Abstract
The incorporation of unnatural amino acids site-specifically is a valuable technique for structure–function studies, incorporation of biophysical probes, and determining protein–protein interactions. THG73 is an amber suppressor tRNA used extensively for the incorporation of >100 different residues in over 20 proteins, but under certain conditions THG73 is aminoacylated in vivo by endogenous aminoacyl-tRNA synthetase. Similar aminoacylation is seen with the Escherichia coli Asn amber suppressor tRNA, which has also been used to incorporate UAAs in many studies. We now find that the natural amino acid placed on THG73 is Gln. Since the E. coli GlnRS recognizes positions in the acceptor stem, we made several acceptor stem mutations in the second to fourth positions on THG73. All mutations reduce aminoacylation in vivo and allow for the selection of highly orthogonal tRNAs. To show the generality of these mutations, we created opal suppressor tRNAs that show less aminoacylation in Xenopus oocytes relative to THG73. We have created a library of Tetrahymena thermophila Gln amber suppressor tRNAs that will be useful for determining optimal suppressor tRNAs for use in other eukaryotic cells.
Keywords: nonsense suppression, frameshift suppression, tRNA, opal suppressor tRNA, unnatural amino acid incorporation
INTRODUCTION
Incorporation of unnatural amino acids (UAAs) site-specifically in proteins biosynthetically is a powerful tool that is increasingly being used. The primary approach has been stop codon (nonsense) suppression using a designed tRNA with an anticodon recognizing the stop codon. In higher eukaryotes, nonsense suppression by a tRNA chemically aminoacylated with an UAA (tRNA-UAA) is primarily limited to Xenopus oocytes, where microinjection of the mutant mRNA and tRNA-UAA is straightforward and electrophysiology allows for sensitive detection of UAA incorporation (Dougherty 2000; Beene et al. 2003). Recently, site-specific UAA incorporation was performed in Xenopus oocytes using frameshift suppression of the quadruplet codons CGGG and GGGU, which allowed for the simultaneous incorporation of three UAAs (Rodriguez et al. 2006).
THG73, an amber suppressor tRNA from Tetrahymena thermophila with a G73 mutation, has been used extensively to incorporate >100 residues in 20 different proteins (Dougherty 2000; Beene et al. 2003). With THG73 and any other suppressor tRNAs, a key issue is orthogonality. An orthogonal tRNA is one that is not measurably aminoacylated with natural amino acids by endogenous aminoacyl-tRNA synthetases (aaRSs). Unlike its mathematical counterpart, there can be degrees of orthogonality when referring to tRNAs. While the orthogonality of THG73 has been evaluated using in vitro translation in Escherichia coli (Cload et al. 1996), wheat germ (England et al. 1999), and rabbit reticulocyte lysate (Rothman et al. 2005), the primary application of THG73 has been for in vivo translation in Xenopus oocytes, which is the focus of the present work. The orthogonality of THG73 is acceptable when injecting low quantities of mutant mRNA and/or tRNA-UAA with incubation times <2 d (Saks et al. 1996; Shafer et al. 2004). However, increasing the amount of mutant mRNA and/or tRNA-UAA or incubation times of 2 d—which we will term “excessive” conditions—led to increased aminoacylation of THG73 in vivo with natural amino acid(s) (Rodriguez et al. 2006). This is highly undesirable, because incorporation of natural amino acids at the suppression site leads to a heterogeneous mixture of proteins containing an UAA or natural amino acid(s) and limits the quantity of protein that can be produced by UAA incorporation.
Previous work from our laboratories showed that yeast Phe frameshift suppressor tRNAs (YFFSCCCG and YFaFSACCC) were aminoacylated much less than THG73 in vivo, but suppression of the quadruplet codons was less efficient than with THG73 and varied greatly with the amount of frameshift suppressor tRNA injected (Rodriguez et al. 2006). Previously, an E. coli Asn amber suppressor (ENAS) tRNA was shown to be highly orthogonal in vivo (Kleina et al. 1990) and in vitro (Cload et al. 1996; Murakami et al. 2003, 2006). Here we show that ENAS has similar amounts of aminoacylation to THG73, suggesting that other well-characterized amber suppressor tRNAs could be prone to aminoacylation under excessive conditions.
Therefore, we chose to determine the amino acid that was being placed on THG73 using the sensitivity of electrophysiology and a well-characterized mutation site of the nicotinic acetylcholine receptor (nAChR), and determined the amino acid to be Gln. Using this knowledge, we created “Knob” mutations on THG73 previously shown to reduce aminoacylation of E. coli tRNA2 Gln by the E. coli glutaminyl-tRNA synthetase (GlnRS) (Liu et al. 1997). The Knob mutations on THG73 resulted in tRNAs that showed drastically reduced suppression efficiency in vivo, making them not viable replacements. The E. coli GlnRS has been shown to interact with the acceptor stem of the tRNA in the crystal structure (Perona et al. 1989), which was subsequently shown to be sensitive to base pairs and/or backbone positioning of the tRNA acceptor stem (Jahn et al. 1991; Ibba et al. 1996; McClain et al. 1998). We created seven unique tRNAs with mutations in the second to fourth positions of the acceptor stem and analyzed their function in vivo relative to THG73. Mutations in the acceptor stem decreased aminoacylation significantly. This library of T. thermophila Gln amber suppressor (TQAS) tRNAs can be used to screen for orthogonality in other eukaryotic cells. To show the generality of the acceptor stem mutations, we also created T. thermophila Gln opal suppressor (TQOpS′ and TQOpS) tRNAs that are also aminoacylated less than THG73. The acceptor stem mutations have general application for the creation of orthogonal suppressor tRNAs for UAA incorporation in higher eukaryotic cells, where random mutagenesis combined with high-throughput screens are not readily applicable.
RESULTS
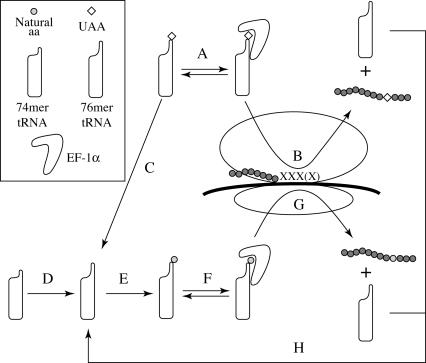
Schematic for site-specific UAA incorporation and aminoacylation
Figure 1 shows a schematic for the steps leading to UAA incorporation in vivo. Upon injection of the tRNA-dCA-UAA, the tRNA can bind to EF-1α and/or other components of the translational machinery in a reversible fashion (Fig. 1A). It has been shown that the prokaryotic ortholog EF-Tu can protect tRNA-dCA-UAA from hydrolysis in vitro (Fig. 1C; Nakata et al. 2006), thus favoring incorporation of UAA by recognition of the suppressor site on the mRNA (Fig. 1B). If tRNA lacking the terminal CA (74 mer) is injected, it can be converted to the full-length tRNA by the addition of CA in vivo (Fig. 1D). The full-length tRNA, either tRNA-dCA or tRNA-CA (76 mer), may then be aminoacylated with a natural amino acid by an endogenous aaRS (Fig. 1E). The tRNA-dCA-amino acid can then bind EF-1α and/or other components of the translational machinery (Fig. 1F) and lead to the incorporation of a natural amino acid at the suppression site (Fig. 1G) in competition with the suppression by tRNA-dCA-UAA (Fig. 1B), a highly undesirable outcome. After translation, both UAA and natural amino acid incorporation causes the tRNA-dCA/CA to be released from the ribosome (Fig. 1H), which can further contribute to the undesired natural amino acid incorporation (Fig. 1E–G).
FIGURE 1.
Site-specific UAA incorporation. (A) tRNA-dCA-UAA binds EF-1α and/or other components of the translational machinery. (B) UAA is incorporated at the suppression site [XXX(X)], resulting in a protein with an UAA. (C) Undesired hydrolysis and/or aaRS editing results in irreversible loss of the UAA, resulting in tRNA-dCA (76 mer). (D) Injection of the tRNA (lacking dCA, 74 mer) is converted to a 76 mer in vivo by the addition of CA (tRNA-CA). (E) Undesired recognition of the suppressor tRNA-dCA/CA by endogenous aaRS(s) can result in aminoacylation of the tRNA with a natural amino acid. (F) tRNA-dCA/CA-amino acid binds EF-1α and/or other components of the translational machinery. (G) Undesired protein translation can occur by placing a natural amino acid at the suppressor site (XXX[X]), rather than termination at the stop codon or a frameshift, and this competes with UAA incorporation. (H) After translation, the tRNA-dCA/CA is released into the cytoplasm and can repeat steps E–H.
Experimental scheme for evaluating aminoacylation and suppression by electrophysiology
All experiments were performed on the nAChR, which has been extensively studied by the site-specific incorporation of UAAs. The muscle-type nAChR is a pentamer composed of α, β, γ, and δ subunits in the ratio of 2:1:1:1, respectively. In order to compare experiments from different batches of oocytes, we normalized the average maximal current in response to 1 mM ACh for each suppressor tRNA to the corresponding average maximal current for THG73. Wild-type mRNA expression would be the most desirable normalization, but injection of 20–60 ng of mRNA (as in the aminoacylation experiments) would produce wild-type currents >100 μA, resulting in current saturation.
To evaluate aminoacylation, we chose a Leu site in the second transmembrane helix (M2) of the β subunit termed β9′. This site has been shown to tolerate placement of many natural amino acids and UAAs (Filatov and White 1995; Kearney et al. 1996; Kosolapov et al. 2000), with most causing characteristic shifts in ACh EC50. Previously, THG73 was shown to incorporate natural amino acid(s) at this site under excessive conditions (Rodriguez et al. 2006). A possible complication for experiments designed to probe aminoacylation is readthrough of the stop codon, which can be tested for by injecting mRNA only into the Xenopus oocyte. We find that, at the β9′ site, such readthrough is insubstantial, producing currents that are ∼3.5% of those seen when THG73 is included.
During the course of this research, we noticed variations in aminoacylation depending on whether the oocytes were acquired from Xenopus Express or Nasco. Xenopus laevis frogs from Xenopus Express are caught in Africa, while Nasco frogs are bred in a laboratory and are from a more restricted gene pool (L. Northey, pers. comm.). For each experiment, the figure legends state the supplier(s) of the oocytes.
Identifying the natural amino acid placed on THG73 using electrophysiology
As noted above, we focus on the β9′ site of the nAChR, which for the wild-type β9′L has an EC50 of 50 μM for ACh. For the well-characterized β9′S the EC50 is lowered ∼33-fold to 1.5 μM ACh (Kearney et al. 1996), and the lowering of EC50 has been shown to scale with the polarity of the residue introduced at the β9′ position (Kearney et al. 1996).
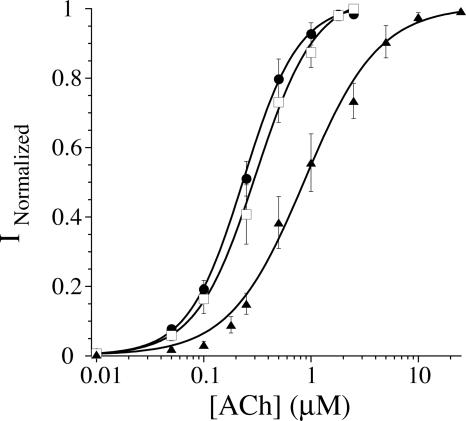
Injection of β9′UAG+THG73 (74 mer) under excessive conditions produces receptors with an EC50 of 0.24 μM ACh (Fig. 2), which is lower than any substitution previously tested at the β9′ position. When β9′UAG+THG73-dCA (76 mer) was evaluated, an EC50 of 0.88 μM was obtained. We chose to screen β9′Q because THG73 evolved to suppress the amber codon with Gln in T. thermophila (Hanyu et al. 1986), and many amber suppressor tRNAs are aminoacylated with Gln in vivo (Normanly et al. 1990). The conventional mutant β9′Q has an EC50 of 0.31 μM ACh (Fig. 2). While the EC50 of β9′Q and β9′UAG+THG73 (74 mer) are comparable, there is approximately a threefold increase in EC50 when β9′UAG+THG73-dCA (76 mer) is injected (Fig. 2). These results suggest that the full-length THG73-dCA (76 mer) is aminoacylated with amino acids other than Gln. Apparently, this is less important for THG73 (74 mer), which must have CA added in vivo to become a full-length tRNA (Fig. 1D). From these results, we conclude that the predominant natural amino acid placed on THG73 is Gln.
FIGURE 2.
Fits to the Hill equation for β9′UAG+THG73 (74 mer)/-dCA (76 mer) and β9′Q. Filled circles are β9′UAG+THG73 (74 mer) (16 ng per oocyte), open squares are β9′Q, and filled triangles are β9′UAG+THG73-dCA (76 mer) (5 ng per oocyte). EC50 values are 0.24 ± 0.006, 0.31 ± 0.02, and 0.88 ± 0.08 μM ACh, respectively. In each experiment n >3 oocytes.
Testing knob mutations on THG73
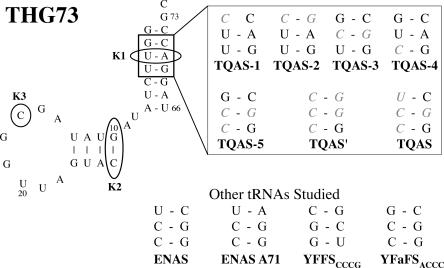
Much is known about the recognition of tRNA by the GlnRS, and key interactions have been assigned to various “knobs” on the tRNA, termed K1, K2, and K3 (Fig. 3). In a study of aminoacylation of E. coli tRNA2 Gln by the E. coli GlnRS (Liu et al. 1997), the mutations that produced the greatest reduction in aminoacylation were K2, K2K3, and K1K2K3. We, therefore, incorporated these mutations into THG73, and tested for orthogonality at the β9′ site.
FIGURE 3.
THG73 mutations and tRNAs studied. Circled positions on THG73 correspond to the knob mutations from Liu et al. (1997), where K1 is C3-G70, K2 is C10-G25, and K3 is G17. The boxed region on THG73 corresponds to the second to fourth positions of the acceptor stem (mutations are shown in gray italics in the right box). Note THG73 K1 and TQAS-3 are the same mutation. Other tRNAs studied are shown at the bottom with only the second to fourth positions of the acceptor stem shown. ENAS and TQAS contain the same nucleotides at these positions.
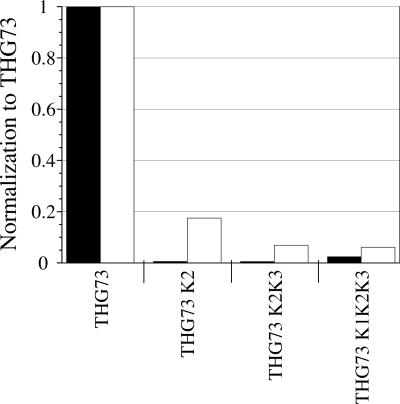
As shown in Figure 4, THG73 K2, THG73 K2K3, and THG73 K1K2K3 all show <20% aminoacylation in vivo when compared to THG73 at β9′UAG (Fig. 4). However, when each tRNA was chemically ligated with dCA-W and injected with α149UAG (a nonpermissive site), <3% of the current of THG73-W was seen (Fig. 4). Therefore, THG73 K2, THG73 K2K3, and THG73 K1K2K3 are nonfunctional in the Xenopus oocyte and are not viable alternatives for UAA incorporation in vivo.
FIGURE 4.
THG73 knob mutations. Individual tRNA average current were normalized by appropriate THG73 average current and bars represent this average ratio (total of oocytes tested is 91, where each bar is 15>n>5 oocytes). Black bars correspond to tRNA-W (21 ng per oocyte) suppressing at α149UAG and white bars correspond to tRNA (74 mer) (17 ng per oocyte) +β9′UAG in Xenopus Express oocytes. THG73 knob mutations are <20% when suppressing at α149UAG and show they are not functional alternatives for UAA incorporation.
We conclude that THG73 is very sensitive to the K2 and K3 mutations in the D-stem and D-loop, respectively, which make tertiary contacts with the variable loop to form the characteristic L-shape, tRNA structure. Previous replacement of the THG73 anticodon with ACCC (Shafer et al. 2004) and CCCG (Rodriguez et al. 2006) also resulted in nonfunctional frameshift suppressors in vivo. THG73 appears to be exceptionally sensitive to mutation within the D domain and the anticodon loop, which limits the regions where this tRNA can be mutated to create functional tRNAs.
Testing aminoacylation of ENAS and ENAS A71 in vivo
With the failure of the knob mutations to produce functional tRNAs for UAA incorporation, we chose to screen the ENAS, which was shown to be orthogonal in vitro (Cload et al. 1996) and in vivo (Kleina et al. 1990). When analyzing the structure of ENAS, we noticed that the second position of the acceptor stem contained the non-Watson–Crick base pair U2-C71. We created the variant ENAS C71A to form the canonical pair U2-A71 (Fig. 3) present in the wild-type tRNA (Kleina et al. 1990).
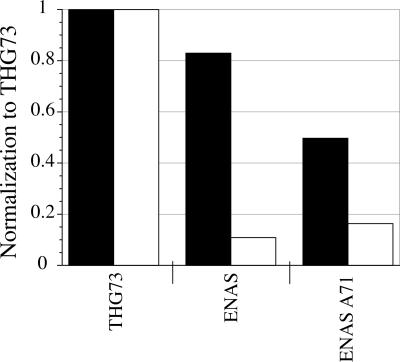
While injection of ENAS or ENAS A71 (74 mer) with β9′UAG resulted in less aminoacylation than THG73 in both Xenopus Express and Nasco oocytes, the effect was much more pronounced with Nasco oocytes (Fig. 5). The EC50s of ENAS (74 mer) or ENAS A71 (74 mer) with β9′UAG were 2.0 ± 0.1 and 1.5 ± 0.1 μM ACh, respectively, in Nasco oocytes. These EC50s are larger than the 0.31 μM ACh for β9′Q (Fig. 2), suggesting that ENAS and ENAS A71 are aminoacylated by another natural amino acid or a mixture of natural amino acids.
FIGURE 5.
ENAS and ENAS A71 aminoacylation tested at β9′UAG. Individual tRNA (74 mer) (17 ng per oocyte) average current was normalized to THG73 (74 mer) average current and bars represent this average ratio (total of oocytes tested is 75, where each bar is 22>n>5 oocytes). Black and white bars correspond to oocytes from Xenopus Express and Nasco, respectively. ENAS and ENAS A71 show a large amount of aminoacylation in Xenopus Express oocytes after a 2-d incubation, which has not been seen in vitro (Cload et al. 1996) or in vivo (Kleina et al. 1990). Aminoacylation is drastically reduced when tested in Nasco oocytes.
While ENAS and ENAS A71 tRNAs do show improved orthogonality compared to THG73, the suppression efficiencies of ENAS-W and ENAS A71-W were <26% of THG73-W in both Xenopus Express and Nasco oocytes (Rodriguez et al. 2007, companion paper, this issue). As such, neither ENAS tRNA is a viable replacement for THG73 for UAA incorporation in Xenopus oocytes.
Testing aminoacylation of THG73 acceptor stem mutations
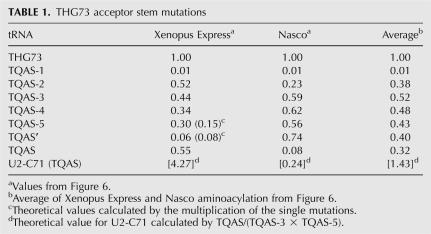
The E. coli GlnRS contacts the tRNA acceptor stem in the crystal structure (Perona et al. 1989), and biochemical experiments have shown specific base pairs and/or positioning of the backbone affect aminoacylation in vivo (Jahn et al. 1991; Ibba et al. 1996; McClain et al. 1998). As such, we decided to screen various mutations in the second to fourth positions of the acceptor stem (shown in Fig. 3) to create a library of T. thermophila Gln amber suppressor (TQAS) tRNAs (note in this nomenclature THG73 would be known as TQAS-0). The mutation G2C on THG73 (TQAS-1) results in the second position having C2 C71, which results in a nonfunctional tRNA and serves as a negative control. The single helix pair mutations C2-G71, C3-G70, and C4 are named TQAS-2, TQAS-3, and TQAS-4, respectively (Fig. 3). We then created the double helix pair mutation C3-G70 and C4 (TQAS-5) and the triple helix pair mutation C2-G71, C3-G70, and C4 (TQAS′). Finally, we placed the ENAS acceptor stem from the second to fourth position, including the U2-C71, on THG73 to create TQAS (Fig. 3).
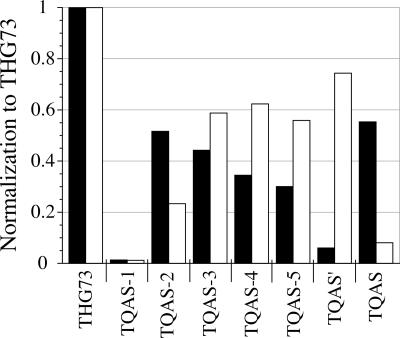
All acceptor stem mutations reduced aminoacylation relative to THG73 in both Xenopus Express and Nasco oocytes when tested at β9′UAG (Fig. 6; Table 1). TQAS-1 shows the same amount of current as β9′UAG mRNA injection alone, but due to the C2 C71 in the acceptor stem TQAS-1-W is unable to suppress α149UAG in both Xenopus Express and Nasco oocytes (Rodriguez et al. 2007, companion paper, this issue). In Xenopus Express oocytes, the single helix pair mutations on THG73 all lower aminoacylation, with orthogonality following the order TQAS-2<TQAS-3<TQAS-4 (Fig. 6). Reduction is also seen for the single helix pair mutations in Nasco oocytes, but the aminoacylation trend is TQAS-4≈TQAS-3>TQAS-2. These results are consistent with previous biochemistry experiments that showed the second and third positions are GlnRS identity elements (Jahn et al. 1991; Ibba et al. 1996), and that removing the wobble (U4-G69) at the fourth position reduces aminoacylation in E. coli (McClain et al. 1998).
FIGURE 6.
THG73 acceptor stem mutations tested at β9′UAG. Individual tRNA (74 mer) (16 ng per oocyte) average current was normalized to THG73 (74 mer) average current. Bar colors are the same as in Fig. 5. Total of oocytes tested is 189 and each bar is 26>n>8 oocytes. All mutations in the acceptor stem lower aminoacylation in vivo relative to THG73. TQAS-1 shows lack of aminoacylation but is not accepted by the translational machinery (Rodriguez et al. 2007, companion paper, this issue). TQAS′ and TQAS are the most orthogonal tRNAs in Xenopus Express and Nasco oocytes, respectively.
TABLE 1.
THG73 acceptor stem mutations
We then combined mutations to reduce aminoacylation further in Xenopus oocytes. In Xenopus Express oocytes, combining the mutations results in a multiplicative reduction in aminoacylation for TQAS-5 and TQAS′ (Table 1), while TQAS, which contains U2-C71, shows the largest amount of aminoacylation (Fig. 6). The overall aminoacylation trend in Xenopus Express oocytes is THG73>>TQAS≈TQAS-2>TQAS-3>TQAS-4>TQAS-5>>TQAS′, making TQAS′ the most orthogonal tRNA (Fig. 6). The same tRNAs tested in Nasco oocytes show no logical trend in combining mutations (Table 1), and TQAS is the most orthogonal tRNA. Nasco oocytes display the overall aminoacylation trend THG73>>TQAS′>TQAS-4≈TQAS-3≈TQAS-5>TQAS-2>>TQAS (Fig. 6). Overall the mutations suggest that in Xenopus Express oocytes, C–G pairs in the acceptor stem reduce aminoacylation. Nasco oocytes prefer the C–G mutation at the second position (Fig. 3, TQAS-2) or the non-Watson–Crick U–C pair at the second position, along with C–G pairs at the third and fourth positions (Fig. 3, TQAS). While differences in the Xenopus Express and Nasco oocytes were unanticipated, Figure 6 shows that the TQAS tRNA library contained an orthogonal tRNA for each genetic background.
Aminoacylation tested at a highly promiscuous site, βA70
The β9′ site can incorporate many natural amino acids and UAAs (Filatov and White 1995; Kearney et al. 1996; Kosolapov et al. 2000), but we chose to further test aminoacylation of the orthogonal tRNAs selected (TQAS′ and TQAS) at another highly permissive nAChR site, βA70. Structural studies place this residue on a highly exposed loop of the receptor (Brejc et al. 2001; Unwin 2005). Previous work on the aligned αD70UAG showed that it can incorporate the large UAA biocytin with little change in the EC50 (Gallivan et al. 1997). Other work has shown that biocytin is a challenging residue for nonsense suppression (Murakami et al. 2006), supporting the notion that αD70 is a promiscuous site. Therefore, we studied the αD70-aligned site in the β-subunit, βA70, considering only Nasco oocytes.
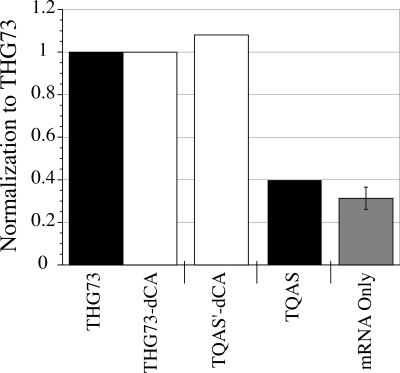
With the injection of β70UAG mRNA only, large amounts of current were obtained, indicating that this readthrough represents ∼30% of the current observed in the aminoacylation experiments (Fig. 7). This large amount of readthrough is not typical of most sites studied (β9′UAG discussed above and α145UAG below) and is most likely caused by the high promiscuity for amino acid incorporation. Injection of THG73-dCA and TQAS′-dCA with β70UAG in Nasco oocytes resulted in approximately the same amount of aminoacylation (Fig. 7, white bars), which is consistent with increased TQAS′ aminoacylation with Nasco oocytes (Fig. 6). However, TQAS shows very little aminoacylation compared to THG73 at this site (Fig. 7), and subtracting the mRNA only results in 9% aminoacylation relative to THG73 (also background subtracted).
FIGURE 7.
tRNA (74 mer/-dCA) aminoacylation tested at a highly promiscuous site, βA70. All experiments were performed in Nasco oocytes. Black bars are tRNA (74 mer), white bars are tRNA-dCA, and the gray bar is mRNA only. Average currents for TQAS′-dCA and TQAS were normalized to THG73-dCA and THG73, respectively. mRNA only was normalized to THG73. Total of oocytes tested is 60, where each bar is 15>n>6 oocytes with 9 ng of tRNA per oocyte. mRNA only shows significant readthrough of the UAG stop codon. THG73, THG73-dCA, and TQAS′-dCA all show significant aminoacylation when assayed at β70UAG. TQAS shows comparable currents to the injection of mRNA only, showing less aminoacylation of TQAS in Nasco oocytes.
The EC50s for mRNA only, THG73, TQAS′-dCA, and TQAS were 71 ± 1, 56 ± 3, 58 ± 3, and 46 ± 2 μM ACh, respectively, and showed little change from the wild-type EC50 of 50 μM ACh. These studies further show that TQAS is the most orthogonal tRNA and that TQAS′ is aminoacylated extensively in Nasco oocytes.
LysRS does not aminoacylate THG73, TQAS′, or TQAS
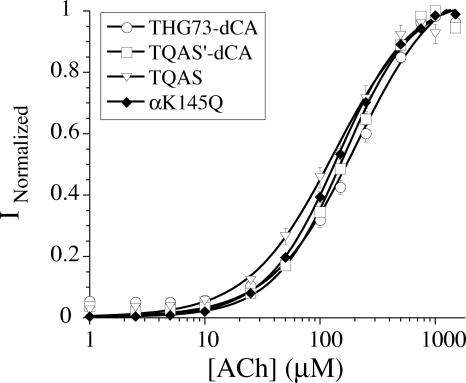
THG73 was shown to be aminoacylated by Gln at the β9′ site (Fig. 2). However, the 9′ position is a conserved Leu in all of the nAChR subunits and forms the hydrophobic gate to the channel (Kearney et al. 1996). Therefore, it is possible that anionic or cationic amino acids such as Asp, Glu, Lys, and Arg may not be incorporated at the β9′ position or may produce nonfunctional receptors if they are. This is a concern because many amber suppressor tRNAs have been shown to be aminoacylated by LysRS and/or GlnRS in vivo (Normanly et al. 1990), and mutations to the anticodon stem of an amber suppressor tRNATyr have shifted the GlnRS or LysRS recognition in vitro (Fukunaga et al. 2006). To evaluate this possibility, we studied a conserved Lys residue in the α subunit of the nAChR, αK145, to test if suppressor tRNAs were aminoacylated by LysRS and/or GlnRS. The conventional mutant αK145Q gave an EC50 of 144 ± 2 μM ACh (Fig. 8), an approximately threefold increase relative to the wild-type EC50 of 50 μM ACh.
FIGURE 8.
tRNA aminoacylation tested at αK145. Total of oocytes tested is 36, where each dose–response is 17>n>5 oocytes. EC50 values are 167 ± 14, 165 ± 14, 134 ± 15, and 144 ± 2 μM ACh in the order listed in the legend, with 7.5 ng of tRNA per oocyte. All tRNAs are still aminoacylated by Gln and not Lys (wild-type EC50 = 50 μM ACh).
Injection of α145UAG mRNA alone resulted in very small currents, such that EC50 could not be determined. Injection of α145UAG with THG73-dCA, TQAS′-dCA, and TQAS resulted in EC50s of 167 ± 14, 165 ± 14, and 134 ± 15 μM ACh, respectively (Fig. 8). The EC50s for all tRNAs are approximately threefold increased relative to the wild-type EC50, and therefore all of the tRNAs are aminoacylated by GlnRS and not by LysRS in Xenopus oocytes.
Analyzing THG73-W and TQAS′-W interactions with the translational machinery in vivo
In Figure 1, we outlined the desired events for the incorporation of UAA and the undesired possibility of incorporation of natural amino acids at the suppression site. THG73 was previously shown to be orthogonal to the translational machinery in vitro and in vivo when using less mRNA and/or tRNA along with incubations <2 d (Cload et al. 1996; Saks et al. 1996; England et al. 1999; Shafer et al. 2004; Rothman et al. 2005). We have shown that with increasing all three of the aforementioned conditions, producing so-called excessive conditions, THG73 can be aminoacylated in vivo. Under these same conditions, TQAS′ shows ∼90% reduction in the aminoacylation product in Xenopus Express oocytes (Fig. 6).
To analyze interactions with the translational machinery we used the temporal control of injection. By injecting tRNA-W first, we analyze the protection of the tRNA-W by EF-1α and/or other components of the translational machinery (Fig. 1A) from the undesired, irreversible loss of the UAA by hydrolysis and/or aaRS editing (Fig. 1C). Subsequent injection of mRNA to assay protein production (Fig. 1B) allows for comparison to what is seen when tRNA-W and mRNA are injected simultaneously. The tRNA–amino acid bond is highly labile at physiological pH. In a recent study, a sample of dCA-Val at pH 7.8 without translational machinery components was 69% hydrolyzed after a 1-h incubation (Wang et al. 2006). The cytoplasm of a Xenopus oocyte is estimated to be between pH 7.3 and 7.7 (Webb and Nuccitelli 1981; Sasaki et al. 1992), and therefore some hydrolysis of tRNA-dCA-W would be expected after 3.5 h, if not protected by other proteins. EF-Tu has been shown to protect YFFSCCCG-UAA from hydrolysis in vitro, but significant loss was seen after 1 h (Nakata et al. 2006).
Simultaneous injection of THG73-W/TQAS′-W (10 ng per oocyte) and α149UAG mRNA gave average maximal currents of −3.9 ± 1 and −4.1 ± 1 μA, respectively, in Xenopus Express oocytes. Surprisingly, we see no loss in current from either THG73-W or TQAS′-W when injecting mRNA 3.5 h after the tRNA-W, with −3.9 ± 0.5 and −3.9 ± 1 μA, respectively. Injection of twice as much THG73-W (20 ng per oocyte) resulted in −6.3 ± 2 μA. This illustrates that the translation system is not saturated, but there is not a linear increase in current with twice as much tRNA-W in a single injection. Therefore, interactions with EF-1α and/or other proteins specifically protect both suppressor tRNA-Ws from deacylation by aaRS(s) and hydrolysis, which is not problematic after 3.5 h in the Xenopus oocytes.
A double injection protocol, in which a second dose of aminoacyl tRNA is injected 24 h after the first, allows us to assay competition between the suppressor tRNA-W and tRNA-Q (generated by aminoacylation in vivo) after 24 h (Fig. 1B,G). Only aromatic amino acids can function at α149UAG, and suppressing with tRNA-W results in wild-type channels. If, however, there is competition with tRNA-Q, then, during the second 24 h, there will be a decrease in the overall protein production or less than double the current. In the event, a double injection with a 24-h interval of THG73-W/TQAS′-W with α149UAG showed currents of −8.9 ± 2 and −8.6 ± 3 μA, respectively, which is greater than twice the protein relative to the single injection. The higher protein production is most likely due to residual mRNA from the first injection, even though nonsense-mediated decay in Xenopus oocytes is expected to remove most mRNA with premature stop codons after 18 h (Whitfield et al. 1994).
The double injection studies establish that aminoacylation of the tRNA-dCA in Xenopus oocytes (Fig. 1E–H) is not problematic for the first 24 or more hours in vivo for either THG73 or TQAS′, because there is no reduction in protein and, therefore, lack of competition for the suppression site. This is surprising because THG73 is aminoacylated ∼90% more than TQAS′ in Xenopus Express oocytes after 2 d (Fig. 6). This does, however, agree with previous investigations of THG73 orthogonality in vivo with incubations of 24 h or less (Saks et al. 1996).
Comparing aminoacylation of amber, opal, and frameshift suppressor tRNAs
Increasing the number of UAAs to be simultaneously incorporated site-specifically requires the use of a unique stop or quadruplet codon for each UAA. Previously we screened two yeast Phe frameshift suppressor tRNAs, YFFSCCCG and YFaFSACCC, in Xenopus oocytes and found that both tRNAs were aminoacylated much less than THG73 when tested at the β9′ site. However, their suppression efficiency was decreased relative to THG73 in vivo, although still adequate for UAA incorporation (Rodriguez et al. 2006). Previous work has established that the opal (UGA) and ochre (UAA) stop codons can be suppressed in mammalian cells when using suppressor tRNAs that are aminoacylated by endogenous aaRSs (Capone et al. 1986) or by the import of an exogenous E. coli aaRSs (Kohrer et al. 2004). The opal codon has also been used to incorporate an UAA in mammalian cells using a tRNA/synthetase pair (Zhang et al. 2004). Also, an opal suppressor was previously created by replacing the anticodon of THG73 with UCA, but this opal suppressor tRNA (not chemically ligated to an UAA) is aminoacylated in vitro and results in protein translation, or aminoacylation product, >90% when compared to wild-type translation. Under the same conditions as the opal suppressor tRNA, THG73 and ENAS showed <5% aminoacylation product (Cload et al. 1996), suggesting that the opal suppressor derived from THG73 would be highly aminoacylated in vivo and not viable for nonsense suppression. In T. thermophila, the ochre suppressor has been shown to suppress both the ochre and amber codons (Hanyu et al. 1986), and so it would not be viable for the simultaneous incorporation of multiple UAAs.
To test the generality of the acceptor stem mutations at reducing aminoacylation in vivo, we created T. thermophila Gln opal suppressor tRNAs (TQOpS′ and TQOpS) by replacing the anticodon of TQAS′ and TQAS with UCA.
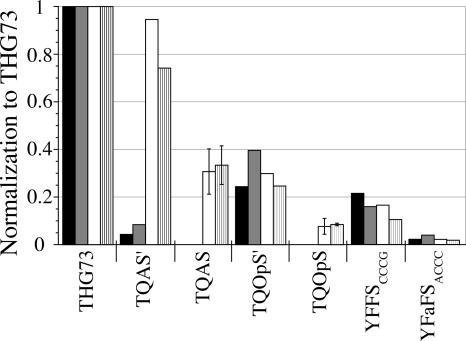
All four subunits of the nAChR terminated with the opal (UGA) stop codon, and these were all mutated to the ochre (UAA) stop codon to avoid suppression by TQOpS′ or TQOpS. The masked construct had an EC50 of 53 ± 3 μM ACh, comparable to the wild-type EC50 of 50 μM. Injection of β9′UGA with no tRNA resulted in 2.8% of the current normalized to THG73 (74 mer), versus 1.7% with β9′UAG. The opal codon thus showed ∼1.6-fold greater readthrough than the amber codon, which has previously been noted in E. coli (Anderson and Schultz 2003). Aminoacylation was tested under identical conditions with THG73, TQAS′, TQAS, TQOpS′, TQOpS, YFFSCCCG, and YFaFSACCC at the β9′ suppression site, stop codon, or quadruplet codon, with tRNA (74 mer) and tRNA-dCA (76 mer). This was done to validate initial screens using only (74 mer) and comparisons between (74 mer) and (76 mer) in Figures 4–7. Comparison of suppressor tRNAs (74 mer) and tRNA-dCAs (76 mer) shows no significant difference for each individual suppressor tRNA in Xenopus Express or Nasco oocytes (Fig. 9). Once again TQAS′ and TQAS showed much less aminoacylation in Xenopus Express and Nasco oocytes, respectively, relative to THG73. TQOpS′ and TQOpS both show less than ∼40% and ∼9% aminoacylation, respectively, compared to THG73 (Fig. 9). This shows that the acceptor stem mutations on THG73 are able to reduce aminoacylation for both amber and opal suppressor tRNAs. YFaFSACCC and YFFSCCCG show a similar trend to that previously reported (Rodriguez et al. 2006) with average aminoacylation products of 19% and 3%, respectively, of THG73. The orthogonality trend is as follows: YFaFSACCC>TQAS′(Xenopus Express)≈TQOpS>YFFSCCCG>TQAS(Nasco)≈TQOpS′>>THG73, which is aminoacylated the most.
FIGURE 9.
Amber, opal, and frameshift suppressor tRNAs tested at β9′. All tRNA 74 mer or -dCA (7.5 ng per oocyte) average currents were normalized to THG73 74 mer or -dCA average currents, respectively. Black bars are tRNA 74 mer+β9′(UAG, UGA, CGGG, or GGGU) and gray bars are tRNA-dCA (76 mer)+β9′(UAG, UGA, CGGG, or GGGU) in Xenopus Express oocytes. White and hatched bars are tRNA 74 mer and 76 mer, respectively, in Nasco oocytes. Total of tested oocytes is 276, where 17>n>5. TQAS′ and TQAS show significantly reduced aminoacylation in Xenopus Express and Nasco oocytes, respectively, when compared to THG73. Both opal suppressor tRNAs (TQOpS′ and TQOpS) show less aminoacylation than THG73. Overall, the frameshift suppressor YFaFSACCC is the most orthogonal tRNA in both Xenopus Express and Nasco oocytes.
DISCUSSION
THG73 is an amber suppressor tRNA that has been used extensively for the incorporation of >100 residues in 20 proteins (Dougherty 2000; Beene et al. 2003). Using conditions such as low quantities of mRNA and/or tRNA with incubation times <2 d, THG73 has been shown to be orthogonal in vitro (Cload et al. 1996; England et al. 1999; Rothman et al. 2005) and in vivo (Saks et al. 1996; Shafer et al. 2004). However, increasing the amount of mRNA and tRNA with incubation of 2 d leads to the aminoacylation of THG73 by an endogenous aaRS in vivo (Rodriguez et al. 2006). We show that the E. coli Asn amber suppressor (ENAS) and ENAS A71 are both susceptible to aminoacylation by aaRSs of the Xenopus oocyte (Fig. 5), even though aminoacylation has not been observed in vitro (Cload et al. 1996; Murakami et al. 2003, 2006) or in vivo (Kleina et al. 1990). This suggests that many well-characterized amber suppressor tRNAs may become aminoacylated in vivo when using increased amounts of mRNA and tRNA along with incubations of 2 d, which is desirable for increased amounts of protein.
By using the sensitivity of electrophysiology and the well-characterized β9′ site, we have been able to show that the natural amino acid placed on THG73 is predominantly, if not exclusively, Gln (Fig. 2). The THG73 (74 mer) and β9′Q show comparable EC50s, but THG73-dCA (76 mer) shows an approximately threefold increase in EC50 (Fig. 2). These experiments suggest that THG73-dCA (76 mer) is more readily aminoacylated by other endogenous aaRSs, perhaps due to the unnatural dCA at the 3′ end, but the predominant amino acid is still Gln. Similar conditions with the THG73 (74 mer) resulted in equivalent EC50 as β9′Q, and therefore the active THG73-CA (Fig. 1D) appears to be more stringently recognized by the GlnRS.
Intriguingly, THG73 is very sensitive to the previously discussed knob mutations meant to disrupt recognition by GlnRS, and the derived tRNAs are not viable for UAA incorporation. The K2 mutation alone destroys activity, and combining it with other knob mutations provides little rescue. The K2 mutation resides within the D domain of THG73, which makes tertiary contacts with the variable loop to form the characteristic L-shape tRNA structure. This mutation may result in misfolding of the THG73, but it apparently has little effect on E. coli tRNA2 Gln (Liu et al. 1997). Interestingly, the K1, or TQAS-3, mutation was shown to be functional by itself on THG73 (Rodriguez et al. 2007), but it is nonfunctional in combination with K2 and K3 (Fig. 4). Replacement of the THG73 anticodon with ACCC (Shafer et al. 2004) or CCCG (Rodriguez et al. 2006) resulted in nonfunctional frameshift suppressors in vivo. THG73 appears to be sensitive to mutation within the D domain and the anticodon loop but tolerates acceptor stem mutations.
ENAS and ENAS A71 aminoacylation in Xenopus Express oocytes is comparable to THG73, but there is a 40% decrease due to the U2-C71 versus U2-A71, respectively (Fig. 5), at the second position of the acceptor stem (Fig. 3). ENAS A71 is more orthogonal, which suggests that the U–A is not recognized by endogenous aaRSs as readily as U–C. The opposite trend is seen in Nasco oocytes, but orthogonality is significantly improved for both ENAS and ENAS A71. Clearly, the identity of the second pair has a strong effect on aminoacylation in vivo.
We hypothesized that similar mutations on THG73 could result in less aminoacylation by the GlnRS, because the identity set includes the second to fourth positions of the acceptor stem (Perona et al. 1989; Jahn et al. 1991; Ibba et al. 1996; McClain et al. 1998). The acceptor stem mutations (shown in Fig. 3) greatly reduced aminoacylation in vivo (Fig. 6; Table 1). In Xenopus Express oocytes, the single helix pair mutations could be combined to further reduce aminoacylation in a multiplicative manner (Table 1). In contrast, Nasco oocytes showed a strong dependence on specific sequences, and there was no logical trend based on the single helix pair mutations. This nonclassical identity set has previously been identified on yeast Phe tRNA, where combining mutations could compensate for deleterious single mutations to increase aminoacylation in vitro (Frugier et al. 1998). While variation due to the Xenopus Express and Nasco oocytes was unanticipated, the library of six functional TQAS tRNAs was able to find an orthogonal suppressor tRNA for each genetic background. We can anticipate that, as other workers move the UAA methodology into other cell types, different orthogonality issues could arise. The library of tRNAs created here contains diverse mutations in the acceptor stem that should allow for orthogonal tRNA selection in other eukaryotic cells, where high-throughput assays for tRNA screening are currently lacking.
Using the temporal control of injection, we were able to evaluate THG73-W and TQAS′-W interactions with the protein translation machinery and competition with the suppressor tRNAs aminoacylated with Gln after 1 d. Shockingly, there is no change in suppression efficiency when the mRNA and tRNA-W are injected simultaneously or when the mRNA was injected 3.5 h after tRNA-W. These experiments show that both suppressor tRNAs are equally protected by EF-1α and/or other components of the translational machinery for at least 3.5 h, which has not been observed in vitro with or without EF-Tu (Nakata et al. 2006; Wang et al. 2006). Most importantly, this shows that aaRS editing and/or hydrolysis (Fig. 1C) are not problematic with 10 ng of tRNA-W injected in oocytes. Injection of 20 ng of THG73-W resulted in a 60% increase in current with a single injection, establishing the lack of saturation at 10 ng, although the response is nonlinear. By injecting mRNA and tRNA-W followed by a second injection of the same after 24 h, we could see if there was competition for the suppression of α149UAG by tRNA-Q (Fig. 1B,G). The double injection for both THG73-W/TQAS′-W showed a slightly greater than twofold increase in current, indicating a lack of competition with tRNA-Q slightly after 24 h. This agrees with previous experiments using <24-h incubation times (Saks et al. 1996; Shafer et al. 2004). Aminoacylation of THG73, therefore, takes place after 24 h and is not different from TQAS′ in Xenopus Express oocytes in the first 24 h. However, TQAS′ is aminoacylated ∼90% less than THG73 after 2 d. Overall, both THG73 and TQAS′ interact with the protein translational machinery similarly in Xenopus Express oocytes.
In order to simultaneously incorporate multiple UAAs, it is necessary to use a unique stop or quadruplet codon for each UAA. Previously, three UAAs could be simultaneously incorporated using amber and frameshift suppression with the UAG, CGGG, and GGGU suppression sites (Rodriguez et al. 2006). A second nonsense site, either opal (UGA) or ochre (UAA), would be valuable for the incorporation of four UAAs. However, the T. thermophila ochre suppressor recognizes both the ochre (UAA) and amber (UAG) stop codons (Hanyu et al. 1986). The opal codon has been used to incorporate an UAA in mammalian cells using a tRNA/synthetase pair (Zhang et al. 2004) and should be feasible for UAA incorporation in Xenopus oocytes. Previous work with an opal suppressor created from THG73 by the replacement of the anticodon with UCA resulted in a large amount of aminoacylation in vitro and production of >90% protein relative to wild-type protein (Cload et al. 1996). These results suggest that the THG73-derived opal suppressor would be aminoacylated much more than the THG73 amber suppressor and would not be able to be evaluated under the conditions tested in this paper. To test the generality of the acceptor stem mutations at reducing aminoacylation, we replaced the anticodon of TQAS′ and TQAS with UCA to create TQOpS′ and TQOpS, respectively. Both TQOpS′ and TQOpS show a great reduction in aminoacylation in vivo relative to THG73 (Fig. 9) and are viable suppressor tRNAs for UAA incorporation at the opal codon (Rodriguez et al. 2007, companion paper, this issue). Intriguingly, there is no significant difference in aminoacylation of TQOpS′ in Xenopus Express and Nasco oocytes (Fig. 9), which suggests the opal suppressor is recognized by a different aaRS(s) and variation in aminoacylation is caused by the GlnRS, which we have shown recognizes THG73, TQAS′, and TQAS (Fig. 8), and the tRNAs show different amounts of aminoacylation depending on the genetic background (Figs. 6, 9).
We have shown that acceptor stem mutations have wide applicability for the creation of orthogonal suppressor tRNAs for the incorporation of UAAs in higher eukaryotic cells. TQAS′ and TQAS still retain slight aminoacylation by the GlnRS in Xenopus Express and Nasco oocytes, respectively. Mutations in the anticodon stem have been shown to decrease GlnRS recognition and may further reduce aminoacylation, but these mutations can also lead to increased LysRS recognition (Fukunaga et al. 2006). The acceptor stem mutations have no adverse effect on the suppression efficiency and can increase the suppression efficiency by as much as 40% (Rodriguez et al. 2007, companion paper, this issue). While aminoacylation was shown to be dependent on the genetic background of the oocytes, there is no effect on the suppression efficiencies of the TQAS tRNAs (Rodriguez et al. 2007, companion paper, this issue).
In summary, we have created a TQAS tRNA library that will aid in the identification of orthogonal tRNAs for the use of higher eukaryotic cells. When combined with the evaluation of suppression efficiencies described in Rodriguez et al. (2007), these tRNAs should significantly expand the applicability of site-specific incorporation of UAAs by chemically aminoacylated tRNAs.
MATERIALS AND METHODS
Materials
All oligonucleotides were synthesized by Integrated DNA Technologies. NotI was from Roche Applied Science. BamHI, EcoRI, FokI, T4 DNA ligase, and T4 RNA ligase were from NEB. Kinase Max, T7 MEGAshortscript, and T7 mMessage mMachine kits were from Ambion. ACh chloride and yeast inorganic pyrophosphatase were purchased from Sigma-Aldrich. dCA and 6-nitroveratryloxycarbonyl protected dCA-W was prepared as reported in Robertson et al. (1989) and Zhong et al. (1998).
tRNA gene preparation and tRNA transcription
THG73, YFFSCCCG, and YFaFSACCC subcloned in the pUC19 vector were previously made (Saks et al. 1996; Rodriguez et al. 2006). Genes for ENAS (sequence from Cload et al. 1996) with flanking EcoRI and BamHI overhangs were phosphorylated using the Kinase Max kit, annealed, and ligated with T4 DNA ligase into EcoRI and BamHI linearized pUC19 vector as described (Nowak et al. 1998). ENAS A71 (original sequence from Kleina et al. 1990 with G1-C72 insertion for T7 polymerase transcription) was created by QuikChange mutagenesis on ENAS in pUC19. Knob mutations from Liu et al. (1997) (K2 is C10-G25; K2K3 is C10-G25 and G17; and K1K2K3 is C3-G70, C10-G25, and G17) on THG73 were created by QuikChange mutagenesis. Acceptor stem mutations on THG73 were created by QuikChange mutagenesis and shown in Figure 3. Replacing the anticodon of TQAS′ and TQAS from CUA to UCA by QuikChange mutagenesis created TQOpS′ and TQOpS, respectively. All mutations were verified by DNA sequencing (California Institute of Technology Sequencing/Structure Analysis Facility). Template DNA for tRNA lacking the 3′CA was prepared by FokI digestion, and tRNA was transcribed using the T7 MEGAshortscript kit with 0.5 μL of yeast inorganic pyrophosphatase (40 U/mL in 75 mM Tris, 10 mM MgCl2, and pH 7.0). tRNA was desalted using CHROMA SPIN-30 DEPC-H2O columns (BD Biosciences), and the concentration was determined by absorption at 260 nm.
nAChR gene preparation and mRNA transcription
The masked α, β, γ, and δ subunits of the nAChR subcloned in the pAMV vector were previously prepared (Rodriguez et al. 2006). All four subunits terminate with the opal (UGA) stop codon and each UGA was mutated to the ochre (UAA) stop codon to avoid possible suppression by TQOpS′ and TQOpS. α149UAG and β9′(UAG, CGGG, and GGGU) were previously prepared on the masked constructs (Rodriguez et al. 2006). β70UAG, α145Q, α145UAG, and β9′UGA were prepared by QuikChange mutagenesis. Mutations were verified by DNA sequencing (California Institute of Technology Sequencing/Structure Analysis Facility). DNA was linearized with NotI, and mRNA was prepared with the T7 mMessage mMachine kit with 0.5 μL of yeast inorganic pyrophosphatase. mRNA was purified by using the RNeasy Mini kit (Qiagen) and quantified by absorption at 260 nm.
dCA and dCA-W ligation to suppressor tRNAs
Seventy-five micromolars (instead of 300 μM, because there was no change in ligation efficiency) of dCA or 6-nitroveratryloxycarbonyl protected dCA-W were coupled to suppressor tRNAs by using T4 RNA ligase for 30 min as previously reported (Nowak et al. 1998; Petersson et al. 2002), desalted using CHROMA SPIN-30 DEPC-H2O columns, and quantified by absorption at 260 nm. tRNA ligation efficiency was qualitatively determined by MALDI mass spectrometry (Petersson et al. 2002), and all tRNA ligations were identical within each prepared group and >80%.
In vivo aminoacylation and suppression experiments
Prior to in vivo aminoacylation and suppression experiments, all tRNAs and mRNAs were simultaneously made and normalized by UV and densiometric analysis using AlphaEaseFC Stand Alone (Alpha Innotech). Stage VI oocytes of Xenopus laevis were prepared as described (Quick and Lester 1994). All tRNAs were refolded at 65°C for 2 min and 6-nitroveratryloxycarbonyl protected dCA-W was deprotected for 5 min by UV irradiation before injection (Saks et al. 1996). Oocytes were injected with 50 nL of mRNA alone or with tRNA and incubated at 18°C for 44–52 h. For EC50 determination of β9′Q (Fig. 2), 3 ng of mRNA in the ratio of 2:1:1:1 for α:β9′Q:γ:δ was injected; 20 ng of mRNA in a subunit ratio of 2:5:1:1 for α:β9′UAG:γ:δ or 10:1:1:1 for α149UAG:β:γ:δ was injected (Figs. 2, 4, 5). Figures 6–8 used the same ratio, but 40 ng to 60 ng of mRNA was injected per oocyte. In Figure 9, 40 ng of mRNA in a subunit ratio of 2:5:1:1 for α:β9′(UAG, UGA, CGGG, or GGGU):γ:δ. Amounts of tRNA injected are listed with each figure.
Electrophysiology
Recordings employed a two-electrode voltage clamp on the OpusXpress 6000A (Molecular Devices). ACh was stored at −20°C as a 1 M stock, diluted in Ca2+-free ND96, and delivered to oocytes by a computer-controlled perfusion system. For all experiments, the holding potential was −60 mV. Dose–response data were obtained from at least eight ACh concentrations and all tRNA aminoacylation or suppression comparisons were tested with a single 1 mM ACh dose. Dose–response relations were fit to the Hill equation to determine the EC50 and Hill coefficient (n H). All reported values are represented as a mean±SE of the tested oocytes (number of oocytes [n] is listed with each figure).
ACKNOWLEDGMENTS
E.A.R. is a National Science Foundation Predoctoral Fellow. This work was supported by the NIH (NS 34407 and NS 11756).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.666807.
REFERENCES
- Anderson, J.C., Schultz, P.G. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- Beene, D.L., Dougherty, D.A., Lester, H.A. Unnatural amino acid mutagenesis in mapping ion channel function. Curr. Opin. Neurobiol. 2003;13:264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- Brejc, K., van Dijk, W.J., Klaassen, R.V., Schuurmans, M., van Der Oost, J., Smit, A.B., Sixma, T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Capone, J.P., Sedivy, J.M., Sharp, P.A., RajBhandary, U.L. Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: Use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol. Cell. Biol. 1986;6:3059–3067. doi: 10.1128/mcb.6.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cload, S.T., Liu, D.R., Froland, W.A., Schultz, P.G. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem. Biol. 1996;3:1033–1038. doi: 10.1016/s1074-5521(96)90169-6. [DOI] [PubMed] [Google Scholar]
- Dougherty, D.A. Unnatural amino acids as probes of protein structure and function. Curr. Opin. Biotechnol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- England, P.M., Lester, H.A., Dougherty, D.A. Mapping disulfide connectivity using backbone ester hydrolysis. Biochemistry. 1999;38:14409–14415. doi: 10.1021/bi991424c. [DOI] [PubMed] [Google Scholar]
- Filatov, G.N., White, M.M. The role of conserved leucines in the M2 domain of the acetylcholine receptor in channel gating. Mol. Pharmacol. 1995;48:379–384. [PubMed] [Google Scholar]
- Frugier, M., Helm, M., Felden, B., Giege, R., Florentz, C. Sequences outside recognition sets are not neutral for tRNA aminoacylation. Evidence for nonpermissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe . J. Biol. Chem. 1998;273:11605–11610. doi: 10.1074/jbc.273.19.11605. [DOI] [PubMed] [Google Scholar]
- Fukunaga, J., Ohno, S., Nishikawa, K., Yokogawa, T. A base pair at the bottom of the anticodon stem is reciprocally preferred for discrimination of cognate tRNAs by Escherichia coli lysyl- and glutaminyl-tRNA synthetases. Nucleic Acids Res. 2006;34:3181–3188. doi: 10.1093/nar/gkl414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan, J.P., Lester, H.A., Dougherty, D.A. Site-specific incorporation of biotinylated amino acids to identify surface-exposed residues in integral membrane proteins. Chem. Biol. 1997;4:739–749. doi: 10.1016/s1074-5521(97)90312-4. [DOI] [PubMed] [Google Scholar]
- Hanyu, N., Kuchino, Y., Nishimura, S., Beier, H. Dramatic events in ciliate evolution: Alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAsGln . EMBO J. 1986;5:1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba, M., Hong, K.W., Sherman, J.M., Sever, S., Soll, D. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl. Acad. Sci. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, M., Rogers, M.J., Soll, D. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kearney, P.C., Zhang, H., Zhong, W., Dougherty, D.A., Lester, H.A. Determinants of nicotinic receptor gating in natural and unnatural side chain structures at the M2 9′ position. Neuron. 1996;17:1221–1229. doi: 10.1016/s0896-6273(00)80252-4. [DOI] [PubMed] [Google Scholar]
- Kleina, L.G., Masson, J.M., Normanly, J., Abelson, J., Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J. Mol. Biol. 1990;213:705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Kohrer, C., Sullivan, E.L., RajBhandary, U.L. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre, and opal suppressor tRNA pairs: Concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004;32:6200–6211. doi: 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosolapov, A.V., Filatov, G.N., White, M.M. Acetylcholine receptor gating is influenced by the polarity of amino acids at position 9′ in the M2 domain. J. Membr. Biol. 2000;174:191–197. doi: 10.1007/s002320001043. [DOI] [PubMed] [Google Scholar]
- Liu, D.R., Magliery, T.J., Schultz, P.G. Characterization of an “orthogonal” suppressor tRNA derived from E. coli tRNA2 Gln . Chem. Biol. 1997;4:685–691. doi: 10.1016/s1074-5521(97)90224-6. [DOI] [PubMed] [Google Scholar]
- McClain, W.H., Schneider, J., Bhattacharya, S., Gabriel, K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Natl. Acad. Sci. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, H., Kourouklis, D., Suga, H. Using a solid-phase ribozyme aminoacylation system to reprogram the genetic code. Chem. Biol. 2003;10:1077–1084. doi: 10.1016/j.chembiol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Murakami, H., Ohta, A., Ashigai, H., Suga, H. A highly flexible tRNA acylation method for nonnatural polypeptide synthesis. Nat. Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- Nakata, H., Ohtsuki, T., Abe, R., Hohsaka, T., Sisido, M. Binding efficiency of elongation factor Tu to tRNAs charged with nonnatural fluorescent amino acids. Anal. Biochem. 2006;348:321–323. doi: 10.1016/j.ab.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Normanly, J., Kleina, L.G., Masson, J.M., Abelson, J., Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 1990;213:719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- Nowak, M.W., Gallivan, J.P., Silverman, S.K., Labarca, C.G., Dougherty, D.A., Lester, H.A. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- Perona, J.J., Swanson, R.N., Rould, M.A., Steitz, T.A., Soll, D. Structural basis for misaminoacylation by mutant E. coli glutaminyl-tRNA synthetase enzymes. Science. 1989;246:1152–1154. doi: 10.1126/science.2686030. [DOI] [PubMed] [Google Scholar]
- Petersson, E.J., Shahgholi, M., Lester, H.A., Dougherty, D.A. MALDI-TOF mass spectrometry methods for evaluation of in vitro aminoacyl tRNA production. RNA. 2002;8:542–547. doi: 10.1017/s1355838202026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick, M.W., Lester, H.A. Methods for expression of excitability proteins in Xenopus oocytes. In: Narahashi T., editor. Ion channels of excitable cells. Academic Press; San Diego, CA: 1994. pp. 261–279. [Google Scholar]
- Robertson, S.A., Noren, C.J., Anthony-Cahill, S.J., Griffith, M.C., Schultz, P.G. The use of 5′-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res. 1989;17:9649–9660. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D.A. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc. Natl. Acad. Sci. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D.A. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: Evaluating suppression efficiency. RNA. 2007 doi: 10.1261/rna.667607. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, D.M., Petersson, E.J., Vazquez, M.E., Brandt, G.S., Dougherty, D.A., Imperiali, B. Caged phosphoproteins. J. Am. Chem. Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- Saks, M.E., Sampson, J.R., Nowak, M.W., Kearney, P.C., Du, F.Y., Abelson, J.N., Lester, H.A., Dougherty, D.A. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. J. Biol. Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- Sasaki, S., Ishibashi, K., Nagai, T., Marumo, F. Regulation mechanisms of intracellular pH of Xenopus laevis oocyte. Biochim. Biophys. Acta. 1992;1137:45–51. doi: 10.1016/0167-4889(92)90098-v. [DOI] [PubMed] [Google Scholar]
- Shafer, A.M., Kalai, T., Bin Liu, S.Q., Hideg, K., Voss, J.C. Site-specific insertion of spin-labeled L-amino acids in Xenopus oocytes. Biochemistry. 2004;43:8470–8482. doi: 10.1021/bi035542i. [DOI] [PubMed] [Google Scholar]
- Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Wang, B., Zhou, J., Lodder, M., Anderson , R.D., III, Hecht, S.M. Tandemly activated tRNAs as participants in protein synthesis. J. Biol. Chem. 2006;281:13865–13868. doi: 10.1074/jbc.C600018200. [DOI] [PubMed] [Google Scholar]
- Webb, D.J., Nuccitelli, R. Direct measurement of intracellular pH changes in Xenopus eggs at fertilization and cleavage. J. Cell Biol. 1981;91:562–567. doi: 10.1083/jcb.91.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, T.T., Sharpe, C.R., Wylie, C.C. Nonsense-mediated mRNA decay in Xenopus oocytes and embryos. Dev. Biol. 1994;165:731–734. doi: 10.1006/dbio.1994.1289. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Alfonta, L., Tian, F., Bursulaya, B., Uryu, S., King, D.S., Schultz, P.G. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, W., Gallivan, J.P., Zhang, Y., Li, L., Lester, H.A., Dougherty, D.A. From ab initio quantum mechanics to molecular neurobiology: A cation-π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]