Abstract
Treatment of mammalian cells with small molecule histone deacetylase (HDAC) inhibitors induces changes in the transcription of specific genes. These changes correlate directly with an increase in the acetylation levels of all four core histones in vivo. Antibodies directed against endogenous HDAC1, HDAC2, or HDAC3 immunoprecipitate histone deacetylase activity that is inhibited in vitro by the small molecule trapoxin (TPX), and all three HDACs are retained by a TPX-affinity matrix. HDAC1 and HDAC2 are associated in HeLa cells in a complex that is predominantly separate from an HDAC3 immune complex. Both Jurkat HDAC1 and HeLa HDAC1/2 immune complexes deacetylate all four core histones and recombinant HDAC1 deacetylates free and nucleosomal histones in vitro. Purified recombinant HDAC1 deacetylates core histones in the absence of protein cofactors. Site-directed mutagenesis was used to identify residues required for the enzymatic and structural integrity of HDAC1. Mutation of any one of four conserved residues causes deleterious effects on deacetylase activity and a reduced ability to bind a TPX-affinity matrix. A subset of these mutations also cause a decreased interaction with the HDAC1-associated proteins RbAp48 and mSin3A. Disruption of histone deacetylase activity either by TPX or by direct mutation of a histidine presumed to be in the active site abrogates HDAC1-mediated transcriptional repression of a targeted reporter gene in vivo.
The genetic information of eukaryotes is packaged into chromatin, a highly organized and dynamic protein-DNA complex. The fundamental subunit of chromatin, the nucleosome, is composed of an octamer of four core histones; an H3/H4 tetramer and two H2A/H2B dimers, surrounded by 146 bp of DNA. The N-terminal tail domains of the core histones contain highly conserved lysines that are sites for posttranslational acetylation. Acetylation and deacetylation are catalyzed by histone acetyltransferases and histone deacetylases (HDACs), respectively. Converting the ɛ-amino groups of lysines into neutral ɛ-acetamido groups results in changes in chromatin structure and gene transcription (1–4). The precise mechanism(s) by which histone acetylation alters transcription is poorly understood, although naturally occurring HDAC inhibitors are beginning to provide useful insights into this cellular phenomenon.
The small molecule HDAC inhibitor trapoxin (TPX) induces cell cycle arrest and differentiation of many cell types in culture (5). These effects correlate with TPX-induced histone hyperacetylation in vivo and inhibition of HDAC activity in vitro (5). A modified synthetic version of TPX, K-trap, was used as an affinity ligand to purify and characterize the first identified HDAC—HDAC1 (6). HDAC1 is highly related to the yeast transcriptional regulator Rpd3p, thus providing a molecular link between histone deacetylation and transcription. The yeast RPD3 and related HDA1 genes encode proteins with HDAC enzymatic activity and disruption of either of these genes causes histone hyperacetylation and changes in transcription (7). The demonstration that HDAC1, -2, and -3 can each interact with the DNA-binding protein YY1 provided evidence that deacetylases associate directly with transcription factors to regulate gene expression in mammals (8, 9). Finally, the observation that HDAC1 and HDAC2 are components of nuclear corepressor complexes and that histone acetyltransferases are contained in coactivator complexes, suggests an intimate relationship between histone acetylation and the cellular transcription apparatus (reviewed in refs. 1–3, 10).
In the present study, we examined the biochemical properties and substrate specificities of three human HDAC-family members. Coimmunoprecipitation and immunoblot experiments indicated the existence of distinct HDAC complexes. In vitro deacetylase assays demonstrated that endogenous HDAC immune complexes deacetylate all four core histones in a TPX-sensitive fashion and recombinant HDAC1 deacetylates both free and nucleosomal histones. Finally, we used site-directed mutagenesis to define a deacetylase consensus motif that links the enzymatic activity of HDAC1 with its ability to mediate targeted transcriptional repression.
MATERIALS AND METHODS
Plasmids, Reporter Constructs, and Mutagenesis.
WT-GAL4-VP16 and H141A-GAL4-VP16 expression plasmids were constructed by subcloning the NotI-EcoRI fragment of pBJ5/HDAC1-F (6) or pBJ5/H141A-F into pSP/GAL(1–147)-VP16. The GAL4-luc reporter and GAL4-VP16 plasmids were described previously (11). Kunkel mutagenesis procedures are outlined in the Muta-Gene system (Bio-Rad). Mutants were subcloned into a C-terminal FLAG-epitope tag encoding-pBJ5 or pBJ5neo vector. Clones were sequenced to confirm expected mutations.
Antibodies, Immunoprecipitations, and K-Trap.
Antibodies against mSin3A (11) and HDAC1 residues 467–482 (6) were described previously. Other antibodies were generated as follows: synthetic peptides corresponding to residues 415–425 of RbAp48, residues 475–488 of HDAC2, or residues 415–428 of HDAC3 were covalently conjugated to keyhole limpet hemocyanin (Pierce) and used to immunize rabbits. Antibodies were affinity purified on peptide columns prepared using the Sulfolink system (Pierce). For HDAC immunoblotting, HDACs were immunoprecipitated by using affinity purified antibodies covalently conjugated to protein A agarose beads (GIBCO) by crosslinking with dimethyl pimelimidate. Cell extracts were prepared by lysis in Jurkat lysis buffer (JLB) (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/10% glycerol/0.5% Triton X-100) containing 2 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin, and 1 μg/ml leupeptin. Lysates were incubated at 4°C for 20 min with inversion and then centrifuged at 10,000 × g for 10 min to isolate supernatants. HDAC1, HDAC2, and HDAC3 immune complexes for deacetylase assays were prepared by incubating extracts with antiserum for 1 h, followed by 45 min precipitation with protein A agarose beads. Immunoprecipitations of recombinant HDAC1-F and HDAC1-F mutants were performed by using anti-FLAG agarose beads (IBI). All immunoprecipitates were washed three times with JLB prior to immunoblot or activity assays. K-Trap matrix preparation and binding assays were as described (12). Specifically bound proteins were eluted from the affinity matrix in SDS loading buffer, separated by SDS/PAGE, and examined by immunoblot analysis.
HDAC Assays.
Immunoprecipitates (endogenous or recombinant HDACs) from HeLa-S3 or simian virus 40 (SV40) large T-antigen (T-Ag) transformed Jurkat T cells were incubated with 1 μl [3H]acetate-labeled HeLa histones (10,000 dpm) for 2 h at 37°C and deacetylase activity was determined as described (6). For HDAC substrate-competition assays, 4 μg [3H]acetate-labeled HeLa histones were incubated simultaneously with enzyme (10 ng) and competitor for 15 min at 37°C and the reaction was stopped on ice and counted (6).
Nucleosomes were prepared from SV40 minichromosomes (gift from G. Sewack and U. Hansen, Dana–Farber Cancer Institute, Boston) by incubating 4.5 μg of minichromosomes with 2 units of micrococcal nuclease (MNase, Worthington) for 8 min at 37°C followed by quenching with 4 mM EDTA on ice. Recovery of mono-, di-, and trinucleosomes was confirmed by examining digested DNA in 2% agarose gels stained with ethidium bromide (data not shown). Anti-FLAG immunoprecipitated HDAC1-F was incubated with nucleosomes or minichromosomes for 2–3 h at 37°C and the reaction was stopped by addition of SDS loading buffer. Deacetylation reactions were separated on an 18% SDS/PAGE nonreducing gel, transferred to Immobilon P, and immunoblotted by using antibodies to acetylated H3 or acetylated H4 (gift from C. D. Allis, Biology Dept., University of Rochester).
HDAC1-F Protein Purification.
Recombinant C-terminally FLAG-epitope tagged HDAC1-F was expressed in Sf9 cells using the baculovirus expression system (PharMingen) described previously (11). Infected Sf9 cells were harvested, washed one time in PBS, lysed in JLB and centrifuged at 20,000 × g. The supernatant was loaded onto a Q-Sepharose Fastflow (Pharmacia) column and washed with several column volumes of Q-Sepharose buffer A (QA; 25 mM Tris, pH 7.1/150 mM NaCl/10% glycerol/0.5% Triton X-100). Proteins were eluted over a salt gradient from 150 mM to 1 M NaCl. Fractions containing HDAC1-F were pooled and loaded onto a hydroxyapatite column (Bio-Rad) and washed with several column volumes hydroxyapatite buffer A (HA; 1 mM NaH2PO4, pH 6.9/150 mM NaCl/10% glycerol); proteins were eluted in a salt gradient from 1 mM to 500 mM NaH2PO4. The fractions containing HDAC1-F were pooled and dialyzed into buffer QA and immunopurified by using anti-FLAG agarose beads (IBI). Specifically bound HDAC1-F was eluted by using excess FLAG peptide. The purified enzyme was stored as a 50% glycerol stock at −20°C.
Histone Isolation and Purification.
[3H]Acetate-incorporated histones were isolated from butyrate-treated HeLa cells by acid extraction as described in ref. 13. Bulk cellular histones (1 mg) were separated on an 18% SDS/PAGE reducing gel and stained with Coomassie blue. Bands corresponding to histones H4 and H3 were excised individually and histones H2A and H2B were excised together. The proteins were electroeluted by using a Bio-Rad 422 protein electroeluter according to established protocols (14).
Cell Culture and Transfections.
SV40 T-Ag Jurkat cells were transfected by electroporation with wild-type or mutant FLAG-epitope tagged pBJ5-HDAC1 expression constructs as described (6), except that T cells were harvested 48 h posttransfection. Human 293 cell transfections and luciferase assays were described (11).
RESULTS
HDAC1 and HDAC2 Are Associated in Vivo.
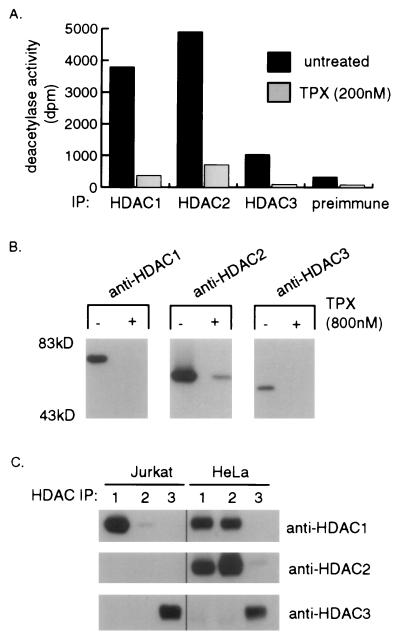
To study the endogenous HDAC enzymes, we generated antibodies to each of the three known human deacetylases, HDAC1, HDAC2, and HDAC3. To eliminate the possibility of antibody cross-reactivity, the antibodies were raised against the C-terminal region of the enzymes, where the amino acid sequences of the three HDACs markedly differ (8). In agreement with results using overexpressed recombinant enzymes (8), all three endogenous HDAC immune complexes possess HDAC activity in vitro (Fig. 1A). To examine whether all or a subset of the HDAC immune complexes are subject to inhibition by TPX, immunoprecipitates were preincubated with 200 nM TPX prior to assaying for deacetylase activity. The activity of all three HDAC immune complexes is significantly reduced by TPX (Fig. 1A). Given the TPX-sensitive deacetylase activity exhibited by the immunoprecipitates, we tested the ability of the three HeLa HDACs to bind a TPX-based affinity matrix (K-trap) (6). Consistent with the inhibitory effect of TPX on the HDAC immune complexes, all three enzymes were retained by K-trap and binding was competed specifically by preincubating with excess soluble TPX (Fig. 1B).
Figure 1.
Characteristics of endogenous HDAC1, -2, and -3 immune complexes. (A) HeLa-S3 cell extract was immunoprecipitated (IP) by using anti-HDAC1, 2, or 3 antibodies or preimmune serum. Each immunoprecipitate was divided in two and one-half of the immunoprecipitate was incubated with 200 nM TPX prior to assaying for deacetylase activity (dpm). (B) Immunoblot of HDAC1, -2, and -3 binding a TPX-based affinity matrix. Equal amounts of HeLa-S3 cell extract were preincubated with (+) or without (−) 800 nM TPX for 20 min at 4°C and then incubated with K-trap affinity resin for 1 h at 4°C. Eluted proteins were separated by SDS/PAGE, transferred and immunoblotted by using the indicated antibody. (C) Anti-HDAC1, -2, or -3 immunoprecipitates (IP) from SV40 T-Ag Jurkat or HeLa-S3 cell extracts were tested for coprecipitation of associated HDACs by immunoblotting with the antibodies indicated.
We next investigated the possibility of an interaction between different HDACs in vivo. Antibodies against each of the three HDACs were used to immunoprecipitate proteins from Jurkat or HeLa cell extract and precipitated proteins were analyzed by immunoblotting. Anti-HDAC1 immunoprecipitates from HeLa cells contained significant amounts of HDAC2 and, conversely, HDAC2 immunoprecipitates contained high levels of HDAC1 (Fig. 1C). Almost no HDAC1 is coimmunoprecipitated with HDAC2 from Jurkat cell extract, consistent with a low level of HDAC2 expression in this cell line (Fig. 1C and data not shown). In contrast to the apparent association between HDAC1 and HDAC2 in HeLa cells, no significant amount of HDAC1 or HDAC2 is immunoprecipitated by anti-HDAC3 antibodies. Likewise, no HDAC3 is detected in anti-HDAC1 or anti-HDAC2 immunoprecipitates (Fig. 1C). Thus, HDAC1 and HDAC2 appear to associate preferentially in cells expressing both proteins, whereas the majority of HDAC3 in both Jurkat and HeLa cells appears to be uncomplexed with either of the two other HDACs. We infer that both differential HDAC expression patterns and HDAC-association preferences may be important in establishing HDAC complexes in vivo.
HDAC1 and HDAC1/2 Immune Complexes Deacetylate All Four Core Histones in Vitro.
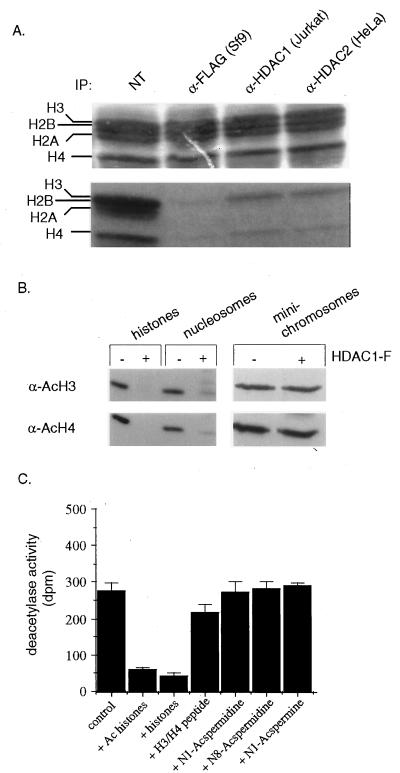
Triton-acid-urea gel analysis of histones from TPX-treated cells indicates that all four core histones become hyperacetylated (5). Based on the above observation that all mammalian HDAC immune complexes tested are inhibited by TPX in vitro and that all core histones are hyperacetylated in response to TPX in vivo, we were prompted to determine the in vitro substrate specificities of the mammalian deacetylases. [3H]Acetate-incorporated HeLa histones were incubated with anti-HDAC1 immunoprecipitates from Jurkat T-cells or anti-HDAC2 immunoprecipitates from HeLa cells. A fluorogram of the electrophoretically separated proteins shows a significant reduction in the extent of acetylation of all core histones following incubation with either the native HDAC immune complexes or with the immunopurified baculovirus-expressed HDAC1-F (Fig. 2A). Anti-HDAC3 immunoprecipitations did not yield sufficient amounts of deacetylase activity to determine accurately the substrate potential of the endogenous enzyme (data not shown).
Figure 2.
HDAC immune complexes deacetylate all four core histones in vitro. (A) 3H-acetate-labeled histones (24 μg) from butyrate-treated HeLa cells were subjected to deacetylation by immunoprecipitates (IP) from the designated cell extracts by using the antibodies indicated. Deacetylase reactions were separated by SDS/PAGE and stained with Coomassie blue (Upper). The gel was treated with En3Hance (National Diagnostics), dried, and placed on Biomax film (Kodak). The developed fluorogram (Lower) shows the extent of deacetylation. NT = no enzyme added. Positions of the core histones are indicated. (B) Immunoblot showing the extent of deacetylation of free, nucleosomal, or minichromosomal histones; (+) indicates incubation with immunopurified HDAC1-F, (−) indicates that no enzyme was added to the reaction. Blots were probed with antibodies against acetylated histones H3 (α-AcH3) and H4 (α-AcH4). (C) Histone substrate-competition assay: deacetylation by HDAC1-F was determined in the presence of the competitor indicated at the bottom of each column. Hyperacetylated histones (Ac histones) and steady-state histones (histones) were added at 5-fold molar excess (20 μg). A mixture of synthetic peptides (H3/H4 peptide) corresponding to H4 (amino acids 1–24) and H3 (amino acids 1–24) were added at ≈30-fold molar excess. Acetylpolyamines (4 μg) were added as indicated.
In order for HDACs to regulate chromosomal gene transcription directly, the enzymes must deacetylate nucleosomal histones. Therefore, we investigated the ability of HDAC1-F to deacetylate histones in the context of a nucleosome. Nucleosomes derived from SV40 minichromosomes were incubated with immunopurified HDAC1-F and significant deacetylation of H3 and H4 was observed by immunoblot analysis (Fig. 2B). We note that deacetylation by HDAC1-F of histones within intact SV40 minichromosomes was undetectable in this assay (Fig. 2B). The significance of this observation is unclear but may indicate that certain forms of higher order chromatin structure confer resistance to HDACs, an idea under current investigation. Minichromosomes stripped of nonhistone chromosomal proteins displayed the same resistance to deacetylation (data not shown).
To address the possibility of a nonspecific deacetylase activity exhibited by HDACs in vitro, we performed a series of substrate-competition experiments. HDAC assays were performed with purified HDAC1-F in the presence of excess nonradiolabeled competitor substrates. As expected, an excess of nonradiolabeled, acetylated histones strongly competed with the radiolabeled histone substrate for deacetylation by HDAC1-F (Fig. 2C). Histones with lower (steady-state) acetylation levels were also effective substrate competitors, while a mixture of nonacetylated H3/H4 N-terminal tail peptides had only a weak inhibitory effect (Fig. 2C). The small molecule polyamines N1-acetylspermine and N1- and N8-acetylspermidine had no effect on HDAC1-F-catalyzed histone deacetylation (Fig. 2C). This agrees with studies demonstrating that the acetylpolyamine amidohydrolase inhibitor 7-[N-(3-aminopropyl)amino]heptan-2-one (APAH) did not induce histone hyperacetylation in vivo (15). The nonhistone chromosomal high mobility group (HMG) proteins are posttranslationally acetylated at specific lysines in vivo (16), making them potential HDAC substrates. Substrate competition experiments using different HMG protein preparations have produced variable results (C.A.H., J.K.T., P.G.G., and S.L.S., unpublished observations).
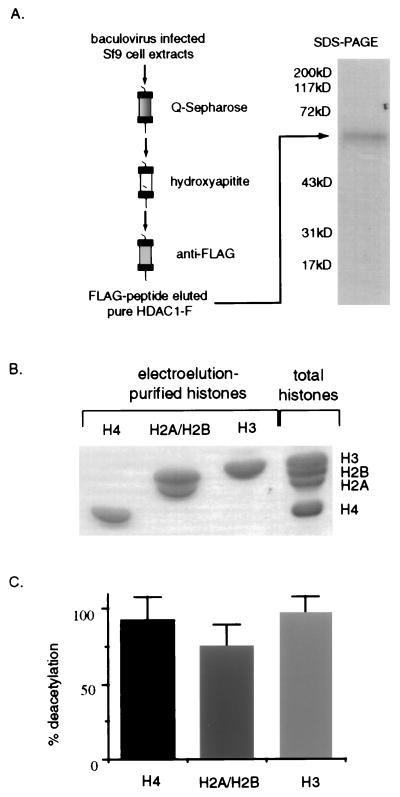
To determine if HDAC1 requires additional protein cofactors for deacetylase activity, we established a protocol to purify the recombinant baculovirus-expressed HDAC1-F to homogeneity (Fig. 3A). We further purified the radiolabeled, acid-extracted histones by gel electroelution to remove potential cofactors that may have copurified with the histone substrate (Fig. 3B). Purified H3, H4, or a mixture of H2A and H2B histones were efficiently deacetylated by pure HDAC1-F (Fig. 3C). These results suggest that HDAC1 possesses intrinsic catalytic activity that does not require protein cofactors for histone deacetylation in vitro.
Figure 3.
Pure HDAC1-F deacetylates histones in the absence of protein cofactors. (A) HDAC1-F was purified by using the protocol shown and the final step FLAG-peptide elution was examined on a 10% SDS/PAGE reducing gel, stained with Coomassie blue. Relative molecular mass is indicated in kilodaltons. (B) [3H]Acetate-labeled histones from butyrate-treated HeLa cells were purified as individual histones (or an H2A/H2B mixture) by electroelution, and a sample from each was run on an SDS/18% polyacrylamide gel and stained with Coomassie blue. Total acid extracted HeLa cellular histones were loaded in the far right lane as a reference and the positions of the individual histones are indicated (Right). (C) Pure HDAC1-F (25 ng) was incubated with the indicated histones (0.5 μg) and the average deacetylation for a given substrate was divided by the total dpm and calculated as the percent deacetylation.
Identification of an HDAC1 Deacetylase Motif.
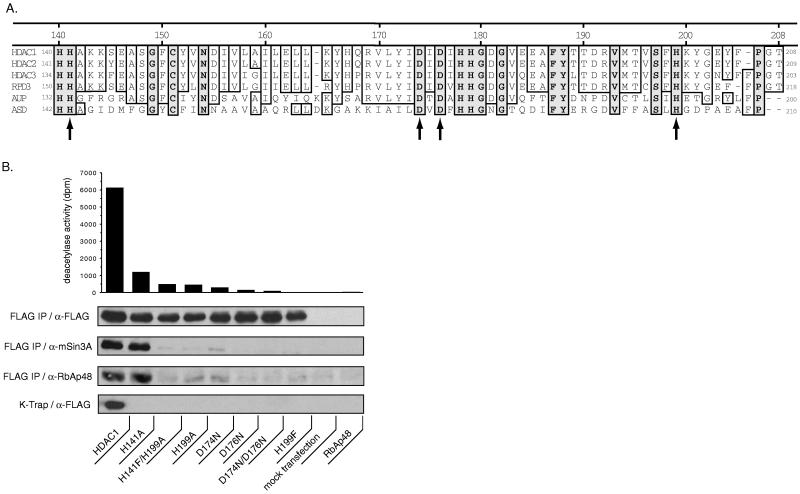
We used site-directed mutagenesis to identify residues in HDAC1 that are required for deacetylase activity. Sequence homology alignment of all known and putative HDACs, using the National Center for Biotechnology Information blast program, revealed general and widespread conservation of residues. Selection criteria for HDAC1 mutagenesis was based on several observations. First, HDAC enzymes are likely to be metalloproteins because the HDAC-inhibitor trichostatin A contains a functionally critical hydroxamic acid (17) that is likely to act via a metal chelation mechanism. Second, the HDAC-related acetylspermine deacetylase is reported to be a zinc metalloenzyme (18). Consistent with the notion that HDAC1 is a metalloenzyme, the zinc chelator 2-mercaptopyridine N-oxide inhibits HDAC1 in the micromolar range (C.A.H., J.K.T., and S.L.S., unpublished observations). Further sequence alignment of HDAC1 with acetylspermine deacetylase and acetoin utilization protein revealed 26 absolutely conserved residues. Among the conserved residues are several histidines and aspartic acids with potential catalytic and/or metal coordinating properties (Fig. 4A). We mutated four of these residues by site-directed mutagenesis, including histidines 141 (H141) and 199 (H199) to alanine (or phenylalanine), and aspartates 174 (D174) and 176 (D176) to asparagine.
Figure 4.
Mutagenesis of HDAC1 identifies important catalytic and/or structural residues in the enzyme. (A) Amino acid sequence alignment of a conserved region of HDAC1 with other eukaryotic HDACs, and with the related prokaryotic acetoin utilization protein (AUP) and acetyl polyamino amidohydrolase (ASD). Completely conserved residues are boxed in gray. Residues targeted for mutation in this study are indicated by an arrow. Other residues with potential roles in catalysis include His-140, Cys-151, His-178, His-179, Tyr-188, and Ser-197. (B) HDAC1 mutant analysis: All wild-type and mutant constructs contained C-terminal FLAG-epitope tags used to immunoprecipitate (FLAG IP) the overexpressed proteins from transfected SV40 T-Ag Jurkat cells. FLAG IPs were tested for deacetylase activity and immunoblotted by using the antibodies indicated to determine immunoprecipitation efficiency (α-FLAG) and coprecipitated proteins (α-mSin3A and α-RbAp48). A background of 444 dpm from the mock transfection (no DNA) IP was subtracted from each IP to normalize deacetylase activity. Extracts from a separate transfection were incubated with K-trap matrix and immunoblotted with α-FLAG antibody. RbAp48 (in pBJ5) is an additional negative control.
HDAC activity from immunoprecipitates of all mutants was reduced by 85–100% relative to wild-type HDAC1-F (Fig. 4B). The residual activity observed may be the result of a partially active enzyme or of an associated deacetylase activity. Blots of the immunoprecipitates were probed with FLAG antibody to confirm equivalent immunoprecipitation efficiencies of wild-type and mutant constructs (Fig. 4B). To determine whether reduced enzymatic activity was due to an altered catalytic site residue or due to a more global change in conformation, blots of the immunoprecipitates were probed for HDAC1-associated proteins. RbAp48 and mSin3A have been demonstrated to associate with endogenous HDAC1 in vivo (2, 6). RbAp48 is a putative adapter protein of chromatin modifying factors and mSin3A is a transcriptional corepressor that bridges HDAC function to DNA-binding transcription factors (2). Immunoprecipitates of enzymes bearing the single or double mutation of D174N, and/or D176N, as well as the double mutation H141F/H199A, or the single mutation H199A, do not efficiently coimmunoprecipitate RbAp48 or mSin3A (Fig. 4B). These residues may serve roles in HDAC1 structure as they appear to be important for maintaining the association of HDAC1 and other proteins. In contrast, the H141A mutant coimmunoprecipitates both RbAp48 and mSin3A to levels identical to wild-type HDAC1, indicating proper native folding of the enzyme and suggesting a primary role for histidine 141 in catalysis (Fig. 4B). Interestingly, pH studies demonstrate that HDAC1-F-mediated histone deacetylation is half-optimal at pH 6.8 (C.A.H., J.K.T., and S.L.S., unpublished observations), which is in the range of pKa’s reported for catalytic histidines in other enzymes.
The HDAC1 mutants were further tested for their ability to bind to the K-trap affinity matrix. It is likely that mutations in the active site might also affect the binding of molecules in the vicinity of the active site. Extract from Jurkat cells overexpressing HDAC1 mutants was incubated with K-trap and specifically bound proteins were analyzed by immunoblotting. Whereas wild-type HDAC1-F was bound by K-trap, none of the HDAC1 mutants showed appreciable binding, suggesting a direct correlation between catalytic activity and TPX binding (Fig. 4B).
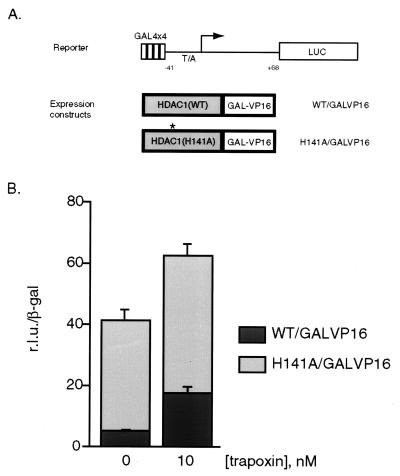
HDAC Activity Is Necessary for Full Repression of an HDAC1-Silenced Reporter Gene.
Previous experiments have shown that the transcription factor Mad functions as a repressor by interacting with mSin3 (19). This repression is sensitive to TPX and trichostatin A, suggesting that Mad recruits an mSin3/HDAC corepressor complex to DNA and represses transcription in part by localized histone deacetylation (11, 20). To extend these observations, we designed a number of chimeric DNA-binding constructs composed of wild-type or mutant HDAC1 fused directly to the DNA-binding transcriptional activator GAL4-VP16 (Fig. 5A). These DNA-binding constructs were cotransfected with a GAL4-driven luciferase reporter plasmid and tested for their ability to repress transcription in vivo (Fig. 5B). Both wild-type and mutant constructs expressed to similar levels (data not shown). In accordance with the previous observation that HDAC1-GAL4 is a repressor (21), the wild-type HDAC1 fusion protein (WT-GAL4-VP16) represses transcription of the reporter gene (Fig. 5B, black column, left) relative to GAL4-VP16, and this repression is reduced ≈4-fold in cells treated with 10 nM TPX (Fig. 5B, black column, right). The partial derepression induced by TPX on the WT-GAL4-VP16 construct agrees with the reduction (≈50%) in deacetylase activity of HDAC1 immunoprecipitates following treatment in vivo with 10 nM TPX (11). The enzymatically compromised HDAC1 mutant chimera (H141A-GAL4-VP16) was a less effective repressor (≈7-fold decreased repression; gray column, left) when compared with WT-GAL4-VP16 (Fig. 5B). Treatment with 10 nM TPX caused only a minor fold-derepression of transcription by H141A-GAL4-VP16 (Fig. 5B, gray column, right), consistent with the reduced TPX-binding activity of the H141A mutant enzyme.
Figure 5.
HDAC activity is important for targeted transcriptional repression by HDAC1 in vivo. (A) GAL4-driven luciferase reporter construct and HDAC1 wild-type (WT) and mutant (H141A) GAL4-VP16 DNA-binding expression constructs. (B) Extracts from transfected 293 cells were prepared and luciferase and β-galactosidase activities were assayed according to manufacturer’s directions (Promega). Luciferase values (relative light units, r.l.u.) were normalized for transfection efficiency by dividing by β-galactosidase activity. Cells were treated with or without 10 nM TPX as described. Between individual experiments, TPX-induced derepression of WT-GAL4-VP16 averaged 7.5-fold as compared with an average of 2.5 for H141A-GAL4-VP16. r.l.u. by GAL4-VP16 are 300–600× higher than WT-GAL4-VP16 and derepression by TPX is generally 1.5–2.0-fold (data not shown).
DISCUSSION
Early studies using HDAC inhibitors suggested a role of histone acetylation in transcriptional control (5). More recent work has identified a series of histone acetyltransferase-coactivator proteins and HDAC-associated corepressor complexes that suggests the enzymatic modulation of histone acetylation is an integral component of gene regulation (1, 2). We performed a series of experiments to characterize the biochemical nature of HDACs in an attempt to directly link the specific enzymatic activity of HDACs to their proposed role in transcriptional regulation.
The TPX-induced hyperacetylation of all four core histones (5) in vivo is consistent with the inhibition of all HDAC-immune complexes by TPX in vitro and with the demonstration that HDAC immune complexes deacetylate all four core histones. Based on the above observations, certain HDACs may not deacetylate specific histones intrinsically, but rather, accessible nucleosomal histones may be deacetylated by a recruited HDAC or HDAC complex. Transcriptional control of specific genes may be orchestrated by distinct HDAC complexes that form depending on the HDAC expression profile of the cell, as well as on the intrinsic associations of HDAC-family members. This does not exclude other HDAC enzymes, such as HDAC3 or the maize nucleolar deacetylases, from exhibiting a more limited histone or lysine substrate specificity (8, 22).
Once recruited to DNA, HDACs must deacetylate nucleosomal histones. HDAC1-F deacetylates histones H3 and H4 from SV40 minichromosome-derived nucleosomes; however, deacetylation of the intact SV40 minichromosome was not detected under these same conditions in vitro. This raises the possibility of a required cofactor to modify or destabilize chromatin in order for HDAC to function on a polynucleosomal template or on higher order chromatin structures. Alternatively, HDAC1 might deacetylate an exclusive site of the minichromosome adjacent to the DNase I hypersensitive region of the promoter. Finally, HDACs may, in certain cases, act during chromatin assembly when histones are potentially more accessible.
We used site-directed mutagenesis to identify residues in HDAC1 that are required for enzymatic activity. Alignment of HDAC1 with other related proteins highlights a putative deacetylase signature motif from which several conserved residues were selected for mutation. Analysis of all mutants made in our study suggests an intimate relationship between enzymatic activity and TPX-binding activity. Furthermore, a single point mutation at a residue presumed to be in the HDAC1 active site can mimic the effect of TPX by inactivating both the deacetylation and transcriptional repression activities of the enzyme. It is worth noting that the acetyltransferase activity of Gcn5p is required for its coactivator function in yeast (C.D. Allis, personal communication). Our studies provide direct evidence that HDAC activity is important for transcriptional repression by HDACs.
Acknowledgments
We thank C. D. Allis, G. Sewack, K. Ebralidse, and U. Hansen for reagents and advice and E. Verdin, D. Kadosh, K. Struhl, F. Dangond, S. Gullans, J. Taunton, and I. Wilson for helpful discussion. We thank the National Institutes of Health (NIH) Cell Culture Center and Sera/Source for technical assistance. This work was supported by a National Institute of General Medical Sciences grant to S.L.S. and NIH Grant GM55668-01 to D.E.A. Predoctoral training grants from the NIH to C.A.H. and from the National Science Foundation and the Harvard-Markey Biomedical Scientist Program to J.K.T. are gratefully acknowledged. S.L.S. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HDAC
histone deacetylase
- TPX
trapoxin
- HMG
high-mobility group
- SV40
simian virus 40
- T-Ag
large T-antigen
References
- 1.Wade P A, Wolffe A P. Curr Biol. 1997;7:82–84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 2.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 3.Hassig C A, Schreiber S L. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 4.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 6.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 7.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W-M, Yao Y-L, Sun J-M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 9.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 11.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 12.Taunton J, Collins J L, Schreiber S L. J Am Chem Soc. 1996;118:10412–10422. [Google Scholar]
- 13.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 14.Ausubel F M, et al., editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. pp. 10.51–10.55. [Google Scholar]
- 15.Marchant P, Dredar S, Manneh V, Alshabanah O, Matthews H, Fries D, Blankenship J. Arch Biochem Biophys. 1989;273:128–136. doi: 10.1016/0003-9861(89)90170-7. [DOI] [PubMed] [Google Scholar]
- 16.Sterner R, Vidali G, Allfrey V G. J Biol Chem. 1981;256:8892–8895. [PubMed] [Google Scholar]
- 17.Yoshida M, Hoshikawa Y, Koseki K, Mori K, Beppu T. J Antibiot. 1990;9:1101–1106. doi: 10.7164/antibiotics.43.1101. [DOI] [PubMed] [Google Scholar]
- 18.Fujishiro K, Ando M, Uwajima T. Biochem Biophys Res Commun. 1988;157:1169–1174. doi: 10.1016/s0006-291x(88)80997-5. [DOI] [PubMed] [Google Scholar]
- 19.Ayer D E, Lawrence Q A, Eisenman R N. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 20.Laherty C, Yang W-M, Sun J-M, Davie J, Seto E, Eisenman R. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 21.Nagy L, Kao H-Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 22.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]