SUMMARY
A remarkable problem in neurobiology is how olfactory receptor neurons (ORNs) select, from among a large odor receptor repertoire, which receptors to express. We use computational algorithms and mutational analysis to define positive and negative regulatory elements that are required for selection of odor receptor (Or) genes in the proper olfactory organ of Drosophila, and we identify an element that is essential for selection in one ORN class. Two odor receptors are coexpressed by virtue of the alternative splicing of a single gene, and we identify dicistronic mRNAs that each encode two receptors. Systematic analysis reveals no evidence for negative feedback regulation, but provides evidence that the choices made by neighboring ORNs of a sensillum are coordinated via the asymmetric segregation of regulatory factors from a common progenitor. We show that receptor gene choice in Drosophila also depends on a combinatorial code of transcription factors to generate the receptor-to-neuron map.
Keywords: Olfaction, Drosophila, odor receptor gene, antenna
INTRODUCTION
One of the most intriguing problems in sensory neurobiology is the problem of odor receptor gene choice. How do individual ORNs select, from among a large repertoire, which receptor genes to express? The problem is formidable in dimension: a large number of receptor genes must be expressed in a large number of distinguishable ORN classes. There is a numerical constraint, in that most ORNs are believed to select a single receptor gene. Moreover, many receptors are expressed in prescribed spatial domains of olfactory fields. The complexity of the conditions presents a challenging problem of regulatory biology (Mombaerts, 2004).
Drosophila provides a particularly interesting system in which to address this problem because its olfactory system has been described in great detail at both cellular and molecular levels and is striking in its complexity and precision of organization (Dahanukar et al., 2005; Hallem et al., 2006). The fly contains two pairs of olfactory organs, the antennae and maxillary palps (Figure 1A). Each organ is covered with olfactory sensilla, ∼400 on the antenna and ∼60 on the maxillary palp, and each sensillum contains the dendrites of up to four neurons, usually two (Shanbhag et al., 1999). Physiological analysis has identified 35 functional classes of ORNs, organized in 17 types of sensillum in stereotyped combinations (Clyne et al., 1997; de Bruyne et al., 1999; de Bruyne et al., 2001; Elmore et al., 2003). Different types of sensilla, and thus the ORN classes they contain, are intermingled on the antennal surface, but each type is restricted to a particular spatial domain.
Figure 1.
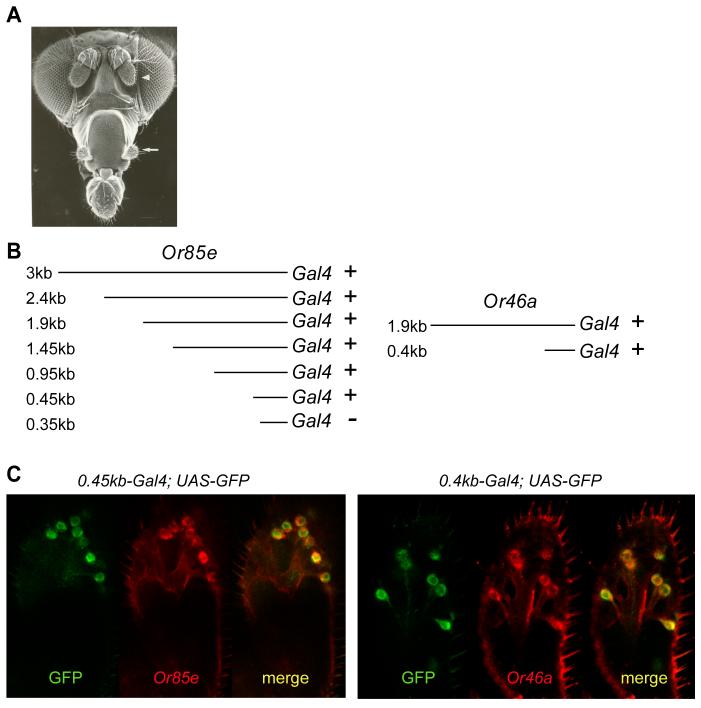
Essential Or regulatory sequences lie within 500 bp of the ATG. (A) Olfactory organs. Arrowhead indicates antenna; arrow indicates maxillary palp. Adapted from (Carlson, 1996). (B) Deletion series of the upstream sequences of Or85e (left) and Or46a (right). X-Gal staining in the maxillary palp is indicated by a “+” or “−”. (C) Confocal micrographs of maxillary palps containing UAS-GFP and Or85e 0.45kb-GAL4 (left) or Or46a 0.4kb-GAL4 (right), which have been co-stained for GFP (green) and their respective RNAs (red). (D) Identification of overrepresented motifs. The positions of two overrepresented motifs upstream of Or genes expressed in the maxillary palp. The numbers of expected and observed occurrences are indicated in the table. Motifs present in the forward strand are shown above each line; those in the reverse strand are shown below each line.
Drosophila contains a family of 60 Or genes (Clyne et al., 1999a; Robertson et al., 2003; Vosshall et al., 1999), each expressed in a subset of ORNs. Genetic analysis has shown that the expression of an Or gene imparts the functional specificity of an ORN in vivo (Dobritsa et al., 2003; Hallem et al., 2004). One subset of Or genes is expressed in the maxillary palp, and another subset is expressed in the antenna (Goldman et al., 2005; Vosshall et al., 2000). Receptor-to-neuron maps were established initially by physiological analysis (Goldman et al., 2005; Hallem et al., 2004), and were subsequently confirmed and extended by molecular studies, which also produced a map of the stereotyped axonal projections into the brain (Couto et al., 2005; Fishilevich and Vosshall, 2005). The maxillary palp contains three types of sensilla, pb1 (palp basiconic 1), pb2, and pb3, housing six classes of ORNs: pb1 contains pb1A and p1B; pb2 contains pb2A and pb2B; pb3 contains pb3A and pb3B (de Bruyne et al., 1999). While five of the six ORN classes express a single Or gene, one class, pb2A, coexpresses two Or genes (Goldman et al., 2005). Further analysis of the entire olfactory system (Couto et al., 2005; Fishilevich and Vosshall, 2005) demonstrated that of 36 ORN classes shown to express Or genes, nine ORN classes (25%) coexpressed two Or genes, and a tenth coexpressed an Or gene and a member of the Gr gene family (Clyne et al., 2000), which encodes a family of taste receptors (Dahanukar et al., 2001). These numbers do not take into account one ubiquitously expressed Or gene, Or83b (Larsson et al., 2004).
Odor coding in Drosophila depends on this diverse population of ORN classes, all of which depend on the choice of particular Or genes (Hallem and Carlson, 2004). The precision of the receptor-to-neuron map implies that the process of receptor gene choice is highly ordered. The division of Or genes between the antenna and maxillary palp implies a mechanism for distinguishing between these two classes of receptor genes. Within an organ, the highly stereotyped organization of ORN classes requires the deployment of a program that is exceptionally rich in information.
Another corollary of the precision of the receptor-to-neuron map in Drosophila is that the logic of receptor gene regulation in the fly is likely to differ from that in mammals. A stereotyped receptor-to-neuron map has not been found in mammals, and in fact a stochastic mechanism of receptor gene choice has recently been identified in the mouse (Lomvardas et al., 2006). Receptor gene regulation is also under different constraints in mammals and flies: the proper expression of receptor genes is required for normal ORN axon targeting in mammals but not in flies (Feinstein et al., 2004; Dobritsa et al., 2003). In mammals a negative feedback mechanism has been proposed, by which the expression of a functional receptor in an ORN would block the expression of a second receptor in that ORN (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). In flies there is evidence against such a model, a model that is systematically tested and refuted here through ectopic receptor expression experiments.
Here we provide a molecular analysis of the logic underlying receptor gene choice in Drosophila. We use computational algorithms, coupled with mutational analysis, to identify essential regulatory elements that dictate in which ORN a particular Or gene is expressed. We focus initially on the maxillary palp on account of its numerical simplicity, and we find that the sequences required for receptor gene choice lie close to the Or genes, in most instances near the 5' end but in some cases near the 3'end. The algorithms identify a dyad element that acts to promote expression of Or genes in the maxillary palp, and a motif that acts to repress their expression in the antenna. We then define a second level of regulation: we identify an element required for expression of a particular receptor gene in its proper ORN class. To test the generality of this finding, we use computational analysis to identify neuron-specific motifs for genes expressed in seven additional ORN classes. Analysis of another maxillary palp Or locus reveals that two coexpressed receptors are produced by alternative splicing of a single gene. We also find instances in which two receptors are encoded on a single dicistronic mRNA; in one case the two receptors are members of different families, the Or family of odor receptors and the Gr family of taste receptors. We next describe systematic Or overexpression experiments, which do not reveal a negative feedback mechanism, as has been proposed in mammals (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). However, systematic analysis reveals that receptor gene choice in neighboring ORNs depends on the asymmetric segregation of regulatory factors in a common progenitor cell. We find that the Lozenge transcription factor is required for the activation of a subset of Or genes, and that different ORNs differ in their dependence on Lz and on another transcription factor, Abnormal chemosensory jump 6 (Acj6). Our results, taken together, support the conclusion that Drosophila, which contains a stereotyped receptor-to-neuron map that has not been found in mammals, does not rely upon a stochastic mechanism such as that identified in mammals (Lomvardas et al., 2006). Rather, it relies on several deterministic mechanisms, including a combinatorial code of transcription factors and regulatory elements, to produce its remarkably complex and precise organization.
RESULTS
Sequences that dictate expression in a specific ORN lie close to an Or gene
In order to identify sequences that dictate the expression of Or genes in particular ORNs, we first sought to define the minimal promoter region that is sufficient to drive faithful expression of an Or gene. We initiated this analysis with the Or85e gene. We had earlier found that the 3.1 kb genomic sequence upstream of the Or85e translational start site was capable of driving faithful expression of the yeast GAL4 gene in pb2A ORNs, as indicated by the expression of UAS-lacZ and UAS-GFP reporter genes that are activated by GAL4 (Goldman et al., 2005).
We generated a series of deletions of this 3.1 kb region, each deletion progressively removing an additional ∼500 bp from the 5' end of the region (Figure 1B). Each truncated Or85e-GAL4 construct was then used to drive expression of UAS-lacZ and UAS-GFP reporters, and at least five independent lines were examined for each deletion construct. We found that a construct containing only 450 bp upstream of the Or85e translational start site, and all larger constructs in the series, gave expression patterns similar to that of the initial 3.1 kb construct (Figures 1B,C). However, a construct containing only 350 bp of upstream DNA showed no expression. To determine whether the cells labeled by the Or85e-GAL4 construct containing 450 bp of upstream DNA are in fact of the pb2A class, we carried out a double-labeling experiment (Figure 1C). We found that all GFP+ cells were also labeled with an in situ hybridization probe for Or85e RNA; moreover, nearly all cells labeled with the Or85e RNA probe were also GFP+. The simplest interpretation of these results is that 450 bp of DNA upstream of the Or85e translational start site contain the information that dictates specific expression in the pb2A neurons.
We were surprised that the information dictating expression in a single ORN class was so economically packed. To determine whether the organization of the Or85e gene is representative of other Or genes, we analyzed another Or gene, Or46a, which is expressed in the ORN that neighbors pb2A, pb2B (Goldman et al., 2005). Consistent with the results found for Or85e, we found that 400 bp upstream of the Or46a translational start site is sufficient to confer specific expression in the pb2B class of ORN (Figures 1B,C).
To determine the distance between the transcriptional start site and the translational start site, we carried out rapid amplification of 5' cDNA ends (5' RACE) from maxillary palp RNA, for both Or85e and Or46a, as well as for two other Or genes, Or59c and Or71a. In all cases, the predicted transcriptional start sites, as determined by the longest RACE products, lie within 50 bp of the predicted translational start sites.
Sequence elements overrepresented upstream of maxillary palp Or genes
To investigate the mechanism by which the selection of an Or gene in an olfactory organ is restricted to an organ-specific subset of Or genes, we asked whether there are regulatory elements that are shared among maxillary palp Or genes, but not antennal Or genes. Such elements might dictate the organ-specific expression of the maxillary palp genes, perhaps by binding maxillary palp-specific transcription factors. We initially focused on the 500 bp upstream of the translational start sites, since regions of this size were sufficient to confer faithful GAL4 expression patterns in the cases analyzed. As a first means of searching for sequence elements shared among maxillary palp Or genes, we generated sequence alignments, but this analysis revealed remarkably little conservation among the 500 bp regions upstream of the 7 maxillary palp Or genes, much less than has been observed upstream of mammalian OR genes (Vassalli et al., 2002). We therefore turned to some powerful computational algorithms to identify sequence motifs shared among maxillary palp Or genes. We used algorithms that detect both of the major types of short DNA elements to which transcription factors bind: unipartite, or oligonucleotide, motifs and bipartite motifs. Specifically, we used OLIGO-ANALYSIS (van Helden et al., 1998), which searches for oligonucleotide motifs of 6-8 nucleotides shared by co-regulated genes, and DYAD-ANALYSIS (van Helden et al., 2000), which searches for shared bipartite sequence motifs consisting of either two trimers or two tetramers, separated by 1-20 nucleotides. We used these algorithms to seek motifs that are overrepresented upstream of maxillary palp Or genes, as compared to upstream of all annotated protein-coding Drosophila genes (n∼13,400; Misra et al., 2002), but that are not overrepresented upstream of antennal Or genes.
We identified a bipartite motif we named Dyad-1 that consists of two trimers, CTA and TAA, separated by nine nucleotides, i.e. CTA(N9)TAA. This motif was of special interest in that all seven maxillary palp Or genes contain at least one iteration of this sequence in the 500 bp region upstream of the predicted translation start site, and most of these genes contain more than one (Figure 1D). The frequency of occurrence upstream of maxillary palp Or genes exceeds that found upstream of the ensemble of Drosophila genes by a factor of 7.2. By contrast, its frequency upstream of 32 Or genes shown to be expressed in the antenna (Vosshall et al., 2000) exceeds that of the ensemble of Drosophila genes by a factor of only 1.2.
We also identified a heptamer sequence, CTTATAA, which we named Oligo-1. This motif attracted our attention, in part because it contains a 6 bp palindromic core sequence, which is characteristic of many transcription factor binding sites. The frequency of occurrence of Oligo-1 in the 500 bp upstream region of maxillary palp Or genes exceeds that of the ensemble of Drosophila genes by a factor of 10.0. By contrast, upstream of the antennal Or genes it is overrepresented by a factor of only 1.3. We also calculated the degree of overrepresentation using a different approach: we compared the iteration frequency in the 500 bp upstream region of each maxillary palp Or gene to that in the entirety of Drosophila noncoding genomic DNA. We found that by this measure, Dyad-1 is overrepresented by a factor of 13.7 upstream of the maxillary palp genes, and Oligo-1 is overrepresented by a factor of 10.0. By contrast, these elements are overrepresented by factors of only 2.0 and 1.0, respectively, upstream of antennal Or genes.
Thus these two sequence elements are present at unexpectedly high frequencies upstream of maxillary palp Or genes, but not antennal Or genes, suggesting the possibility that these sequences play a role in the process by which the choice of Or genes by ORNs is restricted to an organ-specific subset.
Dyad-1 is required for Or expression in the maxillary palp
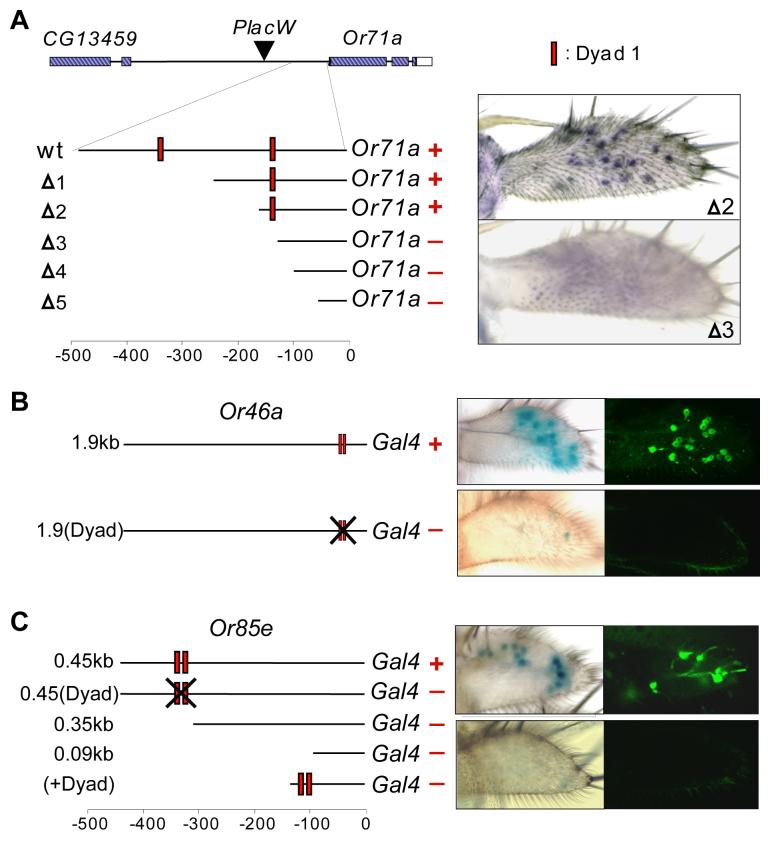
We have tested the function of Dyad-1 elements in the context of three Or genes, using two approaches. First, we generated a series of deletions in the Or71a locus by imprecise excision of a P element located 1.0 kb upstream of the Or71a translational start site (Figure 2A). Chromosomes retaining 170 bp or more of the most proximal upstream DNA sequences (Δ1 and Δ2 in Figure 2A) continued to express Or71a, as determined by in situ hybridization with an Or71a probe. Chromosomes that contain 121 bp or less of this sequence (Δ3, Δ4, and Δ5) did not express Or71a. There are two Dyad-1 elements upstream of Or71a; the chromosomes that retained one of these elements therefore retained Or71a expression, but those that retained no Dyad-1 elements lost expression.
Figure 2.
Dyad-1 is necessary for expression of Or genes in the maxillary palp. (A) Deletions of the Or71a locus were generated by imprecise excision of a P element ∼1 kb upstream. Expression of Or71a (“+”or “−”) was assayed by in situ hybridization. Staining for Or71a mRNA in the maxillary palps of Δ2 and Δ3 is shown. (B) Light and confocal micrographs of maxillary palps containing UAS-lacZ or UAS-GFP and the wild type Or46a 1.9kb-GAL4 (upper panels) or Or46a 1.9(Dyad)-GAL4 (lower panels). (C) Light and confocal micrographs of maxillary palps containing UAS-lacZ or UAS-GFP and wild-type Or85e 0.45kb-GAL4 (upper panels) or mutant Or85e 0.45(Dyad)-GAL4 (lower panels). At least 4 independent transgenic lines were tested for each construct. GFP staining in the maxillary palp is indicated by a “+”or “−”, in flies that contain Or-GAL4 and the UAS-GFP transgene.
Second, we mutated the Dyad-1 elements located upstream of Or46a. Specifically, a 1.9 kb region of DNA upstream of Or46a has previously been shown to drive expression of GAL4 in pb2B ORNs of the maxillary palp (Goldman et al., 2005). Within this 1.9 kb region are two Dyad-1 elements, located 5 bp apart. We tested a construct that carries alterations in the sequences of both elements and the spacing between them, and found that this mutated construct no longer drives expression in the maxillary palp (Figure 2B).
Third, we tested the function of the Dyad-1 elements at Or85e using both ablation and deletion approaches. We used the 0.45 kb Or85e-GAL4 construct described above (Figure 1B) and mutated base pairs of each of its two Dyad-1 elements without altering their relative positions. This ablation abolished expression in the maxillary palp (Figure 2C). Truncated constructs that do not contain Dyad-1 elements did not drive expression (Figures 2C and 1B). The shortest of these constructs contains 90 bp of sequence, which is predicted to contain the basal promoter of the Or85e gene (see www.fruitfly.org/seq_tools/promoter.html for the neural network promoter prediction algorithm). We added the two Dyad-1 sequences to the 90 bp construct; the addition of these Dyad-1 elements did not restore expression in the maxillary palp (not shown; we note that the 90 bp sequence is shown below in Figure 4C to drive expression when different elements are added to it).
Figure 4.
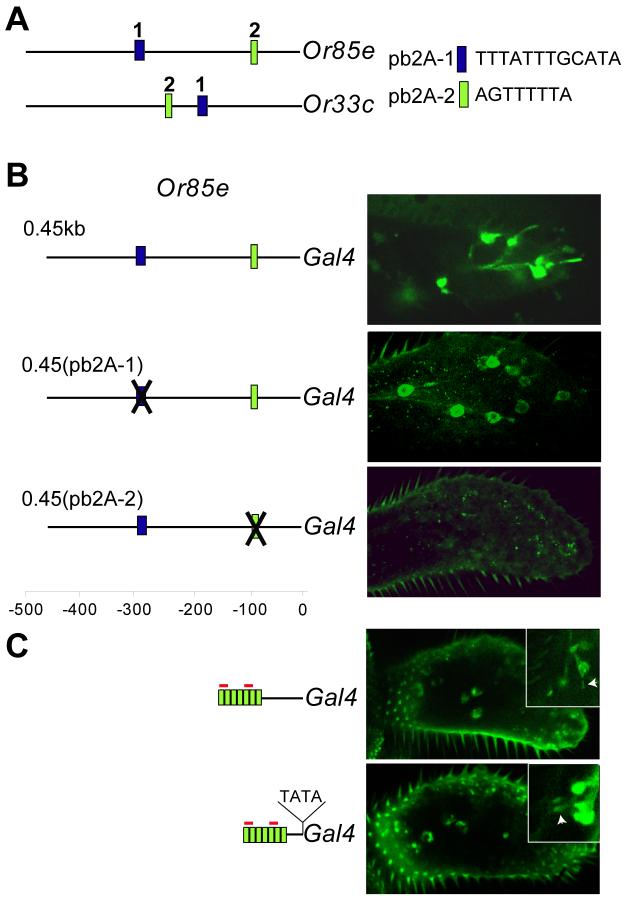
pb2A-2 is a neuron-specific positive regulatory element. (A) The pb2A-1 and pb2A-2 elements, indicated as “1” and “2”, respectively. (B) Mutational analysis of the Or85e promoter. Micrographs show maxillary palps of flies containing UAS-GFP and Or85e 0.45kb-GAL4 (top), Or85e(pb2A-1)-GAL4 (middle), and Or85e(pb2A-2)-GAL4 (bottom). At least 9 independent transgenic lines were tested for each of the mutant constructs. (C) Seven copies of the pb2A-2 element (green boxes) fused to the Or85e 90 bp basal promoter (above) or a TATA box (below) drives expression in the maxillary palp. Dyad-1 sequences in the forward strand are indicated in red. Confocal micrographs show maxillary palps with each GAL4 driver. Insets show images of neurons taken at higher magnification; arrowheads indicate dendrites extending from neuronal cell bodies. Multiple copies of pb2A-2 were used since a single copy was not sufficient to drive expression in a shortened promoter (Figure 2C; 0.35 kb construct).
The simplest interpretation of these results is that Dyad-1 is a positive regulatory element that is necessary but not sufficient for expression in the maxillary palp.
Oligo-1 is required for repression of maxillary palp Or genes in the antenna
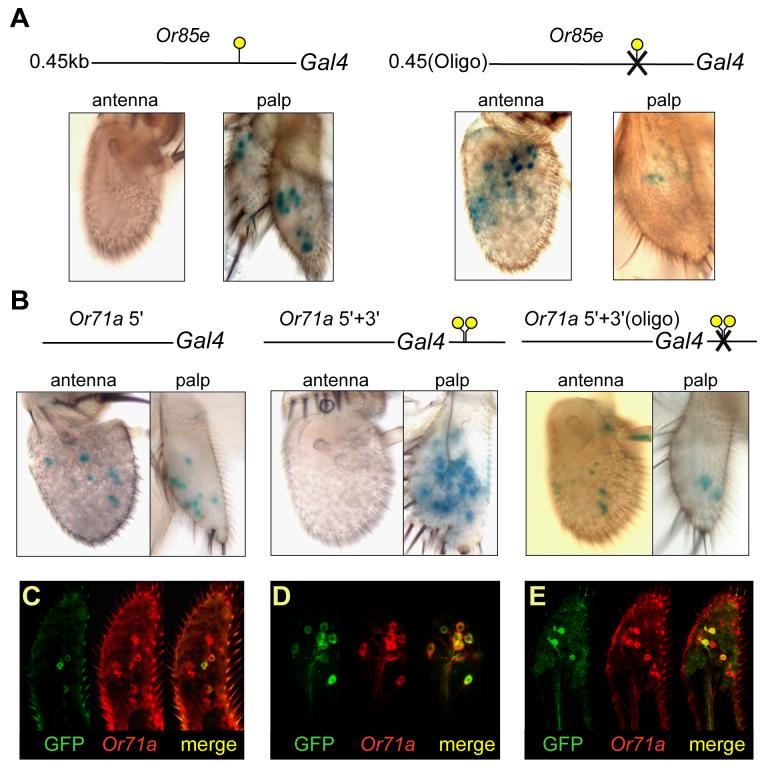
To investigate the function of the Oligo-1 element, we first mutated it in the context of the Or85e promoter. We found that the mutation caused misexpression in the antenna (Figure 3A). We also observed a marked decrease in labeling of maxillary palp ORNs.
Figure 3.
Oligo-1 mediates repression of maxillar palp Or genes in the antenna. (A) Mutation of Oligo-1 in the 450 bp Or85e promoter leads to misexpression in antennal cells (left panels) and a reduction in maxillary palp expression (right panels) in flies containing 0.45(Oligo)-GAL4 and UAS-lacZ. At least 5 independent transgenic lines were tested for both wild-type and mutant constructs. (B) The Or71a 5'-GAL4 construct drives expression of lacZ in the antenna and maxillary palp (left). In Or71a 5'+3'-GAL4 flies, the addition of the 3' region, which contains two Oligo-1 elements, represses expression in the antenna (center). Mutation of the Oligo-1 elements in the 3' region leads to antennal expression (right). At least 2 independent transgenic lines were tested for each construct. Each construct contains 2.3 kb of DNA from the 5' region of Or71a. The distance between the start and stop codons of Or71a is 1.24 kb, compared to 1.6 kb for Or85e. (C-E) Maxillary palps co-stained for GFP (green) and Or71a RNA and from flies containing (C) Or71a 5'-GAL4; (D) Or71a 5'+3'-GAL4; and (E) Or71a 5'+3'(oligo)-GAL4.
We confirmed and extended these results in an analysis of the Or71a gene. There is no Oligo-1 element in the 500 bp upstream of the Or71a gene, but there are two clustered Oligo-1 elements downstream of the gene, located 426 bp and 458 bp downstream from the translation stop site. When we used the 2.3 kb of upstream region alone to drive GAL4 expression, we found misexpression in the antenna (Figure 3B, left). We also observed reduced labeling in the maxillary palp: only 48% of cells labeled with an Or71a in situ hybridization probe were also GFP+ (Figure 3C) and the intensity of the GFP+ labeling appeared weak in a limited experiment (n=9 maxillary palps). When we used both the 2.3 kb of upstream DNA and a 1.4 kb region of downstream DNA (Goldman et al., 2005) that contains both Oligo-1 elements, the antennal misexpression was not observed (Figure 3B, center). Moreover, expression in the maxillary palp was increased: nearly 100% of cells labeled with an Or71a in situ hybridization probe were also GFP+ (n=9 maxillary palps), and the intensity of the GFP+ labeling appeared strong (Figure 3D). To determine whether the repression of antennal labeling and the enhancement of maxillary palp labeling were in fact due to the presence of the Oligo-1 elements in the downstream sequences, we generated a third construct, similar to the second but in which both Oligo-1 sequences were mutated (Figure 3B, right). This construct again produced misexpression in the antenna, and reduced expression in the maxillary palp: again only 48% of cells labeled with an Or71a in situ hybridization probe were also GFP+(Figure 3E). Moreover, expression was highly variable, and the intensity of the GFP+ labeling appeared weak (n=9 maxillary palps).
The simplest interpretation of these results is that the Oligo-1 elements repress expression of maxillary palp Or genes in the antenna and enhance their expression in the maxillary palp. Moreover, Oligo-1 elements appear capable of acting either upstream or downstream of an Or gene.
A regulatory element for a specific ORN class
The Dyad-1 and Oligo-1 elements were identified in a search for sequences that act in the process of receptor gene choice by dictating in which organ an Or gene is expressed. Beyond organ-specific regulation, however, Or genes require an additional level of control to generate the receptor-to-neuron map: they must contain information dictating their precise expression in a single ORN class.
In an effort to identify a neuron-specific regulatory element, we took advantage of the discovery that one maxillary palp ORN, pb2A, co-expresses two unlinked Or genes, Or85e and Or33c (Goldman et al., 2005). We reasoned that since both Or genes are expressed in the same neuron, they are likely to share regulatory elements that dictate expression in this neuron. Accordingly, we examined the 500 bp upstream regions of both genes for sequence elements of at least 6 bp in length that are shared by these two genes, but not by other maxillary palp Or genes.
We identified two elements that meet these criteria: a 12 bp element, TTTATTTGCATA, which we designated the pb2A-1 element, and an 8 bp element, AGTTTTTA, which we designated pb2A-2. pb2A-1 is located at −320 bp relative to the translation start site of Or85e and at −206 bp relative to Or33c; pb2A-2 is located at −102 bp relative to Or85e and −274 bp relative to Or33c (Figure 4A). We note that although these elements were identified by examining the 0.5 kb upstream regions, their specificity extends farther: they are not found in the 1 kb region upstream of any other maxillary palp Or gene, nor in the 0.5 kb downstream region of any maxillary palp Or gene.
To test the function of these elements, we mutated them in the context of the 450 bp Or85e promoter. We found no effect of mutating pb2A-1: the mutant construct produced a pattern of GFP expression that appeared identical to that of the wild-type control construct (Figure 4B). Moreover, we confirmed that these GFP+ cells are in fact pb2A cells by showing that they hybridize to an Or85e probe in a double-label experiment (not shown). Mutation of pb2A-2, however, abolished expression in pb2A cells (Figure 4B), indicating that the pb2A-2 element is necessary for expression in pb2A. We also found that when we fused seven copies of pb2A-2 and two copies of Dyad-1 to the 90 bp basal promoter of Or85e, expression was observed in the maxillary palp (Figure 4C). As a further test, we inserted seven copies of pb2A-2 and two copies of Dyad-1 upstream of a second minimal promoter; in this case the pb2A-2 elements were 45 bp upstream of a TATA box. Again, we found expression in the maxillary palp (Figure 4C). Although expression was too weak to allow double-label in situ hybridization, in each case at least some of the cells could be identified as neurons by the presence of dendrites (Figure 4C, insets), and their pattern of expression and distribution on the maxillary palp are consistent with those of pb2A. These results indicate that not only is pb2A-2 necessary for Or85e expression, but that artificial promoters containing pb2A-2 sequences and Dyad-1 sequences can drive expression in maxillary palp ORNs. We note that two Dyad-1 sequences alone did not drive expression of Or85e, as indicated above.
Two functional odor receptors from one alternatively spliced Or gene
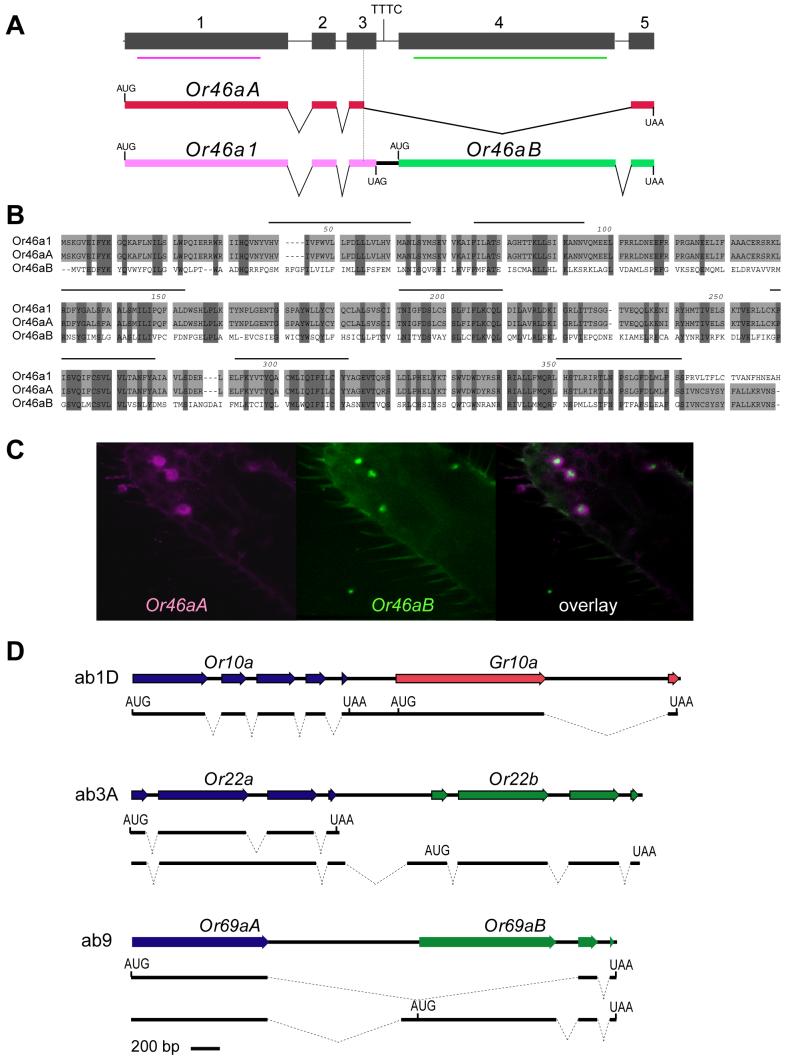
One mechanism by which an ORN may select two Or genes thus appears to depend on the location of common ORN-specific elements, such as pb2A-2, upstream of two different Or genes. We have also found another instance of receptor coexpression that occurs through a different mechanism. This coexpression was revealed by a detailed analysis of the Or46a locus, which was previously shown to be expressed in pb2B cells (Goldman et al., 2005).
The Or46a locus was proposed to contain two coding regions, Or46aA and Or46aB, separated by less than 100 bp and expressed by alternative splicing (Robertson et al., 2003). We confirmed that the locus produces two alternatively spliced mRNAs; both mRNAs were identified in multiple independent experiments. These two splice forms were observed at comparable levels in RT-PCR analysis, and were the major products detected; we cannot exclude the possibility that minor species of functional significance may also be produced. A detailed analysis of the two major mRNAs revealed surprising structures. The shorter mRNA is spliced from an internal position within exon 3 to exon 5, bypassing exon 4, and contains the Or46aA coding region (Figure 5A). The longer mRNA lacks the splice between exons 3 and 5 and is a dicistronic message, containing two coding regions. The first of these two coding regions, which we term Or46a1, is identical to that of Or46aA except at the 3' end, which encodes C terminal residues that lie immediately beyond the seventh predicted transmembrane domain. Or46a1 encodes 18 terminal amino acids that show no identity to the 17 terminal amino acids of Or46aA (Figure 5B). The second of the two coding regions, Or46aB, shares only 36% amino acid identity with Or46aA and 31% identity with Or46a1.
Figure 5.
Two functional odor receptors from one alternatively spliced gene. (A) The Or46a genomic locus and the two alternatively spliced mRNA species detected in the maxillary palp. The red and green bars indicate probes used in panel (C). (B) Amino acid sequence alignment of the three predicted Or46a proteins. (C) Maxillary palps co-stained with RNA probes against Or46aA and Or46aB. (D) Tightly linked antennal receptor genes. Genomic organization and mRNAs detected in the antenna are indicated. The 5' end of the transcript encoding Or22b has been found to lie 5' to the second intron of Or22a but has not been identified precisely; the diagram shows the 5' end predicted by analogy to the shorter mRNA.
This unexpected splicing pattern predicts that multiple receptors from this locus may be coexpressed in the same cell. We carried out a double-label in situ hybridization experiment using probes for Or46aA, consisting of sequences from within exon 1, and Or46aB, consisting of sequences within exon 4 (Figure 5C). We found that all cells labeled by Or46aA sequences were also labeled by Or46aB sequences; thus all cells that express the short transcript also express the long transcript. Likewise, all cells labeled with Or46aB were also labeled by Or46aA, but this latter result is less informative, on account of the expected cross-hybridization between Or46aA and Or46a1. Thus from this experiment alone we cannot determine whether there are any cells that express the long transcript but not the short transcript. In any case, these experiments reveal that pb2B cells express sequences corresponding to more than one Or open reading frame (ORF).
In order to determine whether more than one of the distinct ORFs in fact encodes a functional odor receptor, we used an in vivo expression system, the “empty neuron” system (Dobritsa et al., 2003). Using this method we found that Or46aA and Or46aB both encode functional odor receptors that respond to phenols, while Or46a1 did not impart responses to any odor (data not shown).
The Or46a locus thus encodes three predicted odor receptors. Their expression appears limited to a single class of neuron, pb2B. To investigate the significance of this one receptor gene-multiple receptor organization, we asked whether it is conserved in evolution. We examined the genomes of five additional Drosophila species: D. simulans, D. yakuba, D. erecta, D. pseudoobscura, and D. grimshawi. We found that orthologs of the ORFs encoding Or46aA, Or46a1 and Or46aB are present in all species and thus have been maintained for tens of millions of years (Bergman et al., 2002). Moreover, the organization of the locus is well conserved; for example, the distance between the stop codon of Or46a1 and the start codon of Or46aB is in all cases between 69 bp and 164 bp. Double-label in situ hybridization with D. pseudoobscura sequences representing the orthologs of Or46aA and Or46aB showed coexpression in cells of the D. pseudoobscura maxillary palp (data not shown).
In the mosquito Anopheles gambiae, the genes most closely related to Or46aA and Or46aB are AgOr34 and AgOr37, whose predicted products both show 22% amino acid identity to Or46aB and 19% and 20% identity to Or46aA, respectively. Interestingly, these two mosquito genes are also tightly linked, but in inverted fashion, with only 1.5 kb of intervening noncoding DNA, suggesting the possibility that they may share common regulatory sequences and hence be coexpressed.
We note finally that in order to express a functional Or46aB protein from a dicistronic RNA, an internal ribosome entry site (IRES) is required. Although there is precedent for functional IRES sequences in Drosophila, they have not been well defined. IRES sequences are poorly conserved across phylogeny, but dicistronic RNAs identified from polio virus and the human ornithine decarboxylase gene share a common UUUC sequence approximately 26-34 bp upstream of the distal AUG (Maier et al., 2002). We note the presence of a UUUC sequence 38 bp upstream of the AUG of Or46aB, a sequence that is conserved at the same position in D. yakuba and D. ananassae.
Mechanisms of Or gene coexpression in the antenna
We have presented evidence for two distinct mechanisms by which a single maxillary palp ORN can express two odor receptors, one mechanism that depends on the localization of a common cis-regulatory element upstream of two unlinked Or genes, and one that depends on alternative splicing. To determine whether these mechanisms are singularities, unique to the maxillary palp, we expanded our analysis to include the entire olfactory system. We first examined all pairs of Or coding regions that are separated by <1 kb of intervening DNA: (Or22a, Or22b), (Or33a, Or33b), (Or33b, Or33c), (Or42a, Or42b), (Or59b, Or59c), (Or65b, Or65c), (Or69aA, Or69aB), (Or85b, Or85c); we also examined (Or10a, Gr10a), which are also tightly linked.
We were surprised to find a dicistronic message that encodes both Or10a and Gr10a, in both of two strains analyzed, Oregon-R and w1118 (Figure 5D). Or10a is an odor receptor for methyl salicylate (Hallem et al., 2004) while Gr10a is a member of the gustatory receptor gene family (Clyne et al., 2000). The dicistronic mRNA was identified in each of six independent experiments, each using a different combination of primer pairs. We note that there are two UUUC motifs upstream of the Gr10 translational start codon, at positions −32 and −52, suggesting the possibility that one may act as an IRES. Our results provide a molecular mechanism to explain the coexpression of these two genes in ab1D ORNs, reported recently by Fishilevich and Vosshall (2005).
We found that although Or22a and Or22b are each encoded by independent transcripts, we identified two transcripts that are likely to have the same 5' ends, based on the positions of the longest cDNAs isolated (Figure 5D). These results suggest that Or22a and Or22b can be expressed from the same promoter, which could in principle explain the coexpression of the two genes in ab3A ORNs (Dobritsa et al., 2003). In the longer transcript, encoding Or22b, the second intron is not removed, creating a frameshift mutation and a nonsense codon in Or22a; thus a functional Or22a receptor would be encoded only by the shorter transcript. We note the presence of a UUUC at position −37 upstream of the Or22b translational start codon, suggesting a means by which Or22b could be translated from a long mRNA that also includes an Or22a translational start site. At the same time, there is a TATA box 70 bp upstream of the Or22b translational start codon, and a 1.3 kb region upstream of Or22b was found to drive faithful reporter gene expression (Couto et al., 2005), suggesting the existence of a third, Or22b-specific transcript. In the Canton-S strain, Or22a has been shown to be a functional odor receptor, whereas Or22b is non-functional. However, in an Oregon-R strain both genes encode functional odor receptors (W.vd.G.N and J.C., unpublished data).
Or69a also produces two distinct mRNAs, which contain identical, or nearly identical, 5'ends. The shorter mRNA encodes Or69aA; the longer transcript encodes Or69aB. The mRNA encoding Or69aB contains many of the codons of Or69aA but the Or69aA ORF is terminated by a stop codon following the first splicing event. These results provide a molecular explanation for the coexpression of Or69aA and Or69aB in ORNs of ab9 sensilla; however, a 0.9 kb region upstream of the Or69aB ATG was found to drive reporter gene expression in ab9 sensilla (Couto et al., 2005), suggesting the possibility of an additional Or69aB-specific transcript.
In addition to these three cases of antennal coexpression, recent mapping studies have identified six additional antennal ORN classes that coexpress Or genes, among the 36 antennal ORN classes to which Or genes were mapped (Couto et al., 2005; Fishilevich and Vosshall, 2005). Four of these coexpressed Or gene pairs are unlinked: (Or33b, Or85a); (Or33a, Or56a); (Or33b, Or47a); (Or49a, Or85f), raising the possibility that they might, like (Or33c, Or85e), contain a common regulatory element. Using the same bioinformatics approach used to identify pb2A-1 and pb2A-2, we identified common motifs for each pair (Supplementary Figure 1). These elements were neuron-specific in that they were not found in the 500 bp upstream of any other Or gene in the genome. Shared motifs were likewise identified for (Or22a, Or22b) and (Or69aA, Or69aB), suggesting an additional mechanism by which the upstream and downstream ORFs could be coexpressed. We also found a common element for a pair of Or genes that are coexpressed in a larval ORN (Or94a, Or94b) (Fishilevich et al., 2005). As a control, we carried out the same bioinformatics approach on two genes that are not coexpressed, Or85f and Or56a, and found no elements that met our criteria, i.e. we found no common elements unique to these genes. Or19a and Or19b are coexpressed but appear to have duplicated recently and there is extensive identity in their upstream regions.
Odor receptor expression is permissive: lack of negative feedback regulation in Drosophila
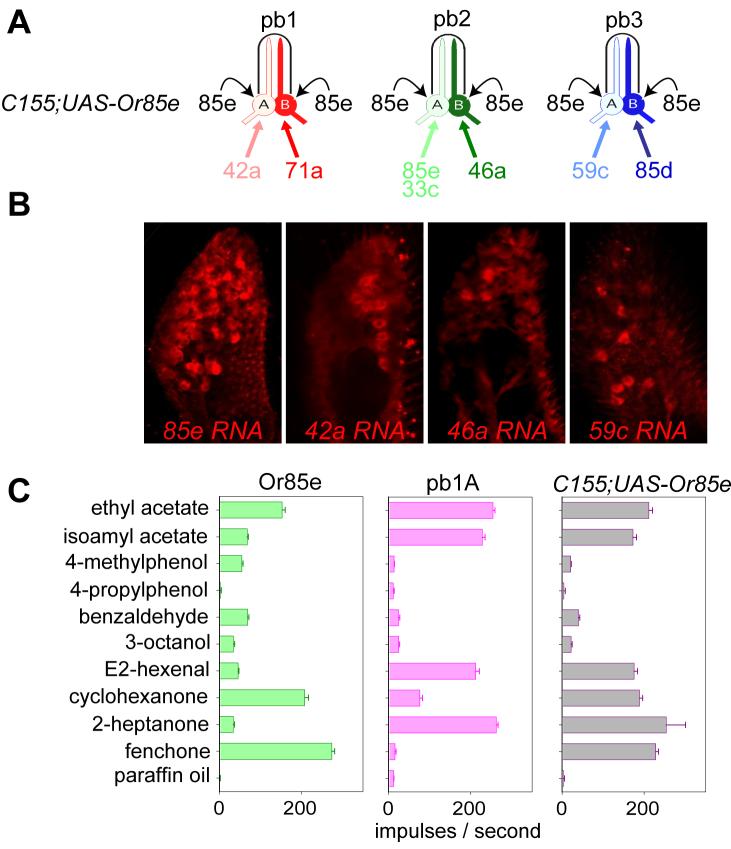
In mammals, analysis of non-functional receptor genes has led to the proposal that the expression of one odor receptor inhibits the expression of others in the same ORN by negative feedback regulation (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). To investigate directly whether expression of an odor receptor inhibits the expression of any others in Drosophila, we ectopically expressed each of three receptors in the maxillary palp and tested the effects on expression of others. We first drove expression of Or85e, which in wild-type is expressed only in pb2A, in all or almost all ORNs of the maxillary palp using the C155-GAL4 driver (Jefferis et al., 2004)(Figure 6A), which initiates expression before the onset of normal Or expression (Clyne et al., 1999a; Komiyama et al., 2004). We then tested the expression of other maxillary palp Or genes by in situ hybridization and electrophysiology.
Figure 6.
Lack of negative feedback regulation. (A) ORN classes in the maxillary palp, indicating the endogenous Or genes they express (color) and the ectopic expression of Or85e (black) in all ORNs of C155-GAL4;UAS-Or85e. (B) In situ hybridizations to maxillary palps of C155-GAL4;UAS-Or85e with probes against the indicated Or RNAs. (C) Odor response profiles of: Or85e in the mutant ab3A empty neuron, showing strong response to fenchone (data from Goldman et al., 2005) (left); pb1A in control C155-GAL4 flies, showing response to 2-heptanone (center); a class of ORNs in C155-GAL4;UAS-Or85e, showing strong responses to both odorants (right). Odorants were tested as vapors of a 10−2 dilution. n=5; error bars=S.E.M.
We confirmed by in situ hybridization that Or85e was in fact expressed in most if not all ORNs of the maxillary palp in C155-GAL4; UAS-Or85e flies (Figure 6B, left panel). We then examined expression of the Or genes that are normally expressed in each of the other five maxillary palp ORN classes. We found that each tested gene was expressed in what appeared to be a normal pattern, in the presence of ectopic Or85e expression (Figure 6B and data not shown). Thus expression of Or85e does not inhibit transcription of other Or genes.
We then asked whether the other Or genes were functionally expressed, by carrying out electrophysiological recordings from C155-GAL4; UAS-Or85e flies, using a diagnostic odor panel that distinguishes between Or85e-expressing ORNs and all other classes of maxillary palp ORNs (Goldman et al., 2005). Or85e responds strongly to fenchone (Figure 6C, left panel); the pb1A neuron, by contrast, responds strongly to 2-heptanone, on account of Or42a expression, but not fenchone (Figure 6C, center panel). When Or85e is overexpressed, we detect neurons that respond strongly to both fenchone and 2-heptanone (Figure 6C, right panel). The simplest interpretation of these results is that misexpression of Or85e in pb1A cells does not inhibit functional expression of Or42a. We found similar physiological evidence to indicate that expression of Or85e does not inhibit the functional expression of Or71a, Or46a, Or59c, and Or85d (not shown). In a more limited experiment, we used C155-GAL4 to drive the early expression of two other receptors, Or42a and Or10a (an antennal gene). By performing a similar electrophysiological analysis we found that in each ORN class in which a novel odor response was conferred by misexpression of Or42a or Or10a, the response conferred by the endogenously expressed Or gene was still present. These results, taken together, indicate that expression of one Or gene does not repress that of others by feedback regulation.
Coordination of receptor choice between two neurons of a sensillum
Each sensillum of the maxillary palp contains two ORNs, combined according to a strict pairing rule. For example, each pb1 sensillum contains a pb1A neuron that expresses Or42a, paired with a pb1B neuron that expresses Or71a; for the present analysis we designate this cellular expression pattern as the (Or42a+;Or71a+) configuration. Such stereotyped pairing of ORNs has been documented in diverse insects but is not observed in mammals. It raises the problem of how the choice of a receptor in one ORN is coordinated with that in a neighboring ORN. In principle, the choice made by one ORN could induce a specific choice in the neighboring ORN; alternatively, the choices of two neighboring maxillary palp ORNs could be made simultaneously and be coordinated by virtue of the asymmetric segregation of regulatory proteins from a common progenitor cell.
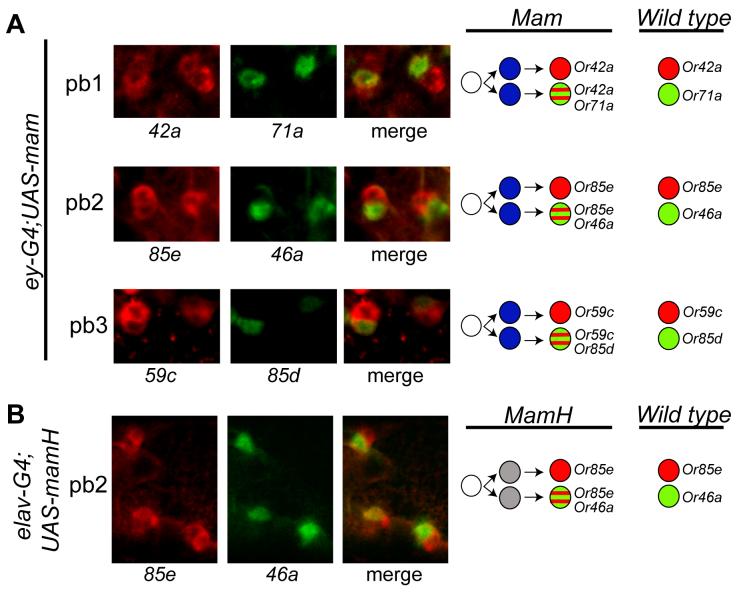
In the development of antennal sensilla there is evidence that a single progenitor, or founder cell, recruits three secondary progenitor cells of which one, PIIc, divides to give rise to two neurons (Sen et al., 2003). Mastermind (Mam), a nuclear protein in the Notch pathway, is essential for asymmetric cell division of embryonic neuroblasts in the developing Drosophila nervous system: both loss and gain of mam function result in altered identities of the daughter cells (Helms et al., 1999; Schuldt and Brand, 1999; Yedvobnick et al., 2004). We have systematically investigated the expression of Or genes by paired ORNs in maxillary palps that misexpress mastermind (mam).
We first expressed UAS-mam in the developing olfactory organs using the eyeless-GAL4 driver (Hummel et al., 2003). We then examined receptor gene expression by double-label in situ hybridization. We found that the coordination of receptor gene expression between neighboring ORNs was abnormal in all three types of sensilla, pb1, pb2, and pb3. A substantial fraction of sensilla expressing Or42a showed expression of this gene in both neurons: these sensilla contained one ORN that expressed Or42a alone and another that expressed both Or42a and Or71a, designated the (Or42a+; Or42a+Or71a+) configuration (Figure 7A). Likewise, we found sensilla in (Or85e+; Or85e+Or46a+) and (Or59c+; Or59c+Or85d+) configurations; in wild-type these genes are expressed only in (Or85e+; Or46a+) and (Or59c+; Or85d+) configurations. We did not identify any other abnormal configurations: those ORNs that misexpressed an Or gene always misexpressed the Or gene of the neighboring ORN.
Figure 7.
Coordination of receptor choice between two neurons of a sensillum. (A) Optical sections from maxillary palps of ey-Gal4;UAS-mam flies co-stained with probes for the indicated Or RNAs. (B) Optical sections from maxillary palps of elav-G4;UASmamH flies co-stained for the indicated Or RNAs.
This abnormal partitioning of receptor expression was confirmed by an independent method, electrophysiological recordings. Specifically, in the wild-type pb2 sensillum, the A cell produces large spike amplitudes and strong responses to fenchone, whereas the B cell produces small spike amplitudes and strong responses to 4-methyl phenol (Goldman et al., 2005). In eyeless-GAL4; UAS-mam, we identified sensilla containing one neuron with a large spike amplitude and a strong fenchone response, paired with a neuron that produces a small spike amplitude and strong responses to both fenchone and 4-methyl phenol, as expected of (Or85e+; Or85e+Or46a+) sensilla (not shown). We have also confirmed the abnormal partitioning of receptor expression in another genotype, by driving a truncated dominant negative form of mam, UAS-mamH (Yedvobnick et al., 2004) with the pan-neuronal driver elav-GAL4. We carried out double-label in situ hybridization with Or85e and Or46a probes and identified (Or85e+; Or85e+Or46a+) sensilla (Figure 7B).
These results are consistent with a model in which the A and B neurons of a maxillary palp sensillum are siblings that derive from a common progenitor by a Mam-dependent asymmetric cell division. Perturbation of Mam function might lead to the abnormal segregation of a regulatory protein,which in turn leads to misexpression of a receptor.
The observation that misexpression in an ORN was restricted to the Or gene of its neighbor, and not to any other Or genes, is consistent with a model in which the progenitor has undergone a restriction that limits its daughter ORNs to the expression of the Or genes of one sensillum type. We note that the data are also consistent with a model in which the progenitor, and both daughter ORNs, contain a positive regulatory factor that binds to a site shared by, and specific to, the Or genes expressed in a particular sensillum type. Computational analysis has in fact revealed such elements – for example, Or42a and Or71a both contain an upstream AAATCAATTA element that is not found adjacent to other Or genes of the maxillary palp or antenna; however, genetic analysis of this element has not revealed a functional requirement for it in receptor gene expression, and we have found no support for the existence of a sensillum-specific determinant of receptor gene expression.
Thus our data, taken together, are consistent with a model in which the coordination of receptor gene expression in a sensillum is achieved through the Mam-dependent segregation of regulatory factors. The results underline the importance of identifying regulatory proteins whose proper distribution in the ORNs of the maxillary palp is essential to the proper distribution of receptor gene expression.
Different Or genes depend on different combinations of the transcription factors Lz and Acj6
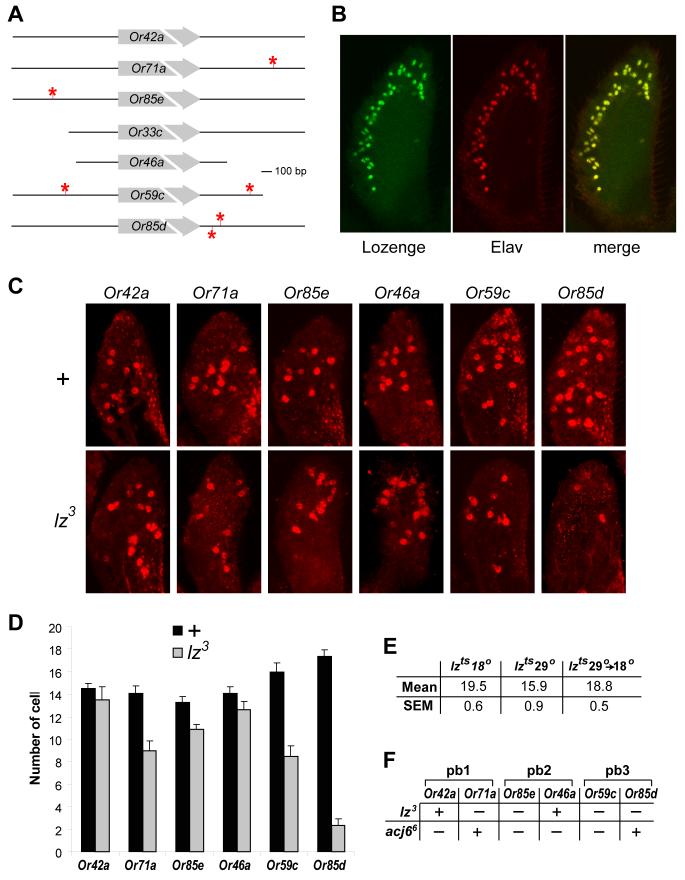
In a complementary analysis of the mechanisms of receptor gene choice, we investigated transcription factors whose expression had been reported in at least one olfactory organ and whose mutations had been shown to cause olfactory defects. We found that one such protein, the Runx domain-containing transcription factor Lozenge, had predicted binding sites (RACCRCA, R=purine; (Flores et al., 2000)) adjacent to four maxillary palp Or genes. Specifically, we found that two maxillary palp Or genes, Or59c and Or85d, had two Lz binding sites, and two genes, Or71a and Or85e, had one Lz binding site, within 1 kb upstream or downstream of the coding region (Figure 8A). Lz is required for the specification of cell fate in the eye (Flores et al., 2000), and for normal numbers of olfactory sensilla in the antenna (Stocker et al., 1993). In the maxillary palp the numbers of sensilla are normal, but electropalpogram recordings showed large reductions in odor responses (Riesgo-Escovar et al., 1997).
Figure 8.
Lz is required for the expression of a subset of Or genes. (A) Lz binding sites are indicated by stars. The diagram depicts either 1 kb upstream and downstream of each Or coding region, or the distance to the coding region of the nearest flanking gene. (B) Confocal micrographs of maxillary palp labeled with antibodies against Lz and Elav. (C) Z-compression of individual maxillary palps from wild-type (top) and lz3 mutant (bottom), hybridized with probes for the indicated RNAs. (D) Numbers of cells labeled with probes for Or RNAs. (n=10 palps). (E) Numbers of cells expressing Or85d RNA in maxillary palps of lzts1 flies cultured at indicated temperatures. (n= 11-18). All flies were examined at age 8d. (F) Expression of Or genes in mutant backgrounds. “-“ indicates reduction in number of cells labeled by in situ hybridization.
To investigate the possibility that Lz is required for normal receptor gene expression, we first asked whether it is expressed in ORNs of the maxillary palp. We found that Lz is coexpressed with Elav, indicating that it is expressed in the nuclei of all maxillary palp ORNs (Figure 8B). We then examined the expression of six maxillary palp Or genes, one from each ORN class, in lz3, a strong hypomorphic mutant (Figure 8C). The four genes that are flanked by predicted Lz binding sites all showed reduced levels of expression (p<0.05); the two genes that contain two Lz binding sites, Or59c and Or85d, showed particularly severe reductions (of 47% and 87%, respectively) in the number of labeled cells (Figure 8D). The mildest reduction, 18%, was observed for Or85e; consistent with this result, a 14% reduction was observed when DNA including the predicted Lz binding site was removed from an Or85e-GAL4 driver (Figure 1B; the construct containing 3 kb of upstream DNA labeled 13.4±0.4 cells whereas the construct containing 0.45 kb labeled 11.5±0.3 cells, n=12). The two genes that did not contain Lz binding sites did not show a reduction in labeling in lz3. These results demonstrate that lz is required for the expression of a subset of Or genes in the maxillary palp.
We next used a weaker, temperature-sensitive allele, lzts1, to investigate the possibility that levels of Or gene expression are susceptible to modulation during the adult stage. We found that Or85d is expressed in 18% fewer cells (p < 0.05) when lzts1 flies are raised at the restrictive temperature (29°) than when raised at the permissive temperature (18°)(Figure 8E). When flies were raised at the restrictive temperature and then shifted to the permissive temperature for 24h, one week after eclosion, the number of Or85d-expressing cells showed an increase of 19% (p < 0.05), to a level indistinguishable from that of flies that had been cultured continuously at the permissive temperature. These results confirm our finding of a functional role for lz in Or expression, provide direct evidence that levels of Or expression can be altered after eclosion, and invite investigation of epigenetic modulation of odor receptor expression in Drosophila.
Only one other transcription factor, the POU domain protein Acj6, has previously been demonstrated to be required for odor receptor expression in Drosophila. Specifically, expression of Or33c, Or42a, Or46a, Or59c, and Or85e was severely reduced by the null allele acj66, whereas expression of Or71a and Or85d was unaffected (Clyne et al., 1999a; Clyne et al., 1999b; Goldman et al., 2005; Komiyama et al., 2004). We have shown here that expression of Or59c, Or71a, Or85e, and Or85d was reduced by lz3, but expression of Or42a and Or46a was not. Thus the maxillary palp Or genes can be divided into three classes based on their sensitivity to these mutations: those sensitive to both acj66 and lz3 (Or59c and Or85e), to acj66 alone (Or42a and Or46a), or to lz3 alone (Or71a and Or85d). These results provide the first experimental evidence to support a model in which Or gene expression depends not only on a combinatorial code of regulatory elements, but also on a combinatorial code of transcription factors.
DISCUSSION
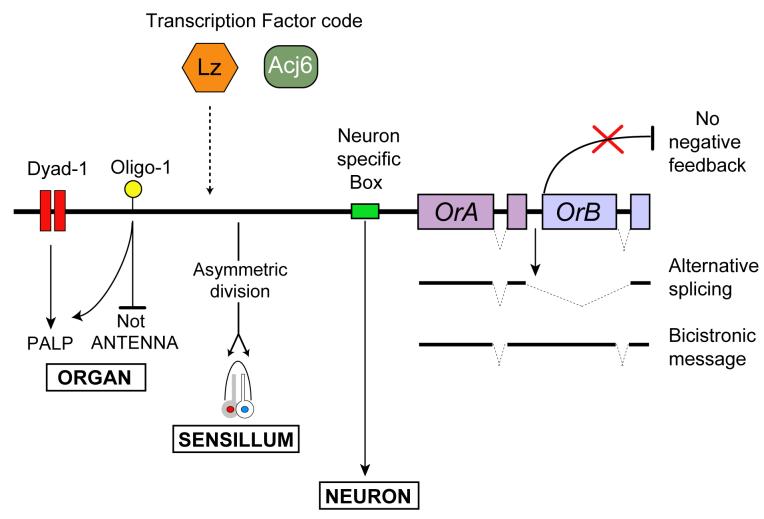
We have investigated how the Drosophila olfactory system has solved a remarkable problem: how to express each of a large repertoire of odor receptor genes in a distinct subset of ORNs. We have analyzed the logic of this process and found that it is accomplished via several mechanisms operating together in concerted fashion (Figure 9).
Figure 9.
Summary of mechanisms of receptor gene choice.
Compact control regions
Given the complexity of the regulatory problem, the economy with which the fly has solved it is striking. We have found that <450 bp of regulatory DNA was sufficient to recapitulate the pattern of Or gene expression in each of two cases analyzed in detail, Or85e and Or46a. The fidelity of expression was determined directly in double-label experiments.
Mammalian OR gene regulation depends both on short-range control operating within ∼160bp (Vassalli et al., 2002; Rothman et al., 2005) and on long-range control exerted by elements as far away as ∼200 kb (Serizawa et al., 2000), or even on another chromosome (Lomvardas et al., 2006). We have found no evidence for mechanisms of long-range control among Drosophila Or genes. We note that the compactness of Or gene regulatory regions, along with the short introns that are characteristic of these genes, may have facilitated the expansion of the Or gene family via duplication and divergence.
Organ specificity: positive and negative elements required for Or expression in the maxillary palp
Dyad-1 is the first dyad motif of its kind shown to be functional in Drosophila, to our knowledge. Certain hormone receptors bind to two identical copies of a hexamer, in either a direct or inverted repeat, separated by up to eight bp. The two trimers of Dyad-1 are distinct and are separated by 9 bp, corresponding approximately to one turn of the DNA double helix; perhaps the two trimers lie on the same side of the double helix and make contact with the same transcription factor.
Oligo-1 plays two roles in the process by which an Or gene is expressed in the correct olfactory organ. It mediates repression in the antenna and promotes expression in the maxillary palp. The dual role of Oligo-1 sites may represent a means of achieving regulatory economy: two functions are fulfilled by a single element. Likewise, the dual role could be a means of expanding the informational capacity of the regulatory system. Rather than existing in either of two states, unbound or bound to a unique regulatory protein, Oligo-1 might exist in three discrete states: unbound, bound to a positive regulatory protein in the maxillary palp, or bound to a negative regulatory protein in the antenna. Such expansion of informational capacity may be particularly important in a regulatory system that must contain sufficient information to direct the precise expression of 60 different receptor genes in a comparable number of distinct ORN classes.
Dyad-1 and Oligo-1 elements lie within 15 bp of each other in three cases. Further analysis will be required to determine whether they bind proteins that are part of the same complex, or whether the binding of a protein to one blocks binding of a protein to the other.
Finally, although our analysis has focused on Or genes expressed in the maxillary palp, the antennal expression observed following mutation of an Oligo-1 site suggests that some of the elements that direct expression in the maxillary palp (elements other than Dyad-1 and Oligo-1) may also be used to direct the expression of other Or genes in the antenna.
Neuron specificity: a neuron-specific regulatory element
We have identified a regulatory element, pb2A-2, that is common to the two Or genes coexpressed in one particular ORN class, pb2A. Functional analysis showed that pb2A-2 is required for expression in pb2A, and that an artificial promoter containing seven concatenated pb2A-2 sequences and two Dyad-1 sequences can drive expression in maxillary palp ORNs, although we have not been able to determine definitively whether they are of the pb2A class. An interesting direction for further analysis will be to determine whether a single pb2A-2 element, in conjunction with Dyad-1 and Oligo-1, is sufficient to drive expression in pb2A.
A second element, pb2A-1, was also found to be unique to these two coexpressed genes, but mutational analysis did not reveal a necessary role in Or gene expression. We do not know whether pb2A-1 has a function that is redundant with that of another, nearby element, or whether it is sufficient to drive expression in the context of a basal promoter.
Or expression is permissive
In order to restrict the number of OR genes expressed in individual ORNs, mammals have been proposed to have evolved a negative feedback mechanism in which expression of one OR gene inhibits the expression of others. We systematically investigated the possibility of a similar mechanism in Drosophila, and found that ectopic expression of Or85e, Or42a, or Or10a does not inhibit the expression of other Or genes in the maxillary palp. The results of these gain-of-function experiments are consistent with observations from loss-of-function experiments: when an individual Or gene is mutated, an “empty neuron” results, i.e. no other Or gene takes its place (Dobritsa et al., 2003; Elmore et al., 2003). Thus the coexpression of Or85e and Or33c in pb2A, and the coexpression of other Or gene pairs in other ORNs, does not require a special mechanism to evade a negative feedback control system.
Coexpression of odor receptors by two mechanisms
One mechanism by which two Or genes can be coexpressed in the same ORN is by sharing common regulatory elements: we have shown that the two unlinked Or genes that are coexpressed in pb2A share at least one specific regulatory element. Surprisingly, we have found that the neighboring ORN also coexpresses odor receptors, but by a different mechanism, based on alternative splicing and a dicistronic mRNA.
Few dicistronic transcripts have been identified in Drosophila, as in most other animals. In fact, a large-scale analysis of cDNAs identified only 31 dicistronic transcripts from the ∼13,400 protein-coding genes of the Drosophila genome (Misra et al., 2002). This study did not report the dicistronic transcript we have identified from the Or46a locus, or the Or10a-Gr10a transcript, perhaps because of the small number of cells in which these transcripts occur, although it did identify one encoding two Gr proteins.
The organization of the Or46a locus has been conserved for tens of millions of years, suggesting that the dicistronic transcript and the coexpression of receptors confer a selective advantage; however, the nature of such an advantage is not clear from the present analysis. It will be interesting to determine whether the products of the Or46a locus form heterodimers, for which there is some precedent among members of the Or family (Neuhaus et al., 2005).
A code of regulatory elements and transcription factors specifying receptor gene choice in Drosophila
Each Or gene must contain regulatory information dictating in which ORN class it will be expressed. Our results support a hierarchical model in which receptor gene choice is governed by two levels of regulation: one dictating the organ in which the gene is expressed, and the second dictating the ORN class, within that organ, in which the gene is expressed. We have identified cis-regulatory elements that are required for expression of Or genes in the maxillary palp and a cis-regulatory element that is required for expression in one specific ORN class within the maxillary palp.
In further support of a level of organ-specific regulation, we note that the olfactory sensilla of the maxillary palp derive from a developmental lineage distinct from those of the antennal sensilla that express Or genes. The maxillary palp sensilla derive from a lineage of sensory organ precursors (SOPs) that depend on the activity of the proneural gene atonal (Gupta and Rodrigues, 1997). Antennal basiconic sensilla arise from SOPs that depend on Amos and high levels of Lozenge, and antennal trichoid sensilla depend on Amos and low levels of Lozenge (Goulding et al., 2000; Gupta et al., 1998); antennal coeloconic sensilla do not express Or genes, with a single exception (Couto et al., 2005; Yao et al., 2005). Thus maxillary palp ORNs may contain a distinct population of transcription factors that promote expression of Or genes containing Dyad-1 and Oligo-1 elements; antennal ORNs may contain regulatory proteins that repress maxillary palp Or genes via Oligo-1 and other undefined elements. As predicted by a model postulating a level of neuron-specific regulation, we have been able to identify candidate regulatory elements that are specific to Or genes of six antennal ORN classes as well (Supplementary Figure 1).
The simplest interpretation of our results, taken together, is that the specificity of Or gene expression in Drosophila is governed in part by a combinatorial code of regulatory elements. We have also provided, for the first time, direct evidence that Or expression is governed by a code of transcription factors. A mutation of the transcription factor Lz has been shown to affect expression of a subset of Or genes, a subset different from that affected by a null mutation of the transcription factor Acj6. These results indicate that in different ORNs, different combinations of transcription factors operate to express individual Or genes. A longterm goal is to achieve a full understanding of how these two combinatorial codes are integrated.
Receptor gene choice in Drosophila and other organisms
In mammals, it is thought that transcriptional regulatory mechanisms direct expression of OR genes in specific zones of the olfactory epithelium, but that within a zone, OR gene choice is based on a stochastic selection mechanism. A third mechanism, negative feedback, could then operate to limit the number of OR genes expressed in individual neurons (Lomvardas et al., 2006; Mombaerts, 2004).
In Drosophila the process of receptor gene choice achieves a conceptually simple end: it produces a highly stereotyped receptor-to-neuron map. However, the large number of receptors and neurons presents a regulatory problem of great complexity. To achieve such a precise and highly ordered organization, Drosophila has evolved a sophisticated suite of regulatory mechanisms. We have documented organ-specific and neuron-specific levels of transcriptional control, including both positive and negative mechanisms. We have also identified a post-transcriptional mechanism, alternative splicing, and the system has even evolved a relatively rare innovation, dicistronic mRNAs.
The worm C. elegans has a much larger repertoire of odor receptor genes than Drosophila, but the number of ORNs to which it allocates them is very limited (Melkman and Sengupta, 2004; Mombaerts, 2004). Thus the number of receptor genes per neuron is increased, but the complexity of the regulatory problem is decreased. In vertebrates, however, the repertoire is very large and the number of receptor genes expressed per neuron is very low. Perhaps as the receptor gene repertoire expanded in vertebrate evolution, the complexity of the regulatory problem eventually exceeded the ability of the system to execute a deterministic plan with sufficient fidelity, and deterministic mechanisms were replaced by a stochastic mechanism and a negative feedback mechanism. In any case, the ultimate result of receptor gene choice in Drosophila is the same as in vertebrates: a spectacular diversity of ORNs that underlie the detection and discrimination of odors.
EXPERIMENTAL PROCEDURES
Stocks
Fly stocks were raised at 25° C unless otherwise indicated. Wild-type flies were Canton-S unless otherwise indicated. w; UAS-mCD8-GFP/CyO;UAS-mCD8-GFP was used as a source of GFP and w;UAS-lacZ/CyO;UAS-lacZ/TM2 was used as a source of lacZ, unless otherwise indicated. w;UAS-Mam;UAS-Mam and w;UAS-MamH were from B. Yedvobnick. w;ey-Gal4 and C155-Gal4 were from the Bloomington stock center.
Molecular analysis
For rapid amplification of 5' cDNA ends and RT-PCR, total RNA was isolated from ∼150 maxillary palps or ∼100 antennae using the QIASHREDDER and RNeasy kits from Qiagen, or mRNA was isolated using the Promega polyATract kit RT-PCR was performed using either the SMART 5'RACE kit (Clontech) or standard oligo dT primers. The PCR products obtained after 35 thermocycles were AT-cloned and confirmed by sequencing. In situ hybridization and immunohistochemical localization were performed as described in Goldman et al (2005). Detection of β–Galactosidase activity was performed by X-Gal staining for up to 6 hours. One copy of the Or-GAL4 driver and one copy of UAS-lacZ, or two copies of UAS-GFP, were used.
The mouse anti-Lz antibody (developed by U. Banerjee) and rat anti-Elav antibody were obtained from the Developmental Studies Hybridoma Bank, University of Iowa.
Generation of the Or85e promoter deletion series, the mutant and minimal promoters, UAS-Or46a derivatives, are described in Supplementary Data.
Bioinformatics
The DYAD ANALYSIS program (van Helden et al., 2000) was used to identify overrepresented bipartite sequences. The OLIGO ANALYSIS program (van Helden et al., 1998) was used to identify overrepresented oligonucleotide sequences. The overrepresented motifs were tested for their presence in 500 bp upstream of all Or genes known to be expressed in the antenna and larva using the PATSER algorithm (http://rsat.ulb.ac.be/rsat/). To identify sequences shared by Or33e and Or85e upstream regions but not the upstream regions of other maxillary palp Or genes, we used a direct DOT-PLOT analysis; the DYAD- and OLIGO-ANALYSIS programs are inefficient in comparing only two sequences.
Drosophila genetics and transformation
Flies containing a transposable P-element, P{lacW}s2172s2172, ∼ 1kb upstream of Or71a were used for an imprecise excision screen. w− progeny were screened by PCR to identify deletions. PCR products were sequenced to identify deletion breakpoints.
All DNA constructs were sequenced and then injected into w1118 flies, unless otherwise indicated. Multiple transgenic lines were generated and tested for each construct.
Electrophysiology
Odors were delivered and action potentials were recorded as described in Dobritsa et al. (2003).
Supplementary Material
Supplementary Figure 1. Shared sequence motifs present in the upstream regions of coexpressed Or genes of the antenna and larva. ORNs are indicated in all cases in which A and B ORNs have been distinguished by physiological analysis; otherwise only the sensillum designation is indicated. L(X) indicates a larval ORN.
ACKNOWLEDGMENTS
We thank Paul Graham for technical assistance, Jacques van Helden and Hugh Robertson for help with bioinformatics, Keita Endo and Chiharo Hama for generously sharing unpublished data and for discussion, Coral Warr and Barry Yedvobnick for reagents and Anupama Dahanukar for critical reading of the manuscript. Supported by NIH DC04729 and DC02174 and a McKnight Investigator Award to J.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bergman C, Pfeiffer B, Rincon-Limas D, Hoskins R, Gnirke A, Mungall C, Wang A, Kronmiller B, Pacleb J, Park S, et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:research0086.1–0086.20. doi: 10.1186/gb-2002-3-12-research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. Olfaction in Drosophila: from odor to behavior. Trends in Genetics. 1996;12:175–180. doi: 10.1016/0168-9525(96)10015-9. [DOI] [PubMed] [Google Scholar]
- Clyne P, Grant A, O'Connell R, Carlson J. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- Clyne P, Warr C, Carlson J. Candidate Taste Receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Clyne P, Warr CG, Freeman M, Lessing D, Kim J, Carlson J. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999a;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Clyne P, Certel S, de Bruyne M, Zaslavsky L, Johnson W, Carlson J. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron. 1999b;22:339–347. doi: 10.1016/s0896-6273(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson B. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Foster K, van der Goes van Naters W, Carlson J. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nature Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Hallem EA, Carlson JR. Insect chemoreception. Curr Opin Neurobiol. 2005;15:423–430. doi: 10.1016/j.conb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne P, Carlson J. Odor coding in a model olfactory organ: the Drosophila maxillary palp. Journal of Neuroscience. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson J. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa A, van der Goes van Naters W, Warr C, Steinbrecht R, Carlson J. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Elmore T, Ignell R, Carlson J, Smith D. Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J Neurosci. 2003;23:9906–9912. doi: 10.1523/JNEUROSCI.23-30-09906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Domingos A, Asahina K, Naef F, Vosshall L, Louis M. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr Biol. 2005;15:2086–2096. doi: 10.1016/j.cub.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall L. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Flores G, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Goldman A, Van der Goes van Naters W, Lessing D, Warr C, Carlson J. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Goulding S, zur Lage P, Jarman A. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Gupta B, Flores G, Banerjee U, Rodrigues V. Patterning an epidermal field: Drosophila lozenge, a member of the AML-1/Runt family of transcription factors, specifies olfactory sense organ type in a dose-dependent manner. Dev Biol. 1998;203:400–411. doi: 10.1006/dbio.1998.9064. [DOI] [PubMed] [Google Scholar]
- Gupta B, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- Hallem E, Carlson J. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Hallem E, Dahanukar A, Carlson J. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho M, Carlson J. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Helms W, Lee H, Ammerman M, Parks A, Muskavitch M, Yedvobnick B. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev Biol. 1999;215:358–374. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos M, Clemens JC, Fishilevich Y, Vosshall L, Zipursky S. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Jefferis G, Vyas R, Berdnik D, Ramaekers A, Stocker R, Tanaka N, Ito K, Luo L. Developmental origin of wiring specificity in the olfactory system of Drosophila. Development. 2004;131:117–130. doi: 10.1242/dev.00896. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Carlson J, Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nature Neuroscience. 2004;7:819–825. doi: 10.1038/nn1284. [DOI] [PubMed] [Google Scholar]
- Larsson M, Domingos A, Jones W, Chiappe M, Amrein H, Vosshall L. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Lewcock J, Reed R. A feedback mechanism regulates monoallelic odorant receptor expression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia D, Mendelsohn M, Kirkland J, Axel A. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Maier D, Nagel A, Preiss A. Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation. Proc Natl Acad Sci U S A. 2002;99:15480–15485. doi: 10.1073/pnas.242596699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkman T, Sengupta P. The worm's sense of smell. Development of functional diversity in the chemosensory system of Caenorhabditis elegans. Dev Biol. 2004;265:302–319. doi: 10.1016/j.ydbio.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Misra S, Crosby M, Mungall C, Matthews B, Campbell K, Hradecky P, Huang Y, Kaminker J, Millburn G, Prochnik S, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Current Opinion in Neurobiology. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Neuhaus E, Gisselmann G, Zhang W, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J, Piekos B, Carlson J. The maxillary palp of Drosophila: ultrastructure and physiology depends on the lozenge gene. J Comp Physiol A. 1997;180:143–150. doi: 10.1007/s003590050035. [DOI] [PubMed] [Google Scholar]
- Robertson H, Warr C, Carlson J. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Schuldt A, Brand A. Mastermind acts downstream of notch to specify neuronal cell fates in the Drosophila central nervous system. Dev Biol. 1999;205:287–295. doi: 10.1006/dbio.1998.9014. [DOI] [PubMed] [Google Scholar]
- Sen A, Reddy G, Rodrigues V. Combinatorial expression of Prospero, Seven-up, and Elav identifies progenitor cell types during sense-organ differentiation in the Drosophila antenna. Dev Biol. 2003;254:79–92. doi: 10.1016/s0012-1606(02)00021-0. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nature Neuroscience. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shanbhag S, Muller B, Steinbrecht A. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int J Insect Morphology & Embryology. 1999;28:377–397. [Google Scholar]
- Shykind B, Rohani S, O'Donnell S, Nemes A, Mendelsohn M, Sun YH, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Stocker R, Gendre N, Batterham P. Analysis of the antennal phenotype in the Drosophila mutant lozenge. J Neurogen. 1993;9:29–53. doi: 10.3109/01677069309167274. [DOI] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream regions of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- van Helden J, Rios A, Collado-Vides J. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Research. 2000;28:1808–1818. doi: 10.1093/nar/28.8.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vosshall L, Wong A, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Yao C, Ignell R, Carlson J. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedvobnick B, Kumar A, Chaudhury P, Opraseuth J, Mortimer N, Bhat K. Differential effects of Drosophila mastermind on asymmetric cell fate specification and neuroblast formation. Genetics. 2004;166:1281–1289. doi: 10.1534/genetics.166.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Shared sequence motifs present in the upstream regions of coexpressed Or genes of the antenna and larva. ORNs are indicated in all cases in which A and B ORNs have been distinguished by physiological analysis; otherwise only the sensillum designation is indicated. L(X) indicates a larval ORN.