 immunolabeled cells among retrovirally infected cells identified by expression of the GFP reporter. D: OP cells infected with retrovirus and then co-cultured with a fibroblast line expressing Jagged1 (+) or the parental L cell line (−). Cells were grown in defined medium with (+) or without (−) FGF2. Co-culture of OP cells with fibroblasts expressing Jagged1 decreased the proportion of OP cells that developed O1 immunoreactivity (a; p < 0.01). FGF2 severely reduced O1 acquisition (b; p < 0.001), which was abrogated by FGFRdn transduction. FGF2 further decreased O1 acquisition as compared to co-culture with Jagged1 expressing cells (c; p < 0.001). E: Notch1IC transduction strongly inhibited OP differentiation (p<0.001), compared with control retrovirus, and was not further reduced with FGF2 treatment. F: Upper panel shows immunostaining for endogenous Maml1 expression in OP cells. Scale bar = 50 μm. Lower panel shows that FGF2 inhibition of OP differentiation was significantly perturbed by Maml1dn transduction (p < 0.05 vs control virus). Values (mean ± standard error of the proportion) are from three independent experiments with at least 300 GFP+ cells counted for each condition.
OP differentiation and acquisition of O1 immunostaining among GFP+ cells was reduced by co-culture with Jagged1-expressing fibroblasts (Figure 1 D, a). This reduction did not require FGF2 treatment and was not altered by expression of FGFRdn to disrupt FGFR signaling. Therefore, although FGF2 has been shown to increase Notch1 expression [4, 5], Notch1 expression may already be sufficient so that FGF2 treatment does not regulate Jagged1 inhibition of OP differentiation in this context. Conversely, FGF2 treatment markedly reduced O1 acquisition among GFP+ cells in OP cell cultures or among OP cells co-cultured with Jagged1-expressing fibroblasts (Figure 1 D, b and c; 1 E). This FGF2 inhibition of O1 acquisition was partially perturbed by expression of Maml1dn (Figure 1 F), indicating an interaction of FGF2 signaling with this co-activator of transcription of notch target genes. The most potent inhibition of OP differentiation into oligodendrocytes was observed with OP expression of Notch1IC to strongly activate the notch pathway (Figure 1 E). These results using O1 acquisition to assess OP differentiation into oligodenddrocytes are supported by the characteristic cell morphologies (Figure 1) and similar results in preliminary studies assessing differentiation with in situ hybridization for proteolipid protein mRNA (data not shown).
To further examine whether FGF2 may interact with downstream components of the notch pathway to inhibit OP differentiation, we assessed transcription of notch target genes with and without FGF2 treatment. Hes1 and Hes5 were chosen as relevant Notch1 target genes involved in glial differentiation. Hes1 activity favors differentiation along the astrocytic pathway over the oligodendrocytic pathway [23]. Hes5 inhibits differentiation along both astrocytic and oligodendrocytic pathways [23]. Cultured OP cells were transfected with the promoter region for Hes1 or Hes5 driving expression of luciferase as a measure of transcriptional activity (Figure 2). FGF2 treatment induced an approximately 4-fold increase in relative transcription from the Hes5 promoter but did not alter Hes1 transcriptional activity.
Figure 2. FGF2 enhances Hes5 promoter activity in OP cells.
OP cells were transfected using plasmids with luciferase reporter transcription driven by the promoter region for Hes1 or Hes5. On the following day, the cultures were transferred to defined medium with (+) or without (−) FGF2 for 10 hrs and transcription activity was then measured as relative luciferase units (RLU). FGF2 treatment increased the Hes5 promoter activity approximately 4-fold (*p < 0.001) but did not alter Hes1 promoter (p > 0.05). Values (mean ± standard error of the mean) are from duplicate wells of three independent experiments.
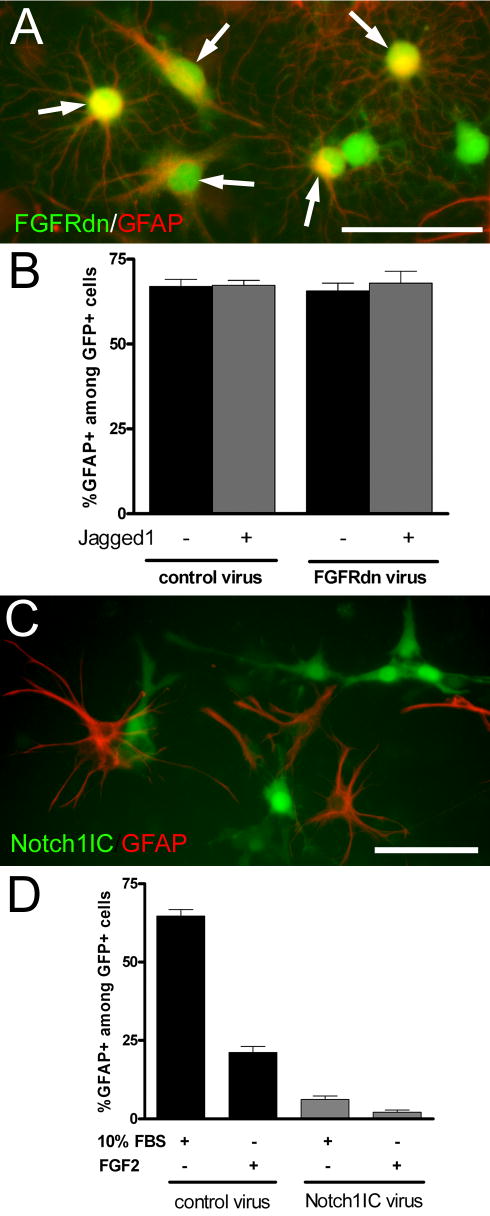
OP cells are bipotential with the ability to differentiate into oligodendrocytes in defined medium, as in the above conditions, or into astrocytes in medium containing 10% FBS [16, 20]. Therefore, we used retroviral infections to monitor OP differentiation into astrocytes relative to modulation of relevant FGF2 and notch signaling components. Astrocytic differentiation of OP cells was indicated by cytoskeletal immunoreactivity for GFAP [20] in cells expressing the GFP retroviral reporter. In contrast to OP differentiation into oligodendrocytes (Figure 1 A, D), co-culture with Jagged1-expressing fibroblasts did not alter OP differentiation into astrocytes (Figure 3 A, B). We next compared OP differentiation into astrocytes in conditions parallel to those of Figure 1E to examine the effect of FGF2 and the Notch1IC domain (Figure 3 C, D). Growth in medium with 10% FBS served as a positive control and resulted in detectable GFAP immunostaining in the majority of GFP+ cells infected with the control retrovirus. In defined medium with FGF2 a much smaller proportion of cells infected with the control retrovirus had detectable GFAP, indicating that FGF2 did not shift OP differentiation toward an astrocytic fate. Expression of the Notch1IC domain dramatically reduced OP differentiation into astrocytes among GFP+ cells grown in medium with 10% FBS. Notch1IC expression also resulted in a very low frequency of GFAP acquisition among GFP+ cells grown in defined medium with FGF2. Therefore, Notch1IC expression potently blocks OP differentiation along both lineages (Figures 1, 3).
Figure 3. OP differentiation into astrocytes.
OP cells were infected with retroviral constructs to modulate receptor and ligand elements of the FGF2 and notch signaling pathways (see Figure 1 legend). OP differentiation into astrocytes was detected by GFAP immunostaining. A, B: OP cells were infected with control retrovirus expressing only the GFP reporter or FGFRdn virus. Fibroblasts expressing Jagged1 or the parental fibroblast line were then added to establish co-cultures with the OP cells. Co-cultures were maintained in medium containing 10% fetal bovine serum (FBS) for 3 days to induce astrocytic differentiation. A: OP cells infected with FGFRdn expressing retrovirus (green) and immunolabeled for GFAP (red; arrows) indicative of differentiation into astrocytes during co-culture with fibroblasts (not fluorescently labeled) expressing Jagged 1. Scale bar = 50 microns. B: Quantitation of GFAP immunolabeling among retrovirally infected cells expressing GFP. OP differentiation into astrocytes was not altered by co-culture with fibroblasts expressing Jagged1, as compared to the parental fibroblast line. In addition, FGFRdn transduction did not alter acquisition of GFAP among the infected cells co-cultured with either fibroblast line. C, D: OP cells infected with the Notch1IC retrovirus grown in medium with 10% FBS or in defined medium with (+) or without (−) FGF2. C: OP cells infected with Notch1IC retrovirus expressing GFP (green) with immunostaining for GFAP (red) following growth in defined medium with FGF2. Scale bar = 50 microns. D: Quantitation of GFAP immunolabeling among retrovirally infected cells expressing GFP. The proportion of infected cells that acquire GFAP immunoreactivity is significantly decreased with Notch1IC transduction (p < 0.001 in DMEM with 10% FBS and p < 0.001 in defined medium with FGF2). B, D: Values (mean ± standard error of the proportion) are from three independent experiments with at least 300 GFP+ cells counted for each condition.
To determine whether the FGF2 signaling with notch pathway components was restricted to OP differentiation as distinct from OP proliferation, BrdU was added to cultures of infected OP cells. BrdU incorporation was similar among GFP+ cells infected with either control or Notch1IC retroviruses with cultures grown in defined medium (control 19.9 ± 0.9%, Notch1IC 21.8 ± 1.7%). Since FGF2 is a potent mitogen for OP cells, BrdU incorporation was greatly increased with FGF2 treatment but was not significantly different among cells infected with the control retrovirus (51.4 ± 6.4%) or the Notch1IC retrovirus (46.9 ± 3.7%). Furthermore, BrdU incorporation was also not significantly altered by Notch1IC transduction using PDGF as the mitogen (control 54.7 ± 5.1%; Notch1IC 49.3 ± 1.9%). Therefore, FGF2 may interact with notch pathway components to have a similar effect on OP differentiation but FGF2 and notch pathways have distinct effects on OP proliferation.
Discussion
The current results demonstrate that FGF2 signaling can interact with down stream components of the notch pathway, including mastermind-like1 and Hes5, to inhibit OP differentiation into oligodendrocytes. This interaction of FGF2 with downstream notch pathway components may suggest a point of dissociation of FGF2 inhibition of OP differentiation as distinct from induction of OP proliferation. Clearly, simply preventing OP differentiation is not sufficient to result in high levels of OP proliferation since Notch1IC expression potently inhibited OP differentiation but did not alter OP proliferation. Furthermore, previously proposed mechanisms of FGF2 and notch signaling synergism in neural stem cells [24] may not apply directly to interactions regulating of OP cell responses.
Expression of FGF2 and notch pathway components in the developing and regenerating CNS are consistent with a potential signaling interaction in vivo. Notch1 receptor is expressed on OP cells while Jagged1 ligand is found on axons prior to myelination [21]. FGF2 expression increases postnatally in the CNS and can inhibit OP differentiation through activation of FGFR1 [14, 25]. Similarly, Notch1 and FGFR1 expression is increased in demyelinated lesions along with Jagged1 and FGF2 [2, 8, 13, 15]. Notch1 and Jagged1 expression in multiple sclerosis lesions correlated with areas of poor remyelination consistent with inhibition of OP cells into remyelinating oligodendrocytes [8]. Furthermore, pharmacological inhibition of notch signaling may enhance remyelination [9]. Our analysis of FGF2 null mice indicated improved remyelination associated with removal of FGF2 inhibition of OP differentiation, while we did not find evidence of an FGF2 effect on OP proliferation in vivo [2, 3, 15].
FGF2 interaction with notch pathway components may be critical for the intricate spatial and temporal regulation of OP differentiation and myelination relative to axonal signals during CNS development. Insights into the mechanisms of FGF2 and Notch1 inhibition of OP differentiation are also relevant to understanding the limited remyelination of chronic lesions in demyelinating diseases.
Acknowledgments
This work was supported by National Institutes of Health grant NS39293, National Multiple Sclerosis (RG3515) and USUHS (RO70QS). We thank Drs. T. Kitamura, R. Kageyama, J. Nye, and J. Aster for providing plasmids and Drs. G. Weinmaster, V. Gallo, and A. Aguirre for help with the Jagged1-expressing fibroblasts. We thank Dr. Q. Yang, N. Flint, and T. Le for technical support and Drs. R. Pannu and R. Moore for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–92. doi: 10.1006/meth.1998.0685. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong RC, Le TQ, Flint NC, Vana AC, Zhou YX. Endogenous cell repair of chronic demyelination. J Neuropathol Exp Neurol. 2006;65:245–56. doi: 10.1097/01.jnen.0000205142.08716.7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong RC, Le TQ, Frost EE, Borke RC, Vana AC. Absence of fibroblast growth factor 2 promotes oligodendroglial repopulation of demyelinated white matter. J Neurosci. 2002;22:8574–85. doi: 10.1523/JNEUROSCI.22-19-08574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongarzone ER, Byravan S, Givogri MI, Schonmann V, Campagnoni AT. Platelet-derived growth factor and basic fibroblast growth factor regulate cell proliferation and the expression of notch-1 receptor in a new oligodendrocyte cell line. J Neurosci Res. 2000;62:319–28. doi: 10.1002/1097-4547(20001101)62:3<319::AID-JNR1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 5.Faux CH, Turnley AM, Epa R, Cappai R, Bartlett PF. Interactions between fibroblast growth factors and Notch regulate neuronal differentiation. J Neurosci. 2001;21:5587–96. doi: 10.1523/JNEUROSCI.21-15-05587.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genoud S, Lappe-Siefke C, Goebbels S, Radtke F, Aguet M, Scherer SS, Suter U, Nave KA, Mantei N. Notch1 control of oligodendrocyte differentiation in the spinal cord. J Cell Biol. 2002;158:709–18. doi: 10.1083/jcb.200202002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Givogri MI, Costa RM, Schonmann V, Silva AJ, Campagnoni AT, Bongarzone ER. Central nervous system myelination in mice with deficient expression of Notch1 receptor. J Neurosci Res. 2002;67:309–20. doi: 10.1002/jnr.10128. [DOI] [PubMed] [Google Scholar]
- 8.John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, Brosnan CF. Multiple sclerosis: re-expression of a developmental pathway that restricts oligodendrocyte maturation. Nat Med. 2002;8:1115–21. doi: 10.1038/nm781. [DOI] [PubMed] [Google Scholar]
- 9.Jurynczyk M, Jurewicz A, Bielecki B, Raine CS, Selmaj K. Inhibition of Notch signaling enhances tissue repair in an animal model of multiple sclerosis. J Neuroimmunol. 2005;170:3–10. doi: 10.1016/j.jneuroim.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–14. [PubMed] [Google Scholar]
- 11.Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–98. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- 12.Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–42. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messersmith DJ, Murtie JC, Le TQ, Frost EE, Armstrong RC. Fibroblast growth factor 2 (FGF2) and FGF receptor expression in an experimental demyelinating disease with extensive remyelination. J Neurosci Res. 2000;62:241–56. doi: 10.1002/1097-4547(20001015)62:2<241::AID-JNR9>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murtie JC, Zhou YX, Le TQ, Armstrong RC. In vivo analysis of oligodendrocyte lineage development in postnatal FGF2 null mice. Glia. 2005;49:542–54. doi: 10.1002/glia.20142. [DOI] [PubMed] [Google Scholar]
- 15.Murtie JC, Zhou YX, Le TQ, Vana AC, Armstrong RC. PDGF and FGF2 pathways regulate distinct oligodendrocyte lineage responses in experimental demyelination with spontaneous remyelination. Neurobiology of Disease. 2005;19:171–182. doi: 10.1016/j.nbd.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25:111–23. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–30. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patten BA, Peyrin JM, Weinmaster G, Corfas G. Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation. J Neurosci. 2003;23:6132–40. doi: 10.1523/JNEUROSCI.23-14-06132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–6. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 22.Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–64. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Liu Y, Levine EM, Rao MS. Hes1 but not Hes5 regulates an astrocyte versus oligodendrocyte fate choice in glial restricted precursors. Dev Dyn. 2003;226:675–89. doi: 10.1002/dvdy.10278. [DOI] [PubMed] [Google Scholar]
- 24.Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24:9497–506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou YX, Flint NC, Murtie JC, Le TQ, Armstrong RC. Retroviral lineage analysis of fibroblast growth factor receptor signaling in FGF2 inhibition of oligodendrocyte progenitor differentiation. Glia. 2006;54:578–90. doi: 10.1002/glia.20410. [DOI] [PMC free article] [PubMed] [Google Scholar]