Abstract
Drosophila eye specification occurs through the activity of the retinal determination (RD) network, which includes the Eyeless (Ey), Eyes absent (Eya), Sine oculis (So) and Dachshund (Dac) transcription factors. Based on their abilities to transform antennal precursors towards an eye fate, the distal antenna (dan) and distal antenna-related (danr) genes encode two new RD factors. Dan and Danr are probable transcription factors localized in nuclei of eye precursors and differentiating eye tissue. Loss-of-function single and double dan/danr mutants have small, rough eyes, indicating a requirement for wild-type eye development. In addition, dan and danr participate in the transcriptional hierarchy that controls expression of RD genes, and Dan and Danr interact physically and genetically with Ey and Dac. Eye specification culminates in differentiation of ommatidia through the activities of the proneural gene atonal (ato) in the founding R8 photoreceptor and Egfr signaling in additional photoreceptors. Danr expression overlaps with Ato during R8 specification, and Dan and Danr regulate Ato expression and are required for normal R8 induction and differentiation. These data demonstrate a role for Dan and Danr in eye development and provide a link between eye specification and differentiation.
Keywords: Drosophila melanogaster, Eye, Dan, Danr, Retinal determination network, Egfr, Atonal, Eyeless, Eyes absent, Dachshund
Introduction
During development, cells become committed to particular fates before differentiating into specialized cell types. The Drosophila eye originates within a monolayer epithelium, the eye–antennal imaginal disc, which invaginates from the epidermis during embryonic stages. During larval stages the eye disc cells propagate but remain undifferentiated until shortly before metamorphosis. However, significant events of specification and determination occur within the eye–antennal disc epithelium prior to differentiation.
An evolutionarily conserved network of transcription factors and cofactors, referred to as the retinal determination (RD) network, has been implicated in commitment to an eye fate in Drosophila and several other organisms including humans (reviewed in Rebay et al., 2005; Silver and Rebay, 2005). In Drosophila the RD factors include the Pax-6 orthologs Eyeless (Ey) and Twin of eyeless (Toy) (Czerny et al., 1999; Quiring et al., 1994), as well as Sine oculis (So), a member of the Six family of homeodomain transcription factors (Cheyette et al., 1994; Li et al., 2003), Eyes absent (Eya), which encodes a conserved protein tyrosine phosphatase (Bonini et al., 1993; Li et al., 2003; Rayapureddi et al., 2003; Tootle et al., 2003), and Dachshund (Dac), a Dach homolog related to the Ski and Sno transcriptional repressors (Hammond et al., 1998; Mardon et al., 1994).
The RD genes are thought to be required for retinal determination for two main reasons: (1) eye-specific loss-of-function mutations result in a loss of eye, and (2) they are capable when ectopically expressed, either singly or in combination, of directing the precursors of the wings, antennae or legs to form eye tissue (Bonini et al., 1993, 1997; Chen et al., 1997, 1999; Cheyette et al., 1994; Czerny et al., 1999; Halder et al., 1995; Mardon et al., 1994; Pignoni et al., 1997; Serikaku and O’Tousa, 1994; Shen and Mardon, 1997).
The RD factors form a complex web of interactions that include both transcriptional regulation and direct protein–protein interactions (Rebay et al., 2005; Silver and Rebay, 2005). A transcriptional hierarchy controls the initiation of RD gene expression during eye development: toy lies at the top of the hierarchy and is required for ey transcription (Czerny et al., 1999). Ey is required for both eya and so expression (Halder et al., 1998; Niimi et al., 1999; Ostrin et al., 2006; Punzo et al., 2002). Finally, Eya and So are required to initiate dac expression (Chen et al., 1997; Pignoni et al., 1997). Although eya, so and dac lie downstream of toy and ey within the RD transcriptional hierarchy, ectopic expression of eya or a combination of eya and so can lead to ectopic ey expression (Bonini et al., 1997; Pignoni et al., 1997), suggesting that feedback loops exist within the hierarchy.
Strikingly, ectopic expression of combinations of the RD genes is much more effective at inducing ectopic eyes than any one individually. This led to the idea that besides the transcriptional hierarchy the RD proteins might also physically interact to form one or more protein complexes necessary for retinal determination. Two-hybrid and in vitro studies have confirmed that Eya and So, Eya and Dac, and Ey and So are capable of direct physical interaction (Bonini et al., 1997; Chen et al., 1997, 1999; Pignoni et al., 1997; Zhang et al., 2006).
The RD genes reduce eye size when misexpressed early in eye precursor development (Curtiss and Mlodzik, 2000). The fact that both loss-of-function mutations and misexpression lead to a loss of eye indicates that the stoichiometry of the RD factors is critical and is in accordance with the idea that they form one or more protein complexes important for eye development.
In the Drosophila eye, the determinative activities of the RD factors culminate in the first sign of eye differentiation: a physical indentation in the disc called the morphogenetic furrow (MF). The MF is first observed at the posterior margin of the eye disc (MF initiation) and is propagated through the disc as a standing wave (MF progression) (Wolff and Ready, 1993). A similar wave of differentiation has been observed in vertebrate eye development (Neumann et al., 2000; Stenkamp et al., 2000). Cells in the MF form evenly spaced clusters that eventually develop into ommatidia containing eight photoreceptors (R1–R8) plus additional support cells (Wolff and Ready, 1993).
The R8 photoreceptor is the first to begin differentiation and its presence is required for recruitment of all other photoreceptors. Hence, correct R8 specification is critical to the spacing and subsequent patterning of the ommatidia. Two factors with interrelated roles in both the spacing of ommatidia and recruitment of photoreceptors other than R8 are the bHLH proneural transcription factor Atonal (Ato) and the Drosophila EGF receptor (Egfr). Ato is required for R8 specification: in null ato- mutants no R8s or any other photoreceptors develop (Jarman et al., 1994). However, weak ato- alleles cause reduction in the number of R8 cells that are recruited, resulting in gaps in the ommatidial arrangement (White and Jarman, 2000). In contrast, the Egfr pathway is not required for R8 specification, but for recruitment of the R1–R7 photoreceptors (Freeman, 1996, 1997).
Although at first glance the functions of Egfr and Ato appear to be distinct, besides its role in R8 precursor specification Ato also seems to be required for aspects of R8 differentiation, including the ability via the Egfr pathway to recruit the R1–R7 photoreceptors into each nascent ommatidium: in weak ato- mutants Egfr-mediated induction of other photoreceptors often fails leading to a phenotype very similar to that observed with a partial loss of Egfr activity (White and Jarman, 2000). Accordingly, Ato is capable of activating the Egfr pathway through its activities in the R8 precursor (Baonza et al., 2001; Chen and Chien, 1999). Egfr also has a potential role in ommatidial spacing. Initial evidence came from the fact that mutants for the dominant Ellipse (Elp) allele of Egfr have fewer ommatidia that are irregularly spaced (Baker and Rubin, 1992). Conversely, reduced levels of Egfr signaling lead to increased numbers of R8 cells that are closer together than normal (Yang and Baker, 2001, 2003).
The Ato expression pattern is highly dynamic, with initial expression in a dorsal–ventral band of cells ahead of the MF called the pre-proneural zone. Ato is then upregulated in the MF in the intermediate groups, which are evenly spaced clusters of approximately 10 cells. Nuclei of 2–3 cells from the intermediate group, each of which is competent to adopt the R8 cell fate, migrate apically to form the R8 equivalence group. Shortly thereafter, Ato becomes restricted to single R8 precursors (Baker et al., 1996; Dokucu et al., 1996; Frankfort and Mardon, 2002; Jarman et al., 1994, 1995).
Recently, a link has been discovered between the RD factors that promote eye specification and Ato at the beginning of differentiation. Ey and So are capable of physical interaction and bind to adjacent sites in the 3′ ato regulatory region, which drives ato expression ahead of the MF as well as in the intermediate groups. Other factors are required for regulation via the 5′ ato regulatory region, which is responsible for ato expression in the R8 equivalence groups and in the R8s themselves (Sun et al., 1998; Zhang et al., 2006). Thus, the Ey and So specification factors are likely to be directly involved in promoting transcription of genes involved in eye differentiation. However, one thing that remains unclear is how transcription factors such as Ey and So interact with the basal transcriptional machinery to initiate and maintain an eye-specific fate.
We have examined the functions of the distal antenna (dan) and distal antenna-related (danr) genes in eye development. These two genes encode related proteins that contain a pipsqueak motif, a DNA-binding motif found in proteins associated with chromatin structure regulation and cellular memory (Couderc et al., 2002; Lehmann, 2004; Siegmund and Lehmann, 2002). Dan and Danr are capable of transforming distal leg structures to an antennal fate. Conversely, in single and double loss-of-function dan and danr mutants the distal antenna is converted to a leg. These results demonstrate that dan and danr are involved in antennal specification (Emerald et al., 2003; Suzanne et al., 2003).
Here we demonstrate that dan and danr also participate in eye specification. Like the RD factors, misexpression of dan or danr in antennal precursors results in their transformation to ectopic eyes. Both dan and danr are required for normal eye development, and misexpression of dan or danr in eye precursors interferes with eye development. Moreover, dan and danr participate in the RD transcriptional hierarchy and the Dan and Danr proteins can physically interact with Ey and Dac. Both dan and danr are required for normal levels of Ato expression. Consequently, danr single and dan danr double mutants have problems with the induction and spacing of the developing R8 photoreceptors, as well as the subsequent recruitment and differentiation of additional photoreceptors in each ommatidium.
Hence, Dan and Danr are unusual in that they seem to function in both antennal and eye specification. One possible explanation for this is that they play a permissive, rather than instructive role in regulating tissue-specific gene expression. Since the Dan and Danr proteins contain a motif associated with chromatin structure regulation (Emerald et al., 2003), we suggest that they may interact with the RD transcription factors or antennal specification factors to help initiate or maintain a chromatin structure conducive to subsequent eye-specific or antennal-specific gene expression.
Materials and methods
Drosophila genetics
Mutations used
ElpB1 (Baker and Rubin, 1989); danrex35, dan danrex56, danemS3 (Emerald et al., 2003); so³ (Cheyette et al., 1994); dac³, dac4 (Mardon et al., 1994);
Transgenes used
Dll-Gal4 (Calleja et al., 1996); ey-Gal4 (Hauck et al., 1999); eyFLP (gift from B. Dickson; (Newsome et al., 2000)); arm-lacZ (Vincent et al., 1994); UAS-ey (Halder et al., 1995); UAS-Dac7C4 (Chen et al., 1997); UAS-dan, UAS-danr (Emerald et al., 2003).
Mosaic analysis
Homozygous danrex35, dan danrex56, danems3, so³ or dac³ clones were generated via the FLP/FRT system (Xu et al., 1993). Genotypes used were:
w,eyFLP/w or Y; FRT82,arm-lacZ,w+/ FRT82,danrex35.
w,eyFLP/w or Y; FRT82,arm-lacZ,w+/ FRT82,dan,danrex56.
w,eyFLP/w or Y; FRT82,arm-lacZ,w+/ FRT82,danems3.
w,eyFLP/w or Y; FRT42, arm-lacZ, w+/ FRT42,so³.
w,eyFLP/w or Y; FRT40,arm-lacZ,w+/ FRT40,dac³.
For making large clones using the Minute technique, the FRT82,arm-lacZ chromosome was replaced with FRT82,arm-lacZ,M (gift from J. Treisman).
Histology
For immunocytochemistry, fixation and treatment of eye discs were performed as described by Tomlinson et al. (1987), using rabbit anti-Ato (1:1000 — gift from A. Jarman), rat monoclonal anti-Elav (1:100 — gift from G. Rubin); rabbit (Cappel) and mouse (Promega) anti-β-galactosidase (1:200); rabbit anti-Ey (Halder et al., 1998) and mouse monoclonals anti-Eya and anti-Dac (1:20 — Developmental Studies Hybridoma Bank). Antibodies against Dan and Danr are described in Emerald et al. (2003). Phalloidin (Molecular Probes) was used to stain the actin cytoskeleton. Confocal microscopy was performed at the Microscopy Shared Research Facility at MSSM.
Adult eyes were fixed, embedded and sectioned as described by Tomlinson et al. (1987).
For SEM, adult flies were dehydrated through an ethanol series: 25%, 50%, 70%, 90%, 100% and 100% ethanol for approximately 12 h each. Following two 30-minute incubations in hexamethyldisilazane (HMDS—Polysciences, Inc.) the flies were allowed to dry in air under normal atmospheric conditions. Fly heads were mounted onto an SEM block then sputter-coated with gold and photographed at the Microscopy Shared Research Facility at MSSM.
Protein–protein interactions
GST-pulldowns
GST fusion proteins were generated from cDNA clones in pGex4T1. 35S-labeled in vitro translated proteins were generated using the TNT Quick Coupled Transcription/Translation kit (Promega), from cDNA clones in pBetaTH. GST-pulldowns were performed as previously described (Jenny et al., 2005). Samples were analyzed by SDS–PAGE followed by autoradiography.
Yeast two-hybrid analysis
Yeast two-hybrid assays were performed as previously described (Jenny et al., 2005). For the bait constructs, full-length cDNAs were subcloned into pAS_mod. Prey constructs were generated using the same cDNAs cloned into pAct_mod. Interactions were evaluated by plating on minimal media lacking adenine.
Results
Ectopic Dan or Danr transforms antennal precursors into eye tissue
We identified dan and danr in a large-scale modular misexpression screen (Mata et al., 2000), in part for their abilities to interfere with eye development when misexpressed in eye precursors (see below). In our initial analysis, we used the Gal4/UAS system (Brand and Perrimon, 1993) to explore the effects of dan or danr misexpression on other tissues. Strikingly, ectopic eyes form in the third antennal segment when Distal-less(Dll)-Gal4 is used to drive dan or danr expression in distal regions of the antenna disc (Figs. 1A–B; Suzanne et al., 2003). Ectopic eyes generated by Dll>dan and Dll>danr in the distal antenna are composed of several ommatidia. Each ommatidium contains a lens and all other cell types found in endogenous ommatidia, all of which appear to have differentiated normally (not shown). Thus, dan and danr are capable of transforming antennal precursors to an eye fate.
Fig. 1.
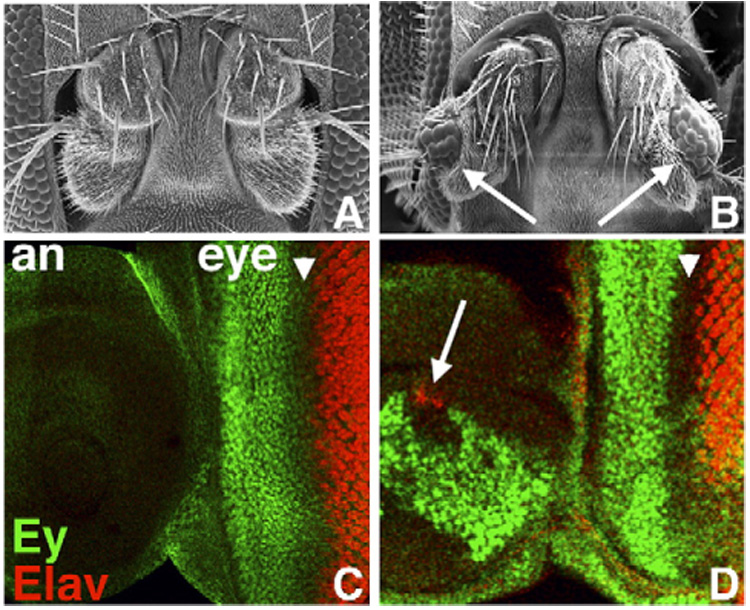
Misexpression of dan or danr during antennal development leads to ectopic eye formation. (A, B) Frontal view of wild-type (A) and Dll>EPg-dan head (B) showing antennae; each antenna in (B) contains ectopic eye tissue (arrows). (C) Wild-type eye–antenna disc. Ey (green) is expressed only in the eye portion of the disc (eye) anterior to the MF (arrowhead); Elav (red) marks developing photoreceptors posterior to the MF. (D) In Dll>EPg-dan discs, in addition to normal patterns in the eye region, Ey (green) is expressed in a large patch in the antennal portion of the disc (an), and Elav (red) is expressed in ectopic photoreceptor precursors (arrow). Anterior is to the left and dorsal up in panels C and D here, as well as in all panels of all subsequent figures.
To determine whether the ectopic eyes generated by Dll>dan and Dll>danr are accompanied by misexpression of Ey, we looked to see whether Ey is ectopically expressed in Dll>dan and Dll>danr antennal discs. Late third instar larval wild-type antennal discs do not express Ey, nor do they express Elav, a marker for neurons including photoreceptors (Fig. 1C). However, in Dll>dan or Dll>danr antennal discs a large proportion of the cells express Ey (Fig. 1D), and differentiating photoreceptors that express Elav are also present (Fig. 1D, arrow). Thus, Dll>dan and Dll>danr promote ectopic eye formation, and they are capable of inducing (or maintaining) expression of the RD factor Ey.
dan and danr are required for eye development
Like the RD factors, dan and danr are capable of transforming antennal tissue to an eye fate, suggesting that they have roles in eye determination. If so, they should also be required during normal eye development. We therefore used antibodies against the Dan and Danr proteins to determine whether they are expressed during normal eye development, and we examined single and double loss-of-function dan and danr mutants for effects on eye development.
Dan and Danr are expressed during eye development
The Dan and Danr proteins are both expressed in nuclei in the developing eye. Dan is first detected in early L3 eye discs in cells surrounding the newly initiated MF (Fig. 2A). As the MF moves across the eye disc from posterior to anterior both Dan and Danr are expressed at high levels anterior to the MF and then at lower levels in differentiating eye precursors posterior to the MF (Figs. 2B–D; Suzanne, 2004; Suzanne et al., 2003). Whereas Dan and Danr expression patterns anterior to the MF look very similar, they differ within and posterior to the MF. In the MF Danr is most strongly expressed in evenly spaced clusters of cells (Figs. 2C, D) that appear to correspond to the “intermediate group” stage of Ato expression (see below). In contrast, Dan expression is maintained at uniform high levels in all cells in the MF (Figs. 2B, D). Posterior to the MF, Dan and Danr are expressed at lower levels in the photoreceptors, with Dan expressed at a higher level than Danr (Figs. 2B–D, compare red and green channels). The Dan and Danr expression patterns resemble those of the RD factors Eya, So and Dac and overlap considerably with the Ey expression pattern (see below). Thus, expression of Dan and Danr in undifferentiated cells anterior to and within the MF is consistent with a role in retinal determination.
Fig. 2.
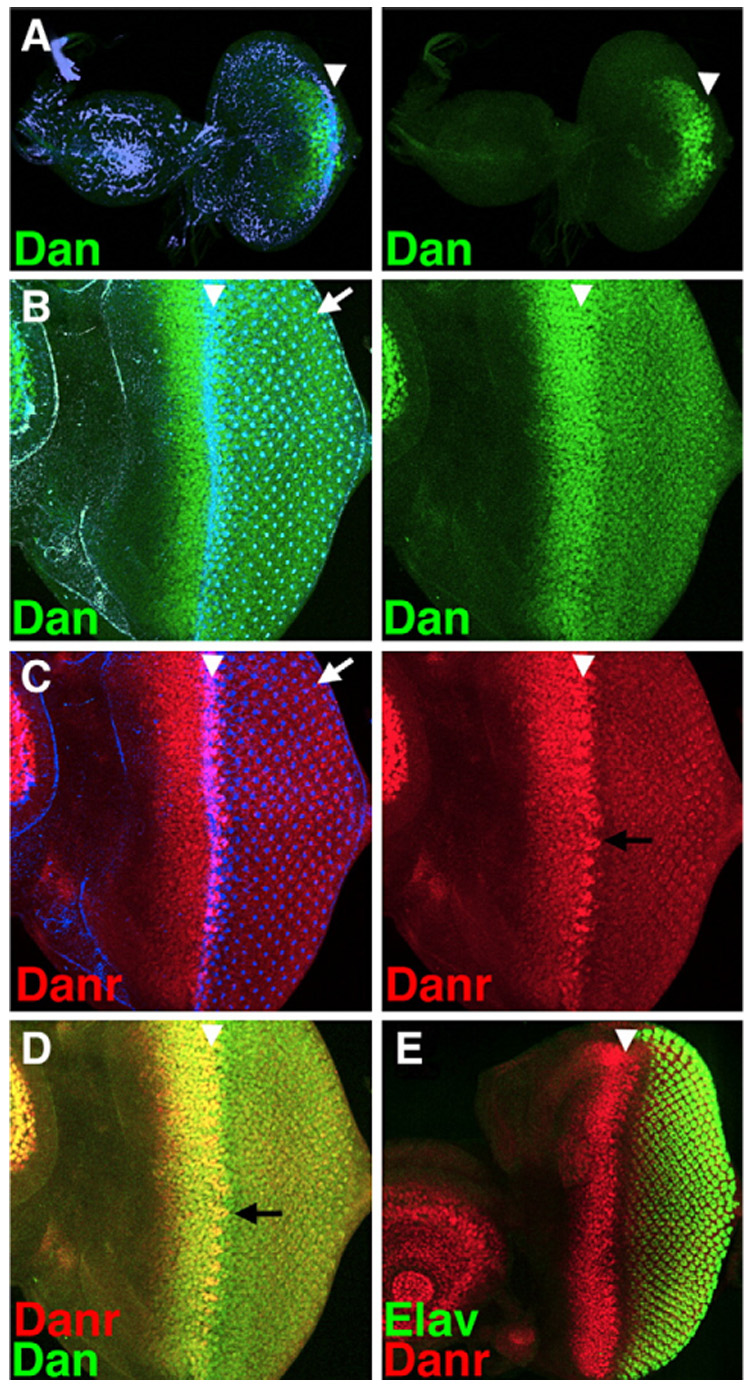
Dan and Danr are expressed during eye and antennal development. (A–C) Left panels show an overlay of Dan (green) or Danr (red) expression with phalloidin staining (blue), which highlights the changes in the actin cytoskeleton that occur within the MF (white arrowhead), as well as the center of each developing photoreceptor cluster (white arrow). Right panels show Dan or Danr expression alone. (A) Dan expression (green) is observed in and around the MF as it initiates at the posterior margin early in the third larval instar. (B–D) All panels show the same disc. In a mid 3rd instar disc, where the MF has progressed a significant distance, Dan (green) is expressed at high levels in all cells anterior to and within the MF and at lower levels in all photoreceptor cells posterior to the MF. Danr (red) is expressed in a similar pattern to Dan anterior to the MF. Its levels drop within and posterior to the MF except in regularly spaced groups of cells within the MF (black arrows). (E) Lower magnification view of an eye–antenna disc showing part of Danr expression in the antenna; also showing the relationship between Danr expression anterior to and within the MF, and Elav expression (green) in the developing photoreceptor cells.
From mid-L3 onward both Dan and Danr are also expressed in the antennal disc (Fig. 2E), and they have redundant functions during antennal specification (Emerald et al., 2003; Suzanne, 2004; Suzanne et al., 2003). This begs the question how misexpression of Dan or Danr can transform antennal precursors to an eye fate when they are already normally expressed during antennal development. However, it should be noted that expression in the eye disc precedes expression of either Dan or Danr in the antennal disc (Fig. 2A). Thus, the timing of Dan/Danr expression appears to be important in defining which tissue is specified.
dan and danr regulate each other’s expression
To determine whether either dan or danr or both are required for eye development, we characterized eye phenotypes of single and double mutants. We first examined expression of Dan and Danr protein in eye discs mosaic for the danrex35, dan danrex56 and danems3 alleles (Emerald et al., 2003) to determine the nature of these alleles with respect to eye development. The danrex35 mutation is a small deletion that removes part of the Danr coding sequence. Danr protein is absent from danrex35 homozygous clones located in the eye disc (Fig. 3B, red), suggesting that danrex35 is a null allele for eye development. Neither the Dan nor the Danr protein is detectable in homozygous clones for dan danrex56, a deletion that removes all of danr, part of dan, and the ~45 kb of DNA between the two genes (Fig. 3C), suggesting that dan danrex56 is a double null mutation. Dan appears to be expressed normally in homozygous clones for the point mutation danems3 (Fig. 3D, green), suggesting that it is not a protein null. Nevertheless, the fact that danems3 was generated as a revertant of the Dll>dan phenotype (Emerald et al., 2003) indicates that danems3 is a loss-of-function dan allele.
Fig. 3.
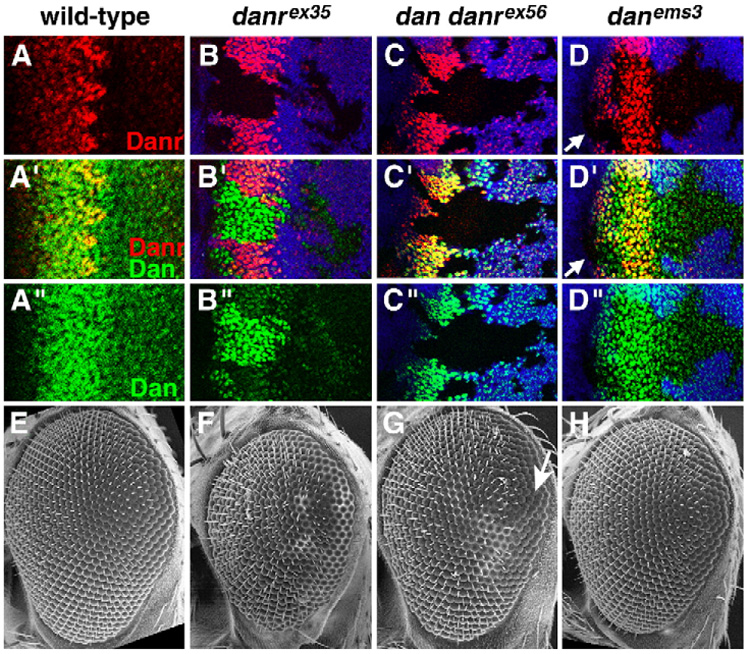
Loss-of-function mutants for dan/danr have small, rough eyes. (A) Close-up of the MF from a wild-type eye–antenna disc. (B–D) Discs containing loss-of-function dan/danr clones. Dan (green), Danr (red) and clonal marker (blue) are shown. Clones are marked by the absence of blue staining (see Materials and methods), except in panel B″, where the clonal marker has been removed for clarity. Overlay between red and blue, green and blue, and red and green appears pink, turquoise and yellow, respectively. (B) danrex35 clones do not express Danr, but express higher levels of Dan. (C) dan danrex56 clones do not express either Danr or Dan. (D) danems3 clones express normal levels of Dan; Danr expression is lost from clones anterior to the MF (arrows). (E) SEM picture of a wild-type eye. Eyes from eyFLP;FRT82, arm-lacZ,M/FRT82,danrex35 (F) or eyFLP;FRT82,arm-lacZ,M/FRT82,dan danrex56 (G) individuals. Most of both eyes consist of homozygous mutant tissue (see Materials and methods). danrex35 and dan danrex56 eyes are small and rough. (H) Eye from a danems3 homozygote is slightly small.
Interestingly, Dan is expressed at higher levels in danrex35 clones than in wild-type tissue (Fig. 3B, green). In addition, Danr protein is expressed at lower levels in danems3 clones that are anterior to the MF (Fig. 3D, red), although Danr expression in danems3 clones within and posterior to the MF appears normal. Thus, Dan and Danr regulate each other’s expression: Danr appears to repress Dan expression, but Dan is required for Danr expression anterior to the MF.
danr single and dan danr double mutants have small, rough eyes
danrex35, dan danrex56 and danems3 mutants are homozygous subviable—approximately 90% die during larval and pupal stages. Homozygous escapers survive for only a few hours and show a transformation of antenna towards leg (Emerald et al., 2003; Suzanne et al., 2003). Using RNAi, Suzanne et al. (2003) showed that reduction of either dan or danr expression results in a mildly rough eye, suggesting a potential role for dan and danr in eye development as well.
We have examined homozygous danrex35, dan danrex56 and danems3 escapers for their effects on eye development. Since escapers may have weaker phenotypes than individuals that die during development, we also examined the phenotypes of flies containing large eye clones of all three mutant phenotypes. When induced in the eye–antennal disc using eyFLP and the Minute technique (Newsome et al., 2000) danrex35, dan danrex56 or danems3 mutant clones comprise >80% of each eye. Both homozygous and mosaic individuals have similar eye phenotypes.
Wild-type eyes have a precise array of ommatidia that gives the eye a smooth surface (Fig. 3E). Both danrex35 homozygous and mosaic eyes are small and rough: ommatidia vary in size, are sometimes fused and are irregularly spaced (Fig. 3F). dan danrex56 eyes appear closer to wild-type size but show areas of roughness, typically at the posterior edge or at the equator (Fig. 3G, arrow). danems3 eyes appear generally normal (Fig. 3H). We quantified the numbers of ommatidia in wild-type eyes and compared this with the danrex35 and dan danrex56 mutant eyes. On average, each wild-type (Oregon R) eye has 533 ommatidia (Table 1). In contrast, danrex35 homozygous eyes are statistically significantly smaller than wild-type, with on average only 409 ommatidia per eye. dan danrex56 eyes are comparable in size to wild-type, averaging 518 ommatidia. In conclusion, loss of danr results in smaller, rougher eyes than wild-type; loss of both dan and danr results in rough eyes. These results indicate that both dan and danr are required for eye development. The fact that the danems3 mutation is unlikely to be a null may explain its relatively weak phenotype (but see below and Discussion).
Table 1.
danrex35 homozygous eyes have fewer ommatidia than wild-type
| Genotype | # of eyes examined | Mean # of ommatidia per eye |
|---|---|---|
| Oregon R | 25 | 533±24 |
| danrex35/danrex35 | 5 | 409±19 |
| dan,danrex56/dan,danrex56 | 5 | 518±25 |
Mean numbers of ommatidia per eye are given±the standard deviation. By the Student’s t-test, the means for the number of ommatidia for wild-type and the danrex35 mutant are significantly different at p=5.9×10−6. There was no significant difference between wild-type and the dan danrex56 mutant.
The dan and danr genes are thought to have redundant roles during antennal development: the Dan and Danr proteins have identical expression patterns in the antenna, and the antennal phenotype of dan danrex56 double mutants is stronger than that of either the danrex35 or the danems3 single mutants (Emerald et al., 2003). In contrast, during eye development Dan and Danr proteins have distinct expression patterns (Fig. 2), and the danrex35 single mutant has a stronger phenotype than the dan danrex56 double mutant. This suggests that dan and danr have a more complicated, and possibly antagonistic, relationship during eye development compared to antennal development (see Discussion).
danrex35 and dan danrex56 mutants affect ommatidial spacing and photoreceptor recruitment
To further explore the roles of dan and danr in eye development, we generated sections of danrex35, dan danrex56 and danems3 adult eye tissue, either in homozygotes or in homozygous mutant clones. In apical sections of wild-type ommatidia, the “outer” photoreceptors, R1–R6, have relatively large rhabdomeres arranged in a trapezoidal pattern. The “outer” photoreceptors surround the “inner” R7 photoreceptor, which has a smaller rhabdomere (Fig. 4A, inset; Wolff and Ready, 1993). The R8 photoreceptor lies beneath R7 and is therefore not visible in apical sections.
Fig. 4.
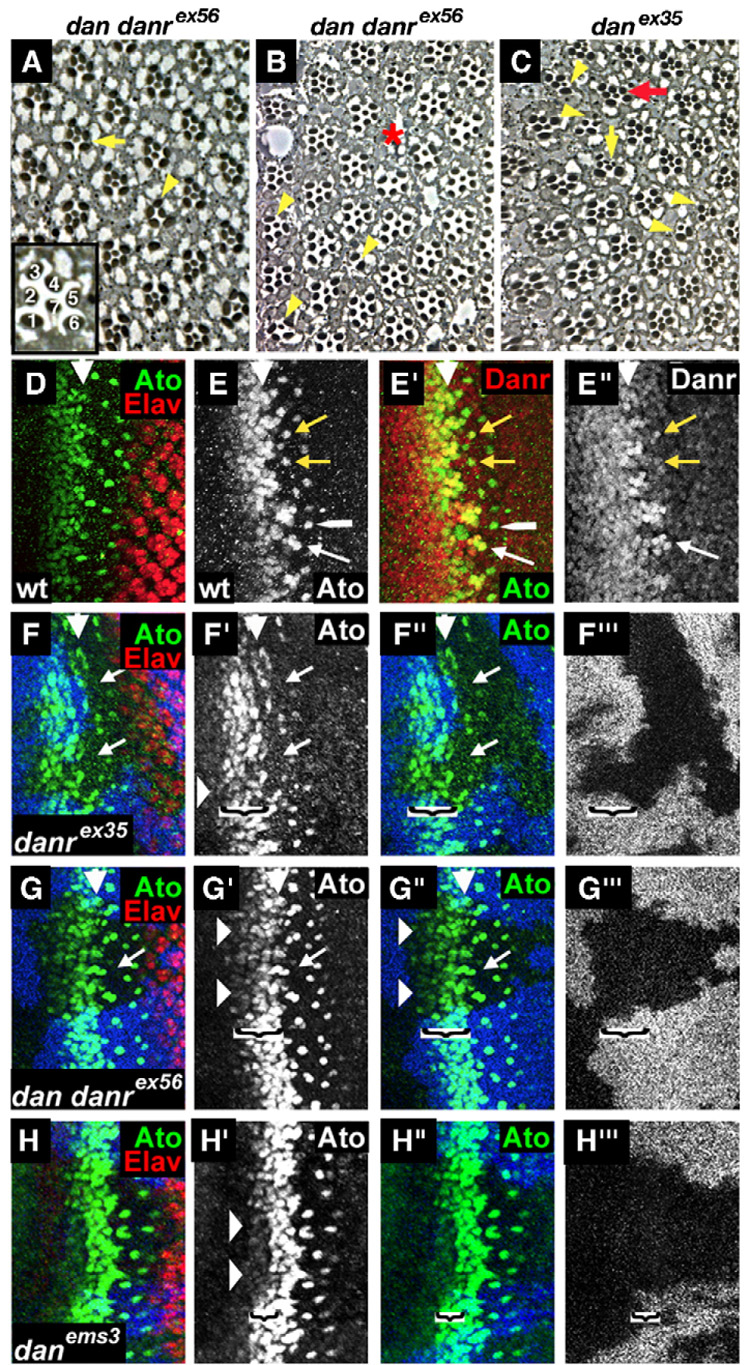
danrex35 and dan danrex56 homozygous eye tissues show defects in ommatidial spacing and photoreceptor recruitment. (A–C) Cross sections of eyes from eyFLP;FRT82,arm-lacZ,M/FRT82,dan danrex56 or eyFLP;FRT82,arm-lacZ,M/FRT82,danrex35 flies. Inset: an enlarged wild-type ommatidium showing the positions of the R1–R7 photoreceptors. The internal structure of dan/danr adult mutant eye tissue is variable, ranging from most ommatidia appearing normal (A) to eyes with more frequent defects (B or C) including a loss of photoreceptors (yellow arrowheads), extra small-rhabdomere photoreceptors (C—yellow vertical arrow), defects in rotation (C—red arrow) or gaps in the ommatidial arrangement (B—red asterisk). (D–G) Third instar eye discs. MF is marked by a white arrowhead. (D) Wild-type eye disc double labeled for Elav (red) and Ato (green). (E) Wild-type eye disc double labeled for Ato (green) and Danr (red). Ato and strong levels of Danr expression co-localize in the intermediate groups, in the R8 equivalence group (white arrow) and for a brief period in newly emerged R8 cells (yellow arrows). Shortly afterwards, the strong levels of Danr expression in R8 fade (white long arrowhead). (F–H) Loss-of-function dan/danr clones marked by absence of the blue staining. (F, G) In danrex35 and dan danrex56 mutant clones the stripe of Ato expression (green) anterior to the MF is reduced (horizontal arrowheads), and the number and spacing of the Ato positive R8 cells are altered (arrows). (H) In danems3 mutant clones a narrower stripe of cells ahead of the MF shows a reduction in Ato expression (horizontal arrowheads). The brackets in panels F–H delineate the width of the region ahead of the MF in which Ato expression is reduced in the clones.
Although sections of danems3 mutant eye tissue appear largely normal, sections of danrex35 and dan danrex56 mutant eyes reveal multiple defects. In general, danrex35 eyes have a greater number of defects compared to dan danrex56 eyes, consistent with their stronger external phenotype. The defects observed include ommatidia that are missing one or more photoreceptors (Figs. 4A–C, arrowheads), with no specific photoreceptor type being consistently affected. In addition, whereas in apical sections wild-type ommatidia have only one “inner” photoreceptor with a small rhabdomere, some danrex35 or dan danrex56 mutant ommatidia contain several photoreceptors with small rhabdomeres (e.g. Fig. 4C, vertical arrow). It is unclear whether this results from recruitment of extra R7/8-like (“inner”) photoreceptors, or the conversion of cells from an “outer”, large-rhabdomere to an “inner”, small-rhabdomere fate. We also have observed gaps (Fig. 4B, asterisk) and other irregularities in the arrangement of ommatidia. In summary, danrex35 and dan danrex56 mutants have defects in ommatidial spacing and also in the recruitment and identity of R1–R7 photoreceptors.
dan and danr are required for Ato expression
The defects observed in danrex35 and dan danrex56 mutant tissue (gaps in the ommatidial arrangement, small eyes, and loss of photoreceptors) are reminiscent of those observed in eyes from weak ato mutants. Although null ato mutants do not develop any R8 cells, weak ato alleles cause a reduction in the number of R8 cells, resulting in gaps in the ommatidial arrangement. In addition, weak ato mutants show photoreceptor loss due to defects in R8 differentiation and recruitment of the R1–R7 photoreceptors (White and Jarman, 2000), similar to the loss of photoreceptors in danrex35 and dan danrex56 mutants. Thus, all of the phenotypes observed in dan/danr mutant eyes could result from a reduction in ato function.
Strikingly, the expression patterns of Ato and Danr overlap precisely. As described above, like Ato, Danr is expressed ahead of the MF and then becomes restricted to evenly spaced groups of cells (Fig. 2C). Co-staining wild-type discs with antibodies against both Danr and Ato reveals that the groups of cells expressing Danr in the MF correspond exactly with Ato expression in the intermediate groups (Fig. 4E). Danr and Ato also overlap in the 2–3 cells that form the R8 equivalence group (Fig. 4E, white arrow), and Danr expression remains strong temporarily in R8 cells (Fig. 4E, yellow arrows), although it fades in R8 before Ato does (Fig. 4E, long white arrowhead).
Based on these observations we asked whether dan or danr regulate Ato expression during eye development and examined Ato expression in danrex35, dan danrex56 and danems3 mutant clones. Ato expression ahead of the MF is reduced in all three types of dan/danr- clones (Figs. 4F–H). However, the region of the Ato expression pattern affected is slightly different among the three alleles. In the danrex35 and dan danrex56 clones, Ato expression appears reduced in all cells ahead of the MF (Figs. 4F, G, arrowheads; width of the effect on Ato expression shown by the brackets) as well as in the intermediate groups, and there are abnormalities in the number and spacing of Ato-expressing cells (Figs. 4F, G, arrows). For danems3 clones, the first few rows of the Ato expression pattern ahead of the MF appear lower than in surrounding wild-type tissue, but a few rows later, and still a few rows anterior to the intermediate groups, Ato expression has recovered to normal levels (Fig. 4H, arrowheads; width of the effect on Ato expression shown by the brackets). The subsequent resolution of Ato into intermediate groups, equivalence groups and a single R8 appears to be normal. This fits well with the very weak phenotype of danems3 adult eyes and suggests that dan participates in initiating Ato expression but that other factors can compensate in its absence. Alternatively, the fact that the danems3 allele is not a null may explain the different effects on the Ato expression pattern.
As with the external appearance of the eye and with the phenotype in eye sections, danrex35 clones have the strongest overall effect on Ato expression, dan danrex56 has a somewhat weaker effect, and danems3 has the weakest effect, although an effect is clearly present. Thus, dan and danr are required for normal levels of Ato expression and for normal R8 spacing, suggesting that both dan and danr promote ommatidial formation/spacing by positively regulating Ato expression.
To confirm roles for dan and danr in the early stages of R8 recruitment and spacing, we tested for genetic interactions between dan/danr mutants and the dominant Ellipse (Elp) allele of Egfr, which results in a moderate increase in Egfr activity (Lesokhin et al., 1999) and has a spacing phenotype: Elp heterozygotes have reduced numbers of ommatidia that are irregularly spaced. Although the ommatidia that form in Elp mutants are often normal (Baker and Rubin, 1989; Baker and Rubin, 1992), we have observed occasional problems with photoreceptor recruitment, leading to loss of photoreceptors (Fig. S1A, arrows). Remarkably, presence of a single copy of any of the danrex35, dan danrex56 or danems3 mutations can enhance the small eye phenotype of Elp heterozygotes (Fig. S1C–H). As expected from its stronger homozygous phenotype, the danrex35 mutation has the strongest effect on Elp, leading to very small eyes that resemble those of Elp homozygotes. However, heterozygous dan danrex56 or danems3 also enhance the Elp heterozygous phenotype, with dan danrex56 having a stronger effect than danems3. Whereas reduction in ommatidial number in Elp heterozygotes is strongly enhanced by dan/danr mutations, there is little effect of single and double dan and danr mutations on the Elp/+ photoreceptor recruitment phenotype (compare Fig. S1B with S1D, F, H). These data substantiate the idea that dan/danr function is required for R8 recruitment and spacing and suggest that dan/danr and Egfr function via a common mechanism during this process.
The effects of both dan and danr mutations on eye morphology and Ato expression, along with the abilities of both Dan and Danr to induce ectopic eye development in the antennae, indicate that both dan and danr are required for normal eye development. Although the danrex35, the dan danrex56 and particularly the danems3 mutants have mild eye phenotypes, all three mutations enhance Elp, and Ato expression is reduced in clones for all three mutations. In conclusion, the experiments described here demonstrate that both the dan and the danr genes function to ensure that sufficient numbers of ommatidia are established and correctly spaced and that all of the photoreceptors within those ommatidia are recruited. dan and danr interact with Egfr during these processes and are required for normal levels of Ato expression.
Dan and Danr participate in the RD hierarchy
The fact that dan and danr are required for normal eye development and can generate ectopic eyes when misexpressed suggests that they are involved in retinal determination. Several observations indicate that Dan and Danr participate in the transcriptional hierarchy responsible for RD factor expression. (1) Dan and Danr are able to induce/maintain Ey expression in the antennal disc (Fig. 1D). (2) Dan/Danr are localized to nuclei and contain a pipsqueak motif (Emerald et al., 2003), which when present in other proteins is known to bind DNA. Therefore Dan and Danr are likely to be transcription factors. (3) Dan and Danr are coexpressed with other RD factors in the undifferentiated cells anterior to and within the MF where retinal determination occurs. For instance, Dan/Danr expression ahead of the MF overlaps with high levels of Ey expression (Fig. S2A). In addition, expression patterns of Dan and Danr closely resemble those of Eya, So and Dac throughout eye development (Fig. S2B, C, not shown). Thus, all five proteins are co-expressed at high levels in the same domain anterior to and within the MF, and Dan and Danr are expressed in a pattern consistent with regulating or being regulated by the RD factors.
To test whether dan or danr is required for RD factor expression we stained eye discs mosaic for loss-of-function dan/danr alleles with antibodies against Ey, Eya and Dac. In danrex35 clones Ey expression is normal, but Eya is present at reduced levels (Fig. 5A, arrows). In contrast, in dan danrex56 and danems3 clones both Ey and Eya are expressed normally (Figs. 5B, C). Dac is expressed normally in danrex35, dan danrex56 and danems3 clones (Figs. 5D–F). Thus, danr is required for high levels of Eya expression. In addition, the fact that Eya expression is affected in danrex35 but not in dan danrex56 clones indicates that dan function is antagonistic to danr function in this context as in others described above.
Fig. 5.
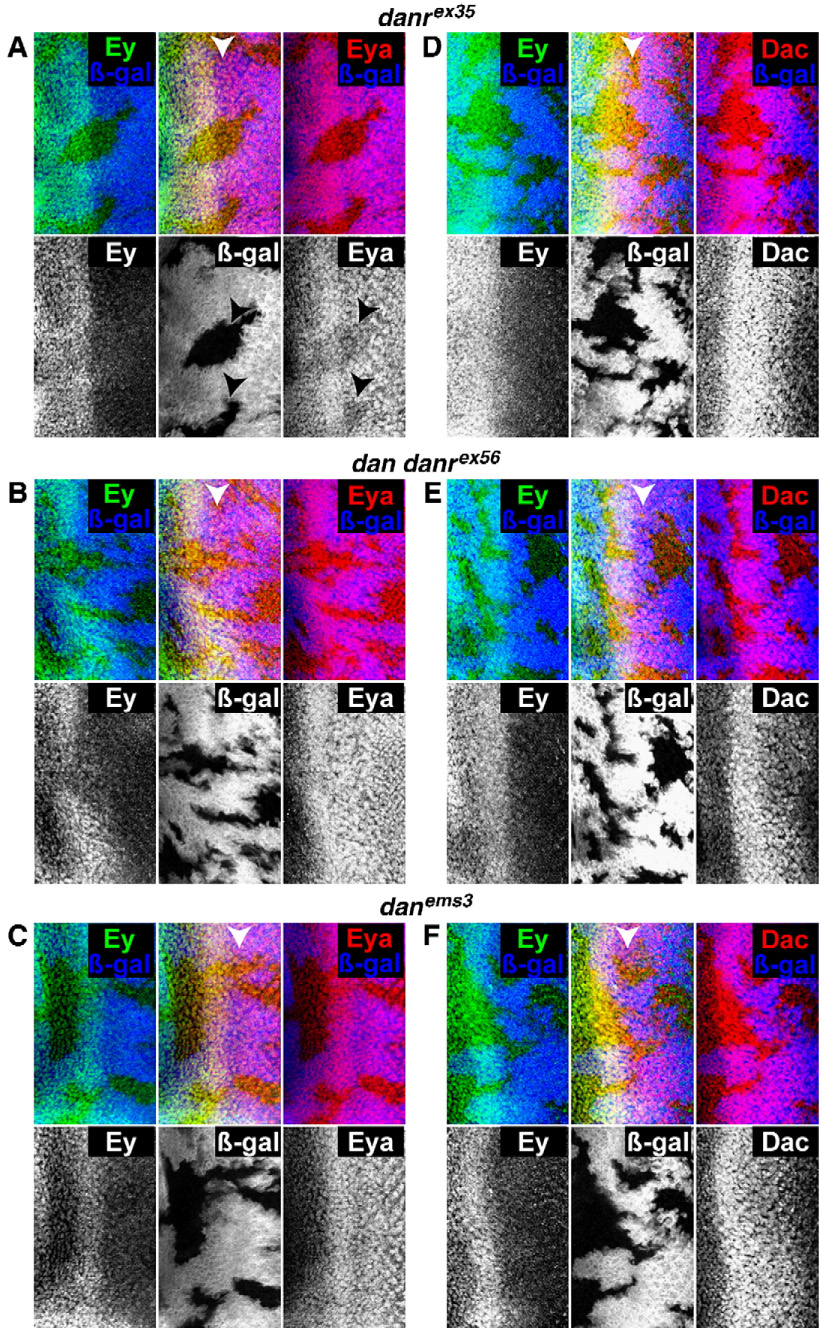
dan/danr and the regulation of RD factor expression. (A–C) Clones marked by the absence of the β-galactosidase (blue) and stained for Ey (green) and Eya (red). (D–F) Clones marked by the absence of the β-galactosidase (blue) and stained for Ey (green) and Dac (red). (A,D) eyFLP; FRT82 arm-lacZ/FRT82 danrex35 discs. Eya is expressed at reduced levels in danrex35 mutant clones (arrows—A), but Ey (A, D) and Dac (D) are expressed normally. (B, E) eyFLP; FRT82 arm-lacZ/FRT82 dan danrex56 discs. (C, F) eyFLP; FRT82 arm-lacZ/FRT82 danems3 discs. Eya (B, C) and Ey (B, C, E, F) and Dac (E, F) are expressed at normal levels in both dan danrex56 and danems3 mutant clones.
We also tested whether So or Dac are required for Dan or Danr expression by examining Dan and Danr expression in homozygous clones of the null alleles so³ and dac³ (Fig. 6). The MF does not initiate in so³ clones that touch the margin of the eye disc. Instead, so³ clones overproliferate, fail to differentiate into photoreceptors and eventually die, causing abnormal folding of the discs. so³ clones can also split the eye field, resulting in MFs on the dorsal or ventral margins that propagate into the center of the disc (Pignoni et al., 1997). Dan and Danr are not expressed in so³ marginal clones (arrow, Fig. 6A). However, the effects of so³ tissue on Dan/Danr expression are nonautonomous in these cases (arrowhead, Fig. 6A), with some so+ cells failing to express Dan and Danr, suggesting that other factors also regulate Dan/Danr expression.
Fig. 6.
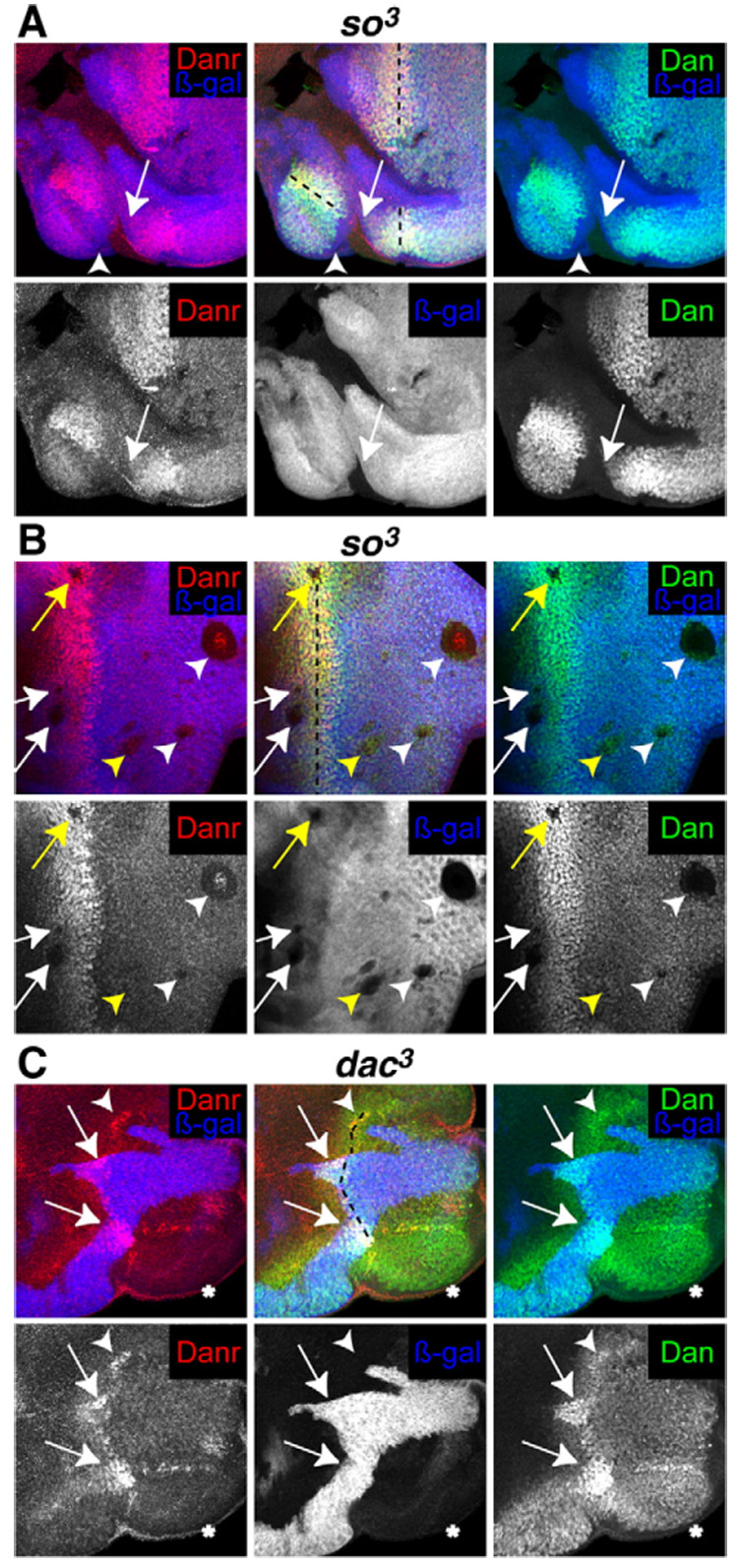
(A, B) Discs from eyFLP; FRT42 arm-lacZ/FRT42 so³. (C) Discs from eyFLP; FRT40 arm-lacZ/FRT40 dac³. (A–C) Null so³ or dac³ clones are marked by the absence of β-galactosidase (blue) and stained with anti-Dan (green) and anti-Danr (red). The position of the MF is marked by a black dotted line. (A) Neither Dan nor Danr are expressed in so³ clones that touch the margin of the disc (arrows), but this effect is nonautonomous (arrowhead). (B) Neither Dan nor Danr are expressed in so³ clones anterior or within the MF (white and yellow arrows), or in large clones posterior to the MF (white arrowheads). Other clones posterior to the MF express Dan and Danr (yellow arrowheads). A single row of cells inside the border of so³ clones located within the MF (yellow arrow) or large clones posterior to the MF (white arrowheads) express low levels of Dan and Danr. (C) Danr is not expressed, and Dan is expressed at low levels in dac³ clones that touch the margin of the disc (asterisk). Dan and Danr are expressed at highest levels in dac+ tissue (arrows) and are expressed at intermediate levels in dac³ tissue through which an MF is progressing (arrowhead).
so³ clones lying entirely within the presumptive eye field block MF progression (Pignoni et al., 1997). Clones ahead of and within the MF invariably fail to express Dan or Danr (white and yellow arrows, respectively, Fig. 6B), as do many of the larger clones behind the MF. The effects of clones ahead of the furrow on Dan and Danr may be autonomous, although in clones within or behind the furrow there appears to be a single row of nuclei, just inside the clonal boundary, that express Dan and Danr, albeit at lower levels than surrounding wild-type cells. Other generally smaller clones express Dan and possibly low levels of Danr (arrowheads, Fig. 6B). Thus, so is required for Dan and Danr expression. Ahead of the furrow so may directly regulate dan and danr expression, but within and behind the furrow other factors are likely to be involved in addition to so.
We next examined expression of Dan and Danr in homozygous clones of the null allele dac³. Clones of dac³ tissue that touch the posterior margin block MF initiation, but the MF can propagate through dac³ clones (Mardon et al., 1994). Both Dan and Danr are expressed in a normal pattern in dac+ tissue, with highest levels in and around the MF (Fig. 6C, arrow). In dac³ clones that touch the posterior margin and therefore lack an MF (Fig. 6C, asterisk), Danr is not expressed and Dan is reduced but is expressed at levels that resemble its pre-MF pattern. This indicates that dac is not required to initiate Dan expression, but is required to initiate Danr expression, and may help to increase Dan levels during MF propagation. Both Dan and Danr are expressed in dac³ tissue in which an MF is propagating, although not at highest levels (Fig. 6C, arrowheads).
Taken together, these results indicate that Dan and Danr lie downstream of so and require dac function for highest levels of expression. However, since Danr is required for highest levels of Eya expression, and since either Dan or Danr misexpression can lead to ectopic Ey expression (Fig. 1D), they are likely to participate in feedback loops that exist within the RD network.
Dan and Danr interact physically with Ey and Dac
Besides regulating each other’s expression, the RD factors Eya, So and Dac interact with one another physically (Chen et al., 1997; Pignoni et al., 1997). To determine whether Dan or Danr might be part of a protein complex (or complexes) containing the RD factors, we tested whether they could interact in vitro in a GST-pulldown assay. As shown in Fig. 7, GST alone or a GST fusion to an unrelated protein do not bind in vitro translated Dan, Danr, Ey, Eya, So or Dac. In contrast, GST fusions of both full-length Dan and full-length Danr bind to in vitro translated Dan, Ey and Dac, but do not bind Danr, Eya or So.
Fig. 7.
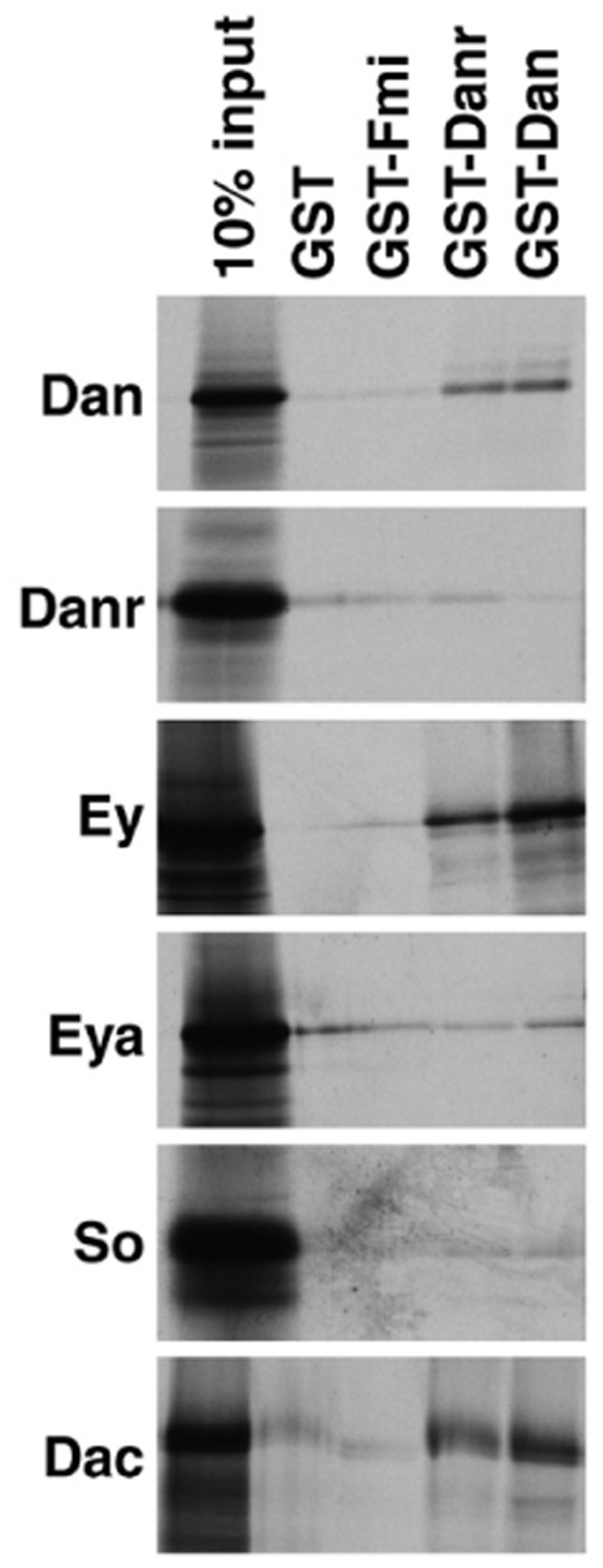
Dan and Danr interact physically in vitro with themselves and with Ey and Dac. Aliquots of in vitro-translated [35S]methionine-labeled Dan, Danr, Ey, Eya, So or Dac were incubated with glutathione-agarose beads containing bound GST, GST fused to an unrelated protein (GST-Fmi), GST-Dan and GST-Danr. Bound proteins were fractionated by SDS–PAGE and visualized by autoradiography. 10% of the input is loaded for each in vitro-translated protein for reference. Both GST-Danr and GST-Dan interact specifically with in vitro-translated Dan, Ey and Dac. For extension of this analysis by yeast two-hybrid data see Table 2.
We have confirmed these observations using yeast two-hybrid assays. We generated fusions of full-length Dan, Danr, Dac or Ey to both the Gal4 activation domain (Act) and the DNA-binding domain (DNA-BD). When transformed into yeast Dan-DNA-BD interacts with itself (Dan-Act), Danr-Act, Dac-Act and Ey-Act to activate transcription, and Danr-DNA-BD also interacts with itself, Dan-Act, Dac-Act and Ey-Act to activate transcription (Table 2). Reverse experiments using Dan-Act and Danr-Act corroborated the results obtained with Dan-DNA-BD and Danr-DNA-BD. Dan-DNA-BD or Danr-DNA-BD do not activate transcription of a Gal4 reporter gene either by themselves or when cotransformed with the Gal4 activation domain. Moreover, neither Dan-Act nor Danr-Act could activate transcription when cotransformed with murine p53-DNA-BD or human lamin C-DNA-BD fusions, confirming that these interactions are specific.
Table 2.
Dan and Danr interact with Ey and Dac in a yeast two-hybrid assay
| Dan-Act | Danr-Act | Dac-Act | Ey-Act | Act alone | |
|---|---|---|---|---|---|
| Dan-DNA-BD | ++++ | ++ | ++ | +++ | − |
| Danr-DNA-BD | + | + | + | + | − |
| Dac-DNA-BD | ++ | + | n.d. | n.d. | − |
| Ey-DNA-BD | +++ | − | n.d. | n.d. | − |
| Murine p53-DNA-BD | − | − | n.d. | n.d. | n.d. |
| Human lamin C-DNA-BD | − | − | n.d. | n.d. | n.d. |
Full-length versions of Dan, Danr, Dac and Ey were fused to both the Gal4 activation domain (Act) and to the DNA-binding domain (DNA-BD). The combinations shown above were transformed into yeast and tested for activation of both his and lacZ reporter constructs. −, no activity; +, activity; n.d., not determined. The Gal4 activation domain along (Act alone) and murine p53 or human lamin C fused to the Gal4 DNA-BD are shown as controls.
Based on these two assays, Dan and Danr are capable of direct physical interaction with two eye specification factors: Ey and Dac. In addition, both Dan and Danr can interact physically with themselves and with each other.
dan/danr misexpression interferes with eye development
A shared characteristic of the RD genes is that they can interfere with normal eye development when overexpressed. The ey-Gal4 line (Hauck et al., 1999) drives expression in a similar pattern to the ey gene: throughout the eye–antennal disc from embryogenesis through early larval stages and at strongest levels ahead of the MF in third instar eye discs. When ey-Gal4 is used to overexpress ey, eya, so or dac the resulting adult flies have eye and head defects, including rough and small eyes (Curtiss and Mlodzik, 2000). The fact that both the loss of RD gene function and RD gene overexpression lead to loss of eye development suggests that the relative amounts of the RD factors are critical for eye development.
The EP line targeting danr was originally identified based on the fact that it generated small, rough eyes when crossed to ey-Gal4. Using UAS-dan and UAS-danr, we examined the effects of dan/danr misexpression on eye development in more detail. Approximately 35% of eyes from ey-Gal4, UAS-danr/+ (ey>danr/+) individuals are rough and/or small (Figs. 8A–D). ey>danr/ey>danr flies have much smaller eyes containing only 10–100 ommatidia (not shown). Thus, misexpression of danr in the developing eye interferes with eye development.
Fig. 8.
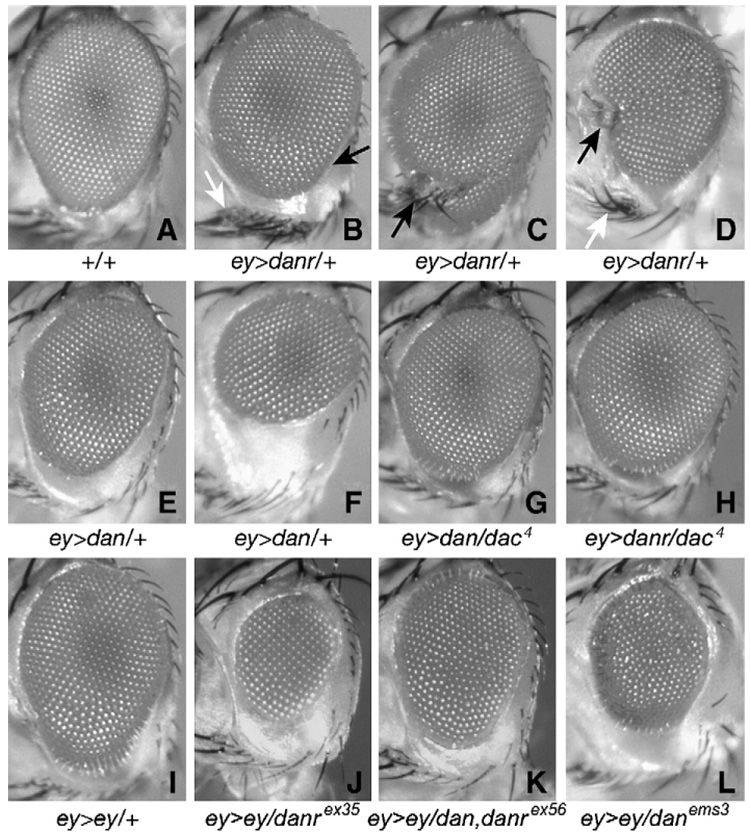
(A–F) Misexpression of dan or danr interferes with eye development. (A) Eye from an Oregon R fly. (B–D) Eyes from ey-Gal4 UAS-danr/+ flies. (E, F) Eyes from ey-Gal4 UAS-dan/+ flies. Eyes from ey>dan or ey>danr flies are small and rough, and ey>danr also causes patterning defects. (G–L) dan and danr can interact in vivo with ey and dac. Eyes from dac/+;ey-Gal, UAS-dan/+ (G) and dac/+;ey-Gal4 UAS-danr/+ (H) flies are larger and less rough than their ey-Gal4 UAS-dan/+ and ey-Gal4 UAS-danr/+ counterparts. (I) Eyes from ey-Gal4 UAS-ey/+ adults are small and rough. Eyes from ey-Gal4 UAS-ey/danrex35 (J), ey-Gal4 UAS-ey/dan danrex56 (K) and ey-Gal4 UAS-ey/danems3 (L) are even smaller and rougher.
The roughness of ey>danr/+ eyes occurs primarily near the equator and in the ventral half of the eye, and when the eye is small the ventral half is more strongly affected (Fig. 8B, arrow). In addition, patterning of other structures of the head is affected in 20% of ey>danr/+ flies. Most often (10% of eyes) this results in an increase in the number of vibrissae (Figs. 8B, D, white arrows) or orbital bristles (not shown). In approximately 5% of cases the eye appears to be split by a wedge of cuticle and bristles (Fig. 8C, black arrow). Rarely (1% of heads) antennal-like structures form in this wedge or in the region of the vibrissae (Fig. 8D, black arrow). Since ey is initially expressed in all eye disc cells at early developmental stages (Kenyon et al., 2003), it is likely that the effects on head structures other than the eye result from early danr misexpression in ventral regions of the eye disc from which these head structures are derived. Misexpression of the other RD genes, especially eya and dac, with ey-Gal4 can have similar effects on head structures other than the eye (not shown). Thus, when misexpressed during eye development, danr gives effects on eye size, smoothness and the development of other head structures that are similar to those observed as a result of RD gene misexpression.
When ey-Gal4 is used to drive UAS-dan (ey>dan/+) approximately 40% of eyes are rough and/or small (Figs. 8E, F). As with danr, disruptions in the ommatidial array in ey>dan/+ eyes occur primarily near the equator and ventrally. Unlike ey>danr/+, head structures other than the eye are rarely affected (<5%) on ey>dan/+ heads.
These results demonstrate that both dan and danr are capable of interfering with eye development in a similar way as other RD genes: both loss of dan/danr function and dan/danr overexpression lead to small eyes. One possible explanation for this is that there are critical levels of Dan and Danr expression for eye development, and deviation from those levels precludes eye development. This might occur, for instance, if Dan and Danr form part of a complex at a certain stoichiometry, and in fact we have shown that Dan and Danr can directly interact with each other and with certain RD factors (Fig. 7). However, it is important to note that Dan expression is upregulated in the absence of danr function (Fig. 3B), and it is possible that the small-eye danrex35 phenotype results from overexpression of Dan. Additional experiments will be required to distinguish among these possibilities.
dan and danr interact genetically with ey and dac
We have used the overexpression phenotypes described above to determine whether dan and danr can interact genetically with ey and dac, as they do physically. A genetic interaction between dan or danr and ey or dac would suggest that their protein products interact in vivo as well as in vitro. To test for genetic interactions between dan/danr and dac we generated both ey>dan/+ and ey>danr/+ flies in which we had reduced dac function by half (in animals heterozygous for the null dac4 allele). Both ey>dan/+ and ey>danr/+ are suppressed by dac heterozygosity (Figs. 8G, H). In ey>danr/dac4 or ey>dan/dac4 eyes the disruptions in the ommatidial array are milder and occur over a smaller percentage of the eye surface area compared to ey>danr/+ or ey>dan/+ eyes. In addition, only 10% of eyes from either ey>danr/dac4 or ey>dan/dac4 animals (Figs. 8G, H) are small, versus 35% for ey>danr/+ and 40% for ey>dan/+. Reducing dac function also strongly reduces the number of patterning defects in the heads of ey>danr/dac4 animals (down to <1% compared to 20% in ey>danr/+ animals). These observations suggest that dan and danr interact functionally with dac.
To test for genetic interactions between dan/danr and ey we generated ey>ey/+ flies in which we had reduced dan and/or danr function using the danrex35, dan danrex56 and danems3 alleles. Eyes from ey>ey/+ flies are 80%–90% the size of WT and show patches of roughness particularly in the ventral half (Fig. 8I); defects in other regions of the head, e.g. antenna, vertex, etc., are observed rarely, <5% of flies (not shown). Reducing dan/danr function in this background (Figs. 8J–L) enhances this phenotype by further reducing eye size and increasing the roughness. Heterozygosity for the danrex35, dan danrex56 and danems3 mutations all enhanced the ey>ey/+ phenotype, with danems3 having the strongest effect. Thus, dan, danr and ey functionally interact during eye development.
Note that whereas danems3 homozygotes have much weaker eye phenotypes than danrex35 and dan danrex56, and the danems3 mutation gives the weakest effect on the Elp phenotype, danems3 shows a stronger enhancement of the ey>ey phenotype (compare Fig. 8L with K). Once again, this suggests that dan and danr have distinct roles in eye development even though they appear to be redundant for antennal development. dan may have a greater function during the process of retinal determination, where its function may overlap with that of other RD factors. In conclusion, dan and danr interact genetically with RD genes, suggesting that their protein products physically interact in vivo as well as in vitro.
Discussion
Dan and Danr function in the RD hierarchy
The abilities of Dan and Danr to induce ectopic eyes, their expression patterns during eye development and the small, rough eyes of single danr- and double dan danr- mutants all point toward roles for Dan and Danr in specifying eye development in Drosophila. In particular, both Dan and Danr are required for expression of Ato, which is in turn required for the correct induction and differentiation of the R8 precursor—the founding cell of each ommatidium. In addition, Dan and Danr participate in the regulatory loops involving the specification factors Ey, Eya, So and Dac, which act at early stages in eye development. Thus, Dan and Danr are two new RD factors.
Small, rough eye dan/danr phenotypes reflect defects in R8 recruitment
Analysis of the Dan and Danr expression patterns and mutant phenotypes suggests that they function in R8 determination and subsequent recruitment of other photoreceptors. High levels of Danr overlap with the Ato expression pattern as it resolves into intermediate groups, which is the earliest patterning event in eye development (Jarman et al., 1995). Two observations suggest that danr and dan participate in R8 determination by regulating Ato expression. First, Ato expression is reduced anterior to the MF in both single and double dan and danr mutant clones. Second, gaps appear in the regular arrangement of R8 precursors in danr single and dan danr double mutant tissue in eye discs, as well as in the array of ommatidia in the adult, that are reminiscent of the phenotype of reduced-function ato alleles, where R8 cells are specified but at lower numbers (White and Jarman, 2000). Other aspects of the dan and danr eye phenotypes, such as the indiscriminate loss of photoreceptors from adult ommatidia, might also be explained by reduction of ato function: although reduced levels of ato function can allow initial specification of R8 cells, the R8 cells that form differentiate imperfectly and recruit reduced numbers of additional photoreceptors to the ommatidium (White and Jarman, 2000).
Another factor with a potential role in ommatidial recruitment/spacing, and a well-established role in photoreceptor recruitment (Freeman, 1997), is Egfr signaling. A number of groups have observed that resolution of Ato expression to single R8 cells is altered in mutant clones that affect Egfr signaling, for instance in Egfr, ras and raf null clones (Baonza and Freeman, 2001; Kumar et al., 1998; Spencer et al., 1998; Yang and Baker, 2001). Moreover, moderately increasing Egfr activity leads to a loss of Ato and an expansion of Ato inhibitors such as Rough (Lesokhin et al., 1999; Spencer et al., 1998). Thus, the fact that dan and danr single and double mutants enhance the Elp/+ phenotype provides support for the idea that both dan and danr are important for ommatidial recruitment/spacing through the regulation of Ato expression.
A number of other factors regulate ato expression, both positively and negatively. These include the Notch and Hedgehog signaling pathways, as well as the transcription factors Hairy, Extramacrochaetae (Emc) and Rough (reviewed in Frankfort and Mardon, 2002). Notch, for instance, does not initiate Ato expression ahead of the MF, but is required for so-called “proneural enhancement”. When Notch activity is lost Ato expression in the pre-proneural zone initiates normally but fails to be upregulated in the MF. In Notch- clones the low levels of Ato expression never resolve into intermediate groups and no R8s ever form (Baker and Yu, 1997). Hairy and Emc repress Ato expression anterior to the MF, and Notch is thought to repress Hairy and Emc, in turn, for proneural enhancement to occur (Baonza and Freeman, 2001). Dan and Danr may directly regulate Ato expression or they may act through these or other factors. Additional experiments are required to distinguish among these possibilities.
The two additional phenotypes observed in dan/danr mutant sections – the presence of more than one “inner” photoreceptor and the mild polarity defects – seem unrelated to the others. Extra “inner” photoreceptors can result from a conversion of “outer” to “inner” photoreceptor fate, as observed in seven-up single and seven-up,rough double mutants (Heberlein et al., 1991; Mlodzik et al., 1990). Alternatively, they might result from effects on later aspect of differentiation of the photoreceptors, including rhabdomere differentiation: opposite effects have been observed in mutants affecting the spalt gene complex, in which “inner” photoreceptors have the large rhabdomeres of “outer photoreceptors” (Mollereau et al., 2001). Currently, we cannot distinguish between these hypotheses. It is possible that Dan/Danr have auxiliary roles in multiple processes, perhaps by acting as chromatin modification factors (see below). Alternatively, they might be partially redundant with other factors.
Dan and Danr have a complex relationship during eye development
The similarity in sequence and expression patterns of Dan and Danr suggests that they are closely related in function. In the antenna Dan and Danr appear to act redundantly: Dan and Danr have identical expression patterns, their mutant phenotypes are similar, and dan danrex56 antennae have a stronger phenotype than either danrex35 or danems3 (Emerald et al., 2003). In the eye, however, Dan and Danr expression patterns differ within and posterior to the MF (Figs. 2B–D) and the danrex35 single mutant phenotype is more severe than the double mutant dan danrex56 phenotype. In addition to being null for both dan and danr, the dan danrex56 deletion removes two uncharacterized potential genes (CG34110 and CG13652). It is formally possible that one or both of these genes is involved in eye development and could affect the double mutant dan danr phenotype. However, given the fact that dan and danr regulate each other’s expression (Fig. 3) and are capable of direct physical interaction, it is much more likely that dan and danr have complex and interrelated functions in eye development.
For instance, Dan and Danr might have different and antagonistic roles in regulating Ato expression. As described above, Ato expression is highly dynamic, with different sets of factors regulating its expression at different stages. The reduction in Ato appears to differ subtly among the dan and danr single and double mutants (Figs. 4F–H): only very early stages of Ato expression are affected in danems3 clones, but danrex35 or dan danrex56 clones show reduction over most of the Ato expression pattern. The fact that the danrex35 phenotype is stronger than the dan danrex56 double could indicate that, following its positive role in initiating Ato, dan switches to a negative role, perhaps to keep Ato levels from getting too high. In the danrex35 null, the phenotype would result not only from loss of danr, but also from the negative effect of dan on later Ato expression. The weaker phenotype of the double dan danrex56 null would reflect the loss of the negative effect of dan on the later stages of Ato expression.
This scenario, in which dan and danr have antagonistic roles in regulating Ato expression, could be mediated in several different ways. For instance, antagonism between dan and danr is consistent with the fact that Danr negatively regulates Dan expression (Fig. 3B) and that misexpression of Dan during eye development leads to a reduction in eye size similar to that observed in danrex35 homozygotes (Figs. 8E, F). On the other hand, Danr expression is reduced in danems3 clones (Fig. 3D) which suggests a positive rather than an antagonistic relationship. However, this effect is limited to the region anterior to the MF and therefore could result from the earliest stages of dan function, when we hypothesize that it is promoting Ato expression/eye development. We have also shown that Dan and Danr are capable of direct physical interaction (Fig. 7 and Table 2), which suggests another mechanism via which Dan and Danr could interact that could be independent of their effects on each other’s expression patterns (see below).
Dan and Danr lie downstream of Dac in the retinal determination network
A number of findings suggest that dan and danr function in the RD transcriptional hierarchy. Dan and Danr are able to transform antennal precursors to an eye fate, are required for normal eye development and can disrupt eye development when misexpressed, phenotypic effects that are similar to those caused by other RD factors. RD factors including Eya, So and Dac are required for Dan and Danr expression. Conversely, Danr is required for strong Eya expression, and both Dan and Danr are able to promote Ey expression, suggesting that they play a role in the RD network feedback loops. Finally, Dan and Danr interact physically and genetically with the RD factors Ey and Dac.
The loss-of-function phenotypes of the single and double dan and danr mutants are not as striking as those of other RD factors. This may reflect a position for dan and danr further downstream in the RD hierarchy. A hypothetical transcriptional cascade would consist of one or a few genes at the top that control expression of multiple genes, which in turn regulate multiple effectors. At each level below the top, function is likely to be split among the effectors and may be overlapping or redundant. Thus, toy and ey at the top of the RD hierarchy appear to govern development of the whole head (Kronhamn et al., 2002). They activate eya and so, which control some if not all aspects of eye development: growth, MF initiation and progression, and neuronal development (Pignoni et al., 1997). Slightly further down the hierarchy, dac controls only a subset of these functions. For instance, growth of the eye field proceeds normally in dac- tissue but the MF does not initiate. If an MF initiates in surrounding wild-type tissue (for instance in a dac- mosaic disc) it can progress through dac- tissue to form ommatidia, but there are defects in ommatidial cell number and organization (Mardon et al., 1994). Moreover, by itself dac induces ectopic eye formation at a much lower frequency than does ey (cf. Chen et al., 1999). This suggests that eya and so control MF initiation and aspects of ommatidial differentiation through dac, but that they also regulate other genes besides dac that carry out their other functions for growth and MF progression. Likewise, dan and danr may control a subset of events in retinal determination that occur downstream of dac and may be redundant with other factors for another subset, as revealed by the fact that the danems3 mutation has little effect on its own but can interact genetically with other mutations that affect eye development (Fig. 5G).
Do Dan and Danr interact with Ey and/or Dac to regulate ato expression?
We have shown that Dan and Danr can physically interact with both Ey and Dac in GST-pulldowns and in yeast two-hybrid assays. What might be the significance of these interactions? The RD protein Eya has previously been shown to interact with Dac and with So (Chen et al., 1997; Pignoni et al., 1997). More recently, Ey and So have been shown to interact with each other in the context of regulating ato expression (Zhang et al., 2006). Hence, existing evidence indicates that the RD factors function in one or more complexes of which Dan and Danr might also be a part. For instance, Dan and Danr might participate with Ey in a complex that regulates ato expression. The members of this complex may require a particular stoichiometry with respect to one another, such that altering the concentration of a particular member results in a loss of ato expression and therefore a detrimental effect on eye development. This could explain why ey, dac, dan or danr all interfere with eye development when misexpressed.
A model based on this type of interaction would still have to explain why the danrex35 phenotype is more severe than that of dan danex56. This could occur, for instance, if Dan and Danr have slightly different and antagonistic functions within the complex depending on the stage of Ato expression, as described above. Alternatively, Dan and Danr could form an obligate heterodimer whose function is partially redundant. In this case if both Dan and Danr are missing, as in the dan danrex56 mutant, then another factor might be able to substitute; if Danr alone is lost, as in the danrex35 mutant, the presence of Dan might interfere with the function of the complex and/or prevent a redundant factor from entering into the complex. The danems3 mutant might have a weaker effect either because it is not a null mutation or because Danr does not interfere with complex function in the absence of Dan or both.
The genetic interactions between ey, dac, dan and danr (Fig. 8) confirm that ey, dan and danr, and dac, dan and danr, whose protein products are capable of direct interaction, are important in the same process(es) during eye development. Although given our current knowledge they do not clarify the relationships among these genes, the nature of these interactions also is not inconsistent with the model presented above. For instance, the fact that a 50% reduction of dac function suppresses the effects of dan or danr misexpression could indicate that Dan, Danr and Dac are all required in the same complex to affect a particular aspect of eye development (such as Ato expression). Reducing levels of any of the three would then be predicted to suppress the effects of misexpressing any of the three. Consistently, we have preliminary evidence that dan and danr mutations also suppress the effects of misexpressing Dac, with dan danrex56 giving the best suppression.
An alternative explanation for why dac mutations suppress ey>dan and ey>danr would be based on the possibility that overexpression of dan or danr leads to an upregulation of dac. In this case, the small, rough eyes of ey>dan and ey>danr would result from an overexpression of Dac, which has already been shown to cause a similar phenotype when overexpressed. Reducing Dac levels with a null mutation would then be expected to suppress the ey>dan or ey>danr phenotypes. However, in fringe-Gal4 UAS-danr individuals, where danr is misexpressed in the ventral half of the eye disc, including ahead of, within and behind the morphogenetic furrow, we can detect no upregulation of Dac protein. This suggests that an increase in Dac expression is not the basis for the small, rough eyes resulting from ey>danr, nor for the genetic interactions observed between dan, danr and dac. In addition, a suppression of dac misexpression by dan and danr mutations is not consistent with this model (see above). For these reasons we favor the former model.
In contrast to the suppression of ey>dan or ey>danr by dac mutations, reducing Dan or Danr levels enhances the effects of ey misexpression, and we have preliminary evidence that reducing Ey levels also enhances the effects of dan or danr misexpression. This suggests that Dan and Danr have a different relationship with Ey than they do with Dac, or that Ey, Dan and Danr participate in a different process than Dac, Dan and Danr or both. It may be informative that the danems3 mutation enhances ey misexpression better than the double dan danrex56 mutation, even though danems3 has the weakest effect in every other context we have observed. ey promotes early stages of ato expression (Zhang et al., 2006). However, if misexpression of ey disrupts the stoichiometry of a complex responsible for early stages of ato expression, the result could be a dominant negative effect and a reduction in ato, leading to a smaller, rougher eye. Reducing dan or danr, which are also required for ato expression, might be expected to reduce ato expression still further, enhancing the ey misexpression phenotype. If both ey and dan are required for the initial stages of ato expression, as described above, whereas danr is required for later stages, then mutations in dan would enhance the ey misexpression phenotype more than the danr single or the double mutation.
Alternatively, it is possible that misexpressing ey upregulates ato. If, as we have speculated above, dan plays a later role in downregulating ato, then reducing dan function with danems3 would increase ato expression even more, enhancing the ey misexpression phenotype. The double dan danrex56 mutation would reduce a potential upregulator (danr) and a potential downregulator (dan), canceling each other out and explaining why the double mutation has a weaker effect on the ey misexpression phenotype. However, this model fails to explain why the danrex35 single mutation also enhances the ey misexpression phenotype, or why ey mutations enhance dan and danr misexpression. Obviously, there are many other possible interpretations of this data. A complete understanding will require both molecular and phenotypic analyses.
dan and danr link retinal determination with differentiation
Although the mild dan/danr mutant phenotypes may make the dan/danr genes appear to be less important for retinal determination, these phenotypes may be especially useful in discovering how retinal determination leads to differentiation. For instance, signaling factors involved in differentiation, like Egfr and others that determine the pattern of R8 formation and induce the recruitment of the R1–R7 photoreceptors, are involved in development of many structures. At some level the RD factors must inform the activities of the signaling factors so that they initiate eye-specific differentiation (as opposed to wing or other structures). However, the mechanisms by which this occurs are unclear. The loss-of-eye phenotypes observed in eya- and so- mutants are so severe that it is difficult to determine how they interact with factors involved in eye differentiation (cf. Pignoni et al., 1997). On the other hand, by analyzing the dan/danr phenotypes we have already discovered connections between the RD network on the one hand and Ato expression and Egfr activity on the other.
One potential mechanism by which dan and danr could connect the RD factors with Egfr and other factors that promote retinal differentiation is suggested by the fact that Dan and Danr contain a “pipsqueak (psq) motif” (Emerald et al., 2003), a helix–turn–helix structure involved in DNA binding found in proteins implicated in chromatin modification (Siegmund and Lehmann, 2002). Once determination of a particular tissue type has occurred, it must be maintained throughout the differentiative period and the rest of the life of the animal. This occurs via changes in the chromatin conformational state that are mediated in all metazoans by the Trithorax and Polycomb groups of proteins (Ringrose and Paro, 2004). Thus, establishment of particular chromatin conformational states is one way to link determination with differentiation.
Notably, several genes encoding Psq domain proteins have homeotic phenotypes and affect gene silencing. These include bric à brac (bab) (Chu et al., 2002; Couderc et al., 2002; Godt et al., 1993) and the eponymous pipsqueak (psq). Psq interacts physically with a complex containing the Polycomb group proteins (Pc-G), and the psq and Pc-G genes interact genetically to cause misexpression of homeotic genes (Huang et al., 2002).
This connection with homeosis and silencing is reminiscent of the effects of Dan/Danr to make what appear to be homeotic changes, converting the fate of antennal precursors to that of an eye (Fig. 1; Suzanne et al., 2003). One possibility is that Dan and Danr are chromatin modifiers that function in eye development. In this model, their expression would be activated by the RD factors. Once expressed, they would bind to Ey and Dac (or other factors in the antenna), perhaps in the context of larger complexes, to contribute to an eye-specific “opening up” of the chromatin to allow easier access to eye-specific genes. Future studies will reveal more about these mechanisms and promote understanding of how specification factors interact with differentiation factors during development.
Acknowledgments
We thank A. Jenny for advice on GST-pulldowns, G. Rubin for anti-Elav, A. Jarman for anti-Ato, U. Walldorf for anti-Ey, B. Dickson for eyFLP, J. Treisman for FRT82,armlacZ,M and G. Mardon for dacP and FRT42,dac³. Confocal laser scanning microscopy was performed at the MSSM-Microscopy Shared Research Facility, supported with funding from NIH-NCI shared resources grant (1 R24 CA095823-01) and NSF Major Instrumentation grant (DBI-972 4504). We thank S. Henderson for help with confocal microscopy. This work was supported in part by a NIH-NEI Grant (R01 EY14597) to M.M., and by NIH-NIGMS Grant S06 GM003136-33.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2007.04.006.
References
- Baker NE, Rubin GM. Effect on eye development of dominant mutations in Drosophila homologue of the EGF receptor. Nature. 1989;340:150–153. doi: 10.1038/340150a0. [DOI] [PubMed] [Google Scholar]
- Baker NE, Rubin GM. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev. Biol. 1992;150:381–396. doi: 10.1016/0012-1606(92)90250-k. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr. Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr. Biol. 1997;7:122–132. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128:3889–3898. doi: 10.1242/dev.128.20.3889. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Chen CK, Chien CT. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5055–5060. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and Eyes Absent proteins form a complex and function synergistically to Induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Chen R, Halder G, Zhang Z, Mardon G. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- Cheyette BNR, Green PJ, Martin K, Hideki G, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chu J, Dong PD, Panganiban G. Limb type-specific regulation of bric a brac contributes to morphological diversity. Development. 2002;129:695–704. doi: 10.1242/dev.129.3.695. [DOI] [PubMed] [Google Scholar]
- Couderc JL, Godt D, Zollman S, Chen J, Li M, Tiong S, Cramton SE, Sahut-Barnola I, Laski FA. The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development. 2002;129:2419–2433. doi: 10.1242/dev.129.10.2419. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog, and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Emerald BS, Curtiss J, Mlodzik M, Cohen SM. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development. 2003;130:1171–1180. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–1306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Godt D, Couderc JL, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech. Dev. 1998;74:121–131. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1999;96:564–569. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U, Mlodzik M, Rubin GM. Cell-fate determination in the developing Drosophila eye: role of the rough gene. Development. 1991;112:703–712. doi: 10.1242/dev.112.3.703. [DOI] [PubMed] [Google Scholar]
- Huang DH, Chang YL, Yang CC, Pan IC, King B. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol. Cell. Biol. 2002;22:6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Jenny A, Reynolds-Kenneally J, Das G, Burnett M, Mlodzik M. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 2005;7:691–697. doi: 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Kronhamn J, Frei E, Daube M, Jiao R, Shi Y, Noll M, Rasmuson-Lestander A. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development. 2002;129:1015–1026. doi: 10.1242/dev.129.4.1015. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lehmann M. Anything else but GAGA: a nonhistone protein complex reshapes chromatin structure. Trends Genet. 2004;20:15–22. doi: 10.1016/j.tig.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Lesokhin AM, Yu SY, Katz J, Baker NE. Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev. Biol. 1999;205:129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1–Dach–Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rorth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, de Celis JF, Desplan C. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C, Greenwood S, Struhl G. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the Eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Punzo C, Seimiya M, Flister S, Gehring WJ, Plaza S. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129:625–634. doi: 10.1242/dev.129.3.625. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. Sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines new family of helix–turn–helix DNA-binding proteins. Dev. Genes. Evol. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Frey RA, Prabhudesai SN, Raymond PA. Function for Hedgehog genes in zebrafish retinal development. Dev. Biol. 2000;220:238–252. doi: 10.1006/dbio.2000.9629. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Suzanne M. Expression analysis of the Drosophila pipsqueak family members fernandez/distal antenna and hernandez/distal antenna related. Dev. Dyn. 2004;230:361–365. doi: 10.1002/dvdy.20046. [DOI] [PubMed] [Google Scholar]
- Suzanne M, Estella C, Calleja M, Sanchez-Herrero E. The Hernandez and fernandez genes of Drosophila specify eye and antenna. Dev. Biol. 2003;260:465–483. doi: 10.1016/s0012-1606(03)00249-5. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Bowtell DD, Hafen E, Rubin GM. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Vincent JP, Girdham CH, O’Farrell PH. A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev. Biol. 1994;164:328–331. doi: 10.1006/dbio.1994.1203. [DOI] [PubMed] [Google Scholar]
- White NM, Jarman AP. Drosophila atonal controls photoreceptor R8-specific properties and modulates both receptor tyrosine kinase and Hedgehog signalling. Development. 2000;127:1681–1689. doi: 10.1242/dev.127.8.1681. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Martinez-Arias MBA, editor. The Development of Drosophila melanogaster. Cold Spring Harbor: Cold Spring Harbor Press; 1993. pp. 1277–1326. [Google Scholar]
- Xu T, Rubin GM, Greenwood S, Struhl G. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]


