Abstract
MicroRNAs (miRNAs) are a novel class of small noncoding RNAs that regulate gene expression at the post-transcriptional level and play a critical role in many important biological processes. Most miRNAs are conserved between humans and mice, which makes it possible to analyze their expressions with a set of selected array probes. Here, we report a simple array platform that can detect 553 nonredundant miRNAs encompassing the entire set of miRNAs for humans and mice. The platform features carefully selected and designed probes with optimized hybridization parameters. Potential cross-reaction between mature miRNAs and their precursors was investigated. The array platform was used to analyze miRNAs in the mouse central nervous system (CNS, spinal cord and brain), and two other non-CNS organs (liver and heart). Two types of miRNAs, differentially expressed organ/tissue-associated miRNAs and ubiquitously expressed miRNAs, were detected in the array analysis. In addition to the previously reported neuron-related miR-124a, liver-related miR-122a, and muscle-related miR-133a, we also detected new tissue-associated miRNAs (e.g., liver-associated miR-194). Interestingly, while the majority of pre-miRNAs were undetectable, miR690, miR709, and miR720 were clearly detected at both mature and precursor levels by the array analysis, indicating a limited cross-reaction between pre-miRNAs and their mature miRNAs. The reliability of this array technology was validated by comparing the results with independent Northern blot analyses and published data. A new approach of data normalization based on Northern blot analysis of one ubiquitously expressed miRNA is introduced and compared with traditional approaches. We expect this miRNA array platform to be useful for a wide variety of biological studies.
Keywords: miRNA, miRNA array, pre-miRNA, miRNA biogenesis, miRNA array normalization, central nervous system (CNS)
INTRODUCTION
MicroRNAs (miRNAs) are recently discovered, abundant small noncoding RNAs that regulate gene expression at the post-transcriptional level (Lai 2003; Nelson et al. 2003; Ambros 2004; Bartel 2004; He and Hannon 2004; Du and Zamore 2005; Berezikov et al. 2006; Nilsen 2007). These small RNA molecules are highly conserved among organisms in both the animal and the plant kingdoms and play critical roles in many important biological processes, including developmental timing (Horvitz et al. 1983; Ambros 1989; Liu et al. 1995; Rougvie and Ambros 1995; Euling and Ambros 1996; Moss et al. 1997; Slack and Ruvkun 1997; Reinhart et al. 2000), neuronal cell fate (Sempere et al. 2004; Johnston et al. 2005; Vo et al. 2005; Li et al. 2006a), apoptosis (Cheng et al. 2005; Cimmino et al. 2005; Tanno et al. 2005; Jovanovic and Hengartner 2006; Thompson and Cohen 2006), proliferation (Brennecke et al. 2003; Hayashita et al. 2005; Lee et al. 2005; Chen et al. 2006; Costinean et al. 2006; Hossain et al. 2006; Hwang and Mendell 2006; Thompson and Cohen 2006), hematopoiesis (Chen et al. 2004; Chen and Lodish 2005; Kluiver et al. 2006; Min and Chen 2006; Ramkissoon et al. 2006; Yu et al. 2006), cancer development (Iorio et al. 2005; Jiang et al. 2005; Akao et al. 2006; Garzon et al. 2006; Lee et al. 2006; Li et al. 2006b; Mattie et al. 2006; Saito et al. 2006; Silveri et al. 2006; Volinia et al. 2006; Yanaihara et al. 2006), neurological disorders (Ding et al. 2005; Mehler and Mattick 2006), insulin secretion or exocytosis of the secretory pathway (Mello and Czech 2004; Poy et al. 2004; Cuellar and McManus 2005; Gauthier and Wollheim 2006; Plaisance et al. 2006), B-cell development (Calin et al. 2004; Costinean et al. 2006), and adipocyte differentiation (Esau et al. 2004; Kajimoto et al. 2006). More than 4500 miRNAs have been identified from 44 organisms and annotated in the miRBase registry (http://microrna.sanger.ac.uk) (Griffiths-Jones 2006; Griffiths-Jones et al. 2006). From these, 475, 377, and 234 miRNAs have been identified in humans, mice, and rats, respectively (Griffiths-Jones 2006; Griffiths-Jones et al. 2006). Bioinformatics analysis predicts that miRNAs comprise ∼5% of the transcriptome (Bentwich 2005), and may regulate the translation of more than one-third of human mRNA species (Lewis et al. 2005). An additional independent analysis predicted that 20% of human genes can potentially be regulated by miRNAs (Xie et al. 2005). One challenge is how to effectively measure the expression of miRNA genes.
The expression levels of individual miRNAs can be determined by quantitative Northern blot analysis (Tang et al. 2003; Tang and Zamore 2004). Northern blot analysis has the advantage of simultaneously detecting the mature miRNAs and miRNA precursors, but it is a very labor-intensive approach. As a result, a number of different miRNA array platforms have been developed (Krichevsky et al. 2003; Babak et al. 2004; Calin et al. 2004; Liu et al. 2004; Nelson et al. 2004; Sempere et al. 2004; Sioud and Rosok 2004; Jiang et al. 2005; Liang et al. 2005; Lim et al. 2005; Monticelli et al. 2005; Shingara et al. 2005; Castoldi et al. 2006; Grundhoff et al. 2006; Lee et al. 2006; Mattie et al. 2006; Tang et al. 2006; Wang and Wang 2006; Zhao et al. 2006; Wang et al. 2007). The earliest prototype array for miRNA expression using isotope labeling was straightforward (Krichevsky et al. 2003). This miRNA array platform was further pursued (Monticelli et al. 2005), but not widely applied due to insufficient optimization with respect to various controls, probe repeats, and the related titration. In contrast, many nonisotope miRNA array platforms have been developed and commercialized. These miRNA array systems have played an essential role in identifying miRNAs that are important for normal development as well as various human diseases (Calin et al. 2005; Monticelli et al. 2005; Watanabe et al. 2005; Wienholds et al. 2005; Calin and Croce 2006; Cummins and Velculescu 2006; Davison et al. 2006; Kim et al. 2006; Martin et al. 2006; Murakami et al. 2006; Nelson et al. 2006; Pallante et al. 2006; Song and Tuan 2006; Thomson et al. 2006; Weber et al. 2006; Weston et al. 2006; Yanaihara et al. 2006).
Unlike array analysis for mRNA expression, miRNA arrays typically involve a complicated procedure due to the small size of miRNAs and the lack of a conserved 3′ end for easy sample labeling. These complicated steps include the ligation of RNA adapters to the miRNAs, RT–PCR amplification, and T7 RNA polymerase transcription (Nelson et al. 2004; Davison et al. 2006). The current nonisotope miRNA array platforms are still expensive and require special skills. Furthermore, the complicated array steps often lead to process-related systematic biases that are inherent to RNA ligation or PCR amplification (Davison et al. 2006). To simplify these steps, we have systematically optimized the conditions for an earlier version of an isotope-labeled miRNA array platform (Krichevsky et al. 2003; Monticelli et al. 2005), and further developed this system demonstrating its use in mouse miRNA analysis. A total of 553 probes were used in this array platform that can detect all annotated miRNAs of humans and mice. This optimized isotope array platform is simple, cost-effective, easy-to-use, high throughput, highly sensitive, and can be easily adapted and utilized for a wide variety of biological studies.
RESULTS
Overall design of a simple miRNA array and its rationale
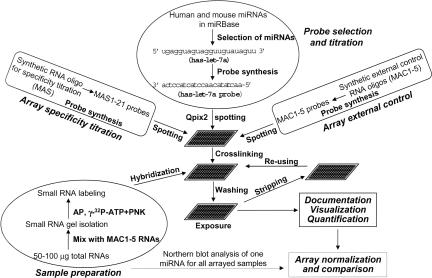
Our objective in the design of the array system was to provide a simple, cost-effective, and high-throughput platform that could be easily adopted in most laboratories. Thus, the overall design of the array platform includes: (1) selecting unique miRNAs for probe design to reduce potential cross-hybridization between different probes; (2) optimizing the probe concentration and specificity; (3) isolating mature miRNAs (∼22 nucleotides [nt]) to avoid the interference of signals from pre-miRNAs (∼70 nt); (4) directly labeling the isolated small RNAs to avoid using adaptors and biased amplification of the miRNAs; (5) introducing a new way of data normalization by using a Northern blot analysis of a constitutively expressed miRNA for initial data adjustment; and (6) introducing a set of external controls for evaluation of process-related loss of signals and quantification of endogenous miRNAs. We used the sensitive isotope 32P to label the small RNAs and nylon membranes for miRNA array analysis to visualize the array results and exclude the nonintuitive results generated from the numeric data of the background. The overall design of the miRNA array platform is outlined in Figure 1.
FIGURE 1.
A scheme of the miRNA array platform. The array design included probe design and specificity titration, external control design and titration, probe spotting, sample preparation, array hybridization, Northern blot analysis for one constitutively expressed miRNA and U6/5S rRNA for initial data normalization, and array normalization and comparison. (Qpix2) A spotting robot; (MAC) miRNA array control; (MAS) miRNA array specificity; (AP) Antarctic phosphatase; (PNK) polynucleotide kinase.
Selection of miRNAs for probe design
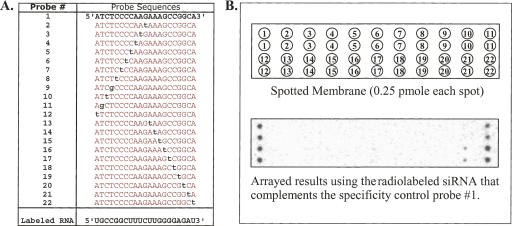
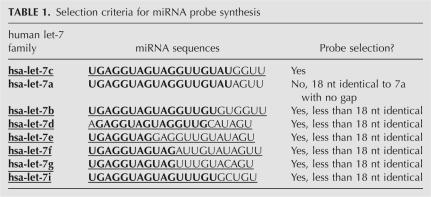
More than 820 miRNAs have been cloned or predicted from humans and mice (Griffiths-Jones 2006; Griffiths-Jones et al. 2006). These miRNAs are divided into different families and some family members (paralogs) only differ by a few nucleotides. Thus, cross-hybridization between the miRNAs and their probes is inevitable. To evaluate the extent of cross-hybridization and to design unique probes that reduce cross-hybridization, we designed and synthesized 22 DNA probes (21 nt): one wild type and the others with a single nucleotide difference from the wild type (Fig. 2A). The 22 DNA probes (0.25 pmol/each) were spotted and immobilized on a membrane by UV cross-linking, and then were hybridized with a radiolabeled synthetic RNA oligo that complements all of these DNA probes. The concentration of the labeled RNA oligo was ∼0.1 pmol, reflecting the upper-limit concentration of endogenous miRNAs in our array platform. The results showed that a 21-nt synthetic RNA that has 18–19 consecutive nucleotides complementary to the probes had a cross-hybridization effect (Fig. 2B). Thus, probes for miRNAs that have less than 18–19 consecutive identical nucleotides were designed and synthesized. For example (Table 1), among the human miRNA let-7 family members, let-7a has 18 consecutive nucleotides identical to let-7c. Therefore, either let-7a or let-7c, but not both, would be a candidate for probe design and synthesis. The reason for such selection is that the probe for let-7a cannot distinguish between let-7a and let-7c and vice versa. Other let-7 paralogs, 7b, 7d, 7e, 7f, 7g, and 7i, will all be selected for probe synthesis, because those let-7 members have less than 18 identical nucleotides in an 18-nt region.
FIGURE 2.
Specificity of single nucleotide differences in miRNA array. (A) The probe sequences of 22 specificity controls. Probe #1 is 100% complementary to the labeled RNA. The others (Probe #2–22) hybridize to the labeled RNA with only a single nucleotide difference at different positions. (B) The labeled RNA was hybridized to the membrane that was spotted in duplicate with the control probes. The array results showed high specificity at the level of a single nucleotide difference.
TABLE 1.
Selection criteria for miRNA probe synthesis
We chose paralogous miRNAs that have less than 18 consecutive identical nucleotides for probe design and synthesis, based on the data of single nucleotide difference experiments (Fig. 2). To date, 847 human and mouse miRNAs, including species conserved, species specific, and identical members of certain miRNA families (Supplemental Table S1), have been annotated and deposited at the miRBase (http://microrna.sanger.ac.uk/). Using the above criteria, 553 miRNAs are eligible for probe design and synthesis. These miRNA probes can detect all annotated human and mouse miRNAs as well as miRNAs from other animal sources (Supplemental Table S2).
Probe concentration titration for array spotting at a linear range
Nonredundant probes (which, as described above, are predicted to not cross-react with each other) were UV cross-linked onto the nylon membrane and hybridization between the probes and isolated miRNAs was performed in a similar fashion as in traditional Northern blot analysis. A total of 553 selected DNA oligonucleotide probes were synthesized by Integrated DNA Technologies (IDT). Subsequently, we determined how much miRNA probes should be spotted on the array using a high-throughput robot, Qpix2, at the Advanced Genetic Technologies Center (AGTC) of the University of Kentucky. We chose three miRNAs that were expressed at high, medium, and low levels, respectively. Their DNA probes were diluted at different concentrations (80, 40, 20, and 10 μM, respectively). These serially diluted probes were printed onto the membrane with one, two, three, four, five, and six prints (each print takes ∼3 nL of the probe) on each spot. Isolated small RNAs were labeled and hybridized to the membrane for titration. Results indicated that three prints (∼9 nL total) of 20-μM probes gave the best array results within the linear range (data not shown).
miRNA array detection range and systematic normalization probes
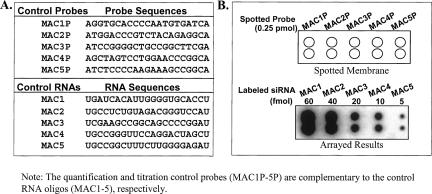
To test the miRNA array linear range for quantification, we designed and synthesized five different synthetic RNA oligos that were different in their sequences to serve as miRNA array controls (MACs) (Fig. 3A). Five MAC probes (MAC1P, MAC2P, MAC3P, MAC4P, and MAC5P), which corresponded to the five synthetic RNA oligos (MAC1, MAC2, MAC3, MAC4, and MAC5) were synthesized and printed on the array membrane at the same concentration (0.25 pmol) as that of the miRNA array probes (Fig. 3A). Different amounts (5, 10, 20, 40, and 60 fmol) of the five synthetic RNA oligos were used to mix with each sample RNA before small RNA isolation and labeling with [γ-32P]ATP and PNK, followed by a hybridization analysis. Results from this analysis (Fig. 3B) demonstrated that the array signals accurately reflected the amount of input of the synthetic RNA oligos in a linear range, indicating that the array platform is highly reliable.
FIGURE 3.
Detection range of miRNA array. (A) The sequences for the quantification and titration RNA oligoss (MAC1, MAC2, MAC3, MAC4, and MAC5) and their corresponding DNA probes (MAC1P, MAC2P, MAC3P, MAC4P, and MAC5P). (B) The detection range analysis. The mixture of synthetic RNAs (MAC1–5) with different amounts (5–60 fmol) were radiolabeled and hybridized to the membrane that was spotted with the same amounts (0.25 pmol) of the five DNA probes. The range of 5–60 fmol of synthetic RNA reflects the array linear range that is the average amount of most endogenous miRNAs in our preliminary array analysis.
miRNA array probe duplicate printing and UV cross-linking
Based on the above probe concentration optimization experiments, 0.2 pmol of each of the 553 miRNA probes were printed in duplicate. The array map was designed in a 32 row × 18 column format so that the 553 probes in duplicate could be spotted in a reasonable area to detect high, medium, and low levels of endogenous miRNAs simultaneously, as well as for easy quantification. Each pair of probe duplicates was arranged vertically inside a square of the array. The five external control probes (MAC1, MAC2, MAC3, MAC4, and MAC5) were also included at the lower-left corner of the array for array system normalization (Table 2). To generate array membranes in high quantity and with high quality, the 553 miRNA probes and the five external control probes of the above-designed array were spotted in duplicate onto Hybond N membranes by using the Genetix Qpix2 robot. After probe printing, the spotted membranes were wetted in 2X SSPE and the probes were permanently linked to the membrane by UV cross-linking. The efficiency and reliability of the cross-linking have been verified by the stable array results using the stripped membranes for many times without a decrease of miRNA detecting ability.
TABLE 2.
Human/mouse array map
miRNA array analysis of mouse CNS and non-CNS organs/tissues
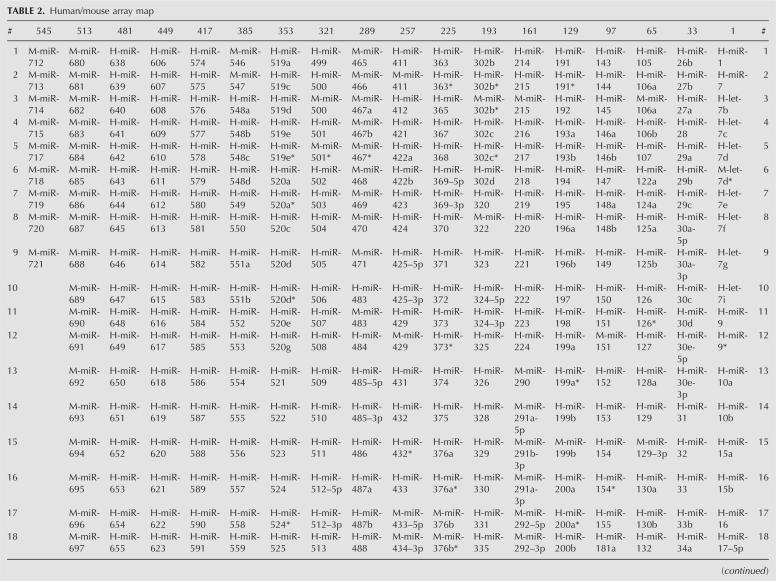
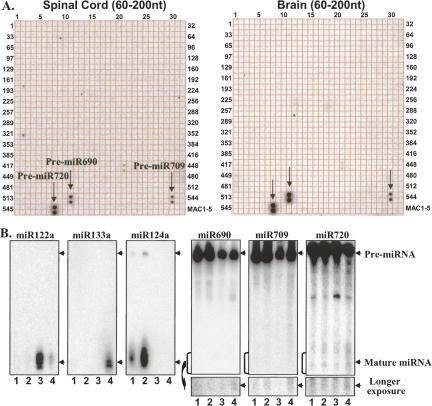
We applied the array platform to analyze mouse miRNAs in two CNS (brain and spinal cord) and two non-CNS (liver and heart) tissues. Small RNAs (15–28 nt) were isolated from the total RNAs of mouse brain, spinal cord, liver, and heart using a 15% urea– polyacrylamide gel electrophoresis (PAGE) sequencing gel. These small RNAs were dephosphorylated with Antarctic phosphatase (AP, New England Biolabs) and then labeled with [γ-32P]ATP and polynucleotide kinase (PNK). These radiolabeled small RNAs were hybridized with the miRNA membrane array and subsequently detected using a PhosphorImager (Typhoon 9400, Amersham). The representative array images are shown in Figure 4.
FIGURE 4.
An array analysis of miRNA global expression in mouse CNS and non-CNS tissues. Total RNA (50 μg) from mouse brain, spinal cord, liver, and heart was separated on a 15% PAGE and 15–28 nt small RNAs were isolated from the gel and treated for the array as described in the Materials and Methods. Of the four organs, CNS, spinal cord (A) and brain (B), contain the most abundant miRNAs. Liver (C) contains the lowest number of miRNAs of the four organs. Heart (D) also contains abundant miRNAs. The five external array controls (MAC1–5) are located at the bottom row on the right.
A total of 148 unique miRNAs were detectable in the four tissues altogether and they are summarized in Table 3. For an easy description, the 148 unique miRNAs were divided into two categories: tissue-specific and tissue-shared-specific miRNAs. Tissue-specific miRNAs are those that were only expressed in one of the arrayed tissues, while the tissue-shared-specific miRNAs were conserved in two or more tissues and are only detectable in these tissues. The tissue-shared-specific miRNAs were further divided into several subcategories according to their expression patterns among different tissues. Among the 148 unique miRNAs, 112, 93, 44, and 114 miRNAs were detected in the mouse brain, spinal cord, liver, and heart, respectively, and 36 miRNAs were ubiquitously expressed in all of the four arrayed organs. The 36 ubiquitous miRNAs include let-7b, let-7c, let-7d, let-7e, let-7f, miR17-5p, miR21, miR22, miR23b, miR24, miR26a, miR30a-5p, miR30c, miR30d, miR92, miR99a, miR103, H-miR106a, M-miR106a, miR107, miR125b, miR143, miR145, miR191, miR222, miR296, miR320, miR324-3p, miR361, miR423, miR484, miR486, miR652, miR690, miR709 (this one was determined by Northern blot; see cross-reaction between mature miRNAs and their precursors below), and miR720. The expression levels of the 36 ubiquitous miRNAs varied among tissues.
TABLE 3.
miRNA expression in mouse CNS and non-CNS organs
There are 18 miRNAs specifically expressed in both brain and spinal cord (defined as “CNS-specific”): miR34a, miR129, miR129-3p, miR212, miR298, miR300, miR326, miR330, miR346, miR370, miR381, miR383, miR409-3p, miR431, miR433, miR485-5p, miR487b, and miR541. Furthermore, the brain showed its own tissue-specific miRNAs: miR-9, miR-96, miR-134, miR-331, miR-341, miR-344, miR-409-5p, miR-434-3p, miR-485-3p, miR-491, miR-532, and miR-543, while the spinal cord showed no tissue-specific miRNA of its own (Table 3; Fig. 4A). MiR-124a was the most abundant in the mouse brain and spinal cord, which is in agreement with previous array results (Krichevsky et al. 2003; Saba and Booth 2006). MiR-124a was previously considered as neuron-specific miRNA. However, our array data showed that a low level of miR-124a was also expressed in the heart (Fig. 4D). Thus, the miR-124a is categorized as one of 32 “CNS-heart-shared”-specific miRNAs as shown in Table 3 and Supplemental Figure S1.
Among the 44 liver-expressed miRNAs (Table 3, Note 1), only three (miR192, miR215, and miR194), were liver specific. The specific expression of miR194 was further validated in this study (see the Northern blot validation section). Liver showed an expression of fewer miRNAs than other arrayed tissues. Liver also showed less shared-specific miRNAs with other arrayed tissues (Table 3).
There were 114 miRNAs expressed in the mouse heart (Table 3, Note 1). Among them, 25 were heart specific: miR-1, miR-15a, miR-15b, miR-27a, miR-27b, miR-29b, miR-29c, miR-31, miR-32, miR-208, miR-210, miR-339, miR-365, miR-373*, miR-378, miR-451, miR-487a, miR-488, miR-493-3p, miR-496, miR-499, miR-504, miR-513, miR-612, miR-689, and miR-714 (Table 3). MiR-133a, a muscle-enriched miRNA, is highly expressed in the mouse heart. This miRNA was also expressed in the CNS, and thus categorized into CNS heart-shared-specific miRNAs (Table 3). A detailed analysis of miRNAs for their organ specificity and conservation among two, three, or four mouse organs is provided in Table 3 and Supplemental Figure S1. As shown in Table 3 and Supplemental Figure S1, the brain and spinal cord are the most similar, sharing 87 miRNAs, with 18 miRNAs expressed specifically. Interestingly, the heart not only expresses as many as 114 miRNAs, but also shares many miRNAs with the brain (81 miRNAs) and the spinal cord (74 miRNAs). In contrast, the liver shares much fewer common or shared-specific miRNAs with the CNS than the heart, with no specific miRNAs shared with brain or spinal cord alone (Supplemental Fig. S1).
Northern validation of the miRNA array results
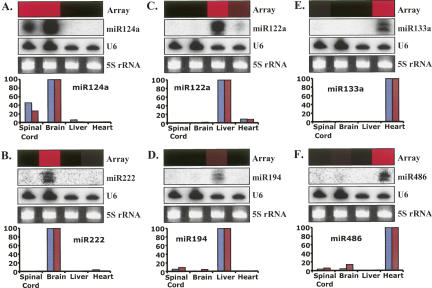
Our miRNA array is based on DNA (array-based probe printed on membrane)–RNA (miRNA labeled in solution) hybridization, which is different from the traditional Northern blot technique of labeling the DNA probe (in solution). To confirm that the array results are reliable, we chose miRNAs with low, medium, and high abundances and validated the array results independently using Northern blot techniques. The array and Northern blot validation data are shown in Figure 5. MiR-124a is considered to be a brain-specific miRNA (Conaco et al. 2006), but we found that miR-124a was not only expressed at high levels in brain tissue, but also was expressed at high levels in the spinal cord (Figs. 4A, 5A). Interestingly, miR-124a was also detectable at very low levels in the heart by our array analysis (Fig. 4D), but was not apparent by the Northern blotting analysis (Fig. 5A). This might be due to the lower sensitivity of Northern blotting or an inaccurate processing of the mature miR-124a in this tissue (see the Discussion section). MiR-222, which was reported to be detected in blood (Marsit et al. 2006), was expressed in brain tissues with medium abundance (Fig. 5B). MiR-122a was thought to be a “liver-specific” miRNA with high abundance (Chang et al. 2004; Jopling et al. 2005; Krutzfeldt et al. 2005), but we found it to be detectable in the heart (Fig. 5C). MiR-194 was expressed in the liver with low abundance (Fig. 5D). MiR-133a (Fig. 5E) and miR-486 (Fig. 5F) were both clearly detected in the heart with medium abundance. The array data are all in excellent agreement with the Northern blot results, indicating that the miRNA array technology is reliable, highly sensitive, and accurate.
FIGURE 5.
Northern blot validation of some tissue-associated miRNAs. Total RNA (10 μg per sample) from mouse spinal cord, brain, liver, and heart were electrophoresed in 15% polyacrylamide gels under denaturing conditions, blotted, and hybridized with 32P-labeled miRNA probes. 5S rRNA stained with ethidium bromide was used for loading control. U6 (light blue column) or 5S rRNA (purple column) was used for normalization separately and the signal corresponding to the miRNA with the highest signal intensity was set to 100. The expression patterns of miR-124a (A), miR-222 (B), miR-122a (C), miR-194 (D), miR-133a (E), and miR-486 (F) were consistent with the array data. The array bar was exported individually from the clustering tree (see Fig.8). Red indicates positive signal and black indicates no signal in the array.
Cross-reaction between mature miRNAs and their precursors
The biogenesis of miRNAs is a process of multistep reactions (Lee et al. 2002, 2003). Long primary transcripts (pri-miRNAs) are first processed into short miRNA precursors (pre-miRNAs) by Drosha complex in the nucleus (Han et al. 2004). Pre-miRNAs are transported into cytoplasm, where mature miRNAs of ∼22 nt are generated by Dicer (Lee et al. 2002, 2003; Yi et al. 2003; Lund et al. 2004). Only mature miRNAs are the active players that are assembled into effecter complexes or RISCs. Thus, it is important for an array platform to detect only the levels of mature miRNAs without a cross-reaction with pre-miRNAs. To prevent such cross-reactions, we specifically isolated small RNAs of 15–28 nt using a 15% sequencing gel in our array analysis. Since pre-miRNAs are generally in stem–loop structures, they may not be able to unwind and hybridize with the array probes. However, the stem region of the pre-miRNAs generally contains mismatches or bulges that destabilize the stem region, making it possible to form a relatively stable base pairing between the probes and the pre-miRNAs. This consequently may lead to a cross-reaction between the probes and the pre-miRNAs. To evaluate the extent of such cross-reactions, we specifically isolated RNAs of ∼60–200 nt, containing enriched pre-miRNAs, using a 15% sequencing gel, and subjected these longer RNAs to the array analysis. Three pre-miRNAs, pre-miR-690, pre-miR-709, and pre-miR-720, were able to cross-react with their miRNA probes. Pre-miR-594 also showed a cross-reaction signal in the array analysis. However, miR-594 was recently found to be from a tRNA (Helvik et al. 2007) and excluded from the bona fide miRNA category (Fig. 6A).
FIGURE 6.
Cross-reaction of miRNA precursors with miRNA probes. (A) Array analysis for mouse spinal cord and mouse brain using 60–200 nt small RNAs. Some miRNA (miR-690, miR-709, and miR-720) precursors of over 60 nt cross-reacted with miRNA probes (Fig. 4, cf. A and B). (B) Northern blotting analysis of pre-miR-690, pre-miR-709, and pre-miR-720, which are highly abundant and cross-react with the probes of their mature miRNAs, and pre-miR-124a, pre-miR-122a, and pre-miR-133a, which are very little or none and have no cross-reaction with the probes of their mature miRNAs. Signals from pre-miRNAs are highlighted by arrows. (Lanes 1,2,3,4) Samples from spinal cord, brain, liver, and heart, respectively.
To evaluate the abundance of these pre-miRNAs and their mature miRNAs, we conducted a Northern blot analysis of total RNA from different tissues using probes that hybridize with these miRNAs or their precursors. In agreement with the earlier array (Fig. 4) and Northern blot (Fig. 5) analyses, miR-122a, miR-124a, and miR-133a showed robust mature miRNA signals in corresponding tissues, but none or minimal pre-miRNA signals (Fig. 6B). The Northern blot analysis of 10 μg of total RNAs showed significant amounts of pre-miR-690, pre-miR-709, and pre-miR-720, but barely detectable signals corresponding to mature miR-690, miR-709, and miR-720. Longer exposure images showed a faint broad band corresponding to each mature miRNA (Fig. 6B). The relative low abundances of these mature miRNAs are in contrast to their robust miRNA signals detected in the array analysis (Fig. 4). This may be due to the fact that small RNAs were enriched from greater amounts of total RNAs (50–100 μg) in the miRNA array analysis. Alternatively, the relative abundances of pre-miRNAs and their mature miRNAs detected in this study potentially reveal the difference of endogenous processing between long primary RNAs to pre-miRNAs and to mature miRNAs (see more details in the Discussion section). In conclusion, the majority of pre-miRNAs is at much lower levels and show no cross-reaction with miRNA array probes. Isolation of small RNAs (15–28 nt) to exclude the potential cross-reaction of pre-miRNAs, such as the pre-miRNAs of miR-690, miR-709, and miR-720, is a safeguard step for specific miRNA array analysis and proper data interpretation.
Array data adjustment, normalization, and clustering
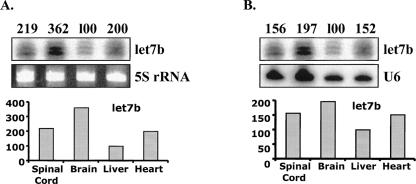
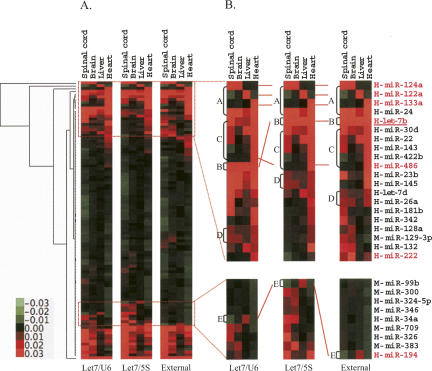
The raw array data were first corrected by subtracting the background, adjusted with external and internal controls by Northern blot analysis, and then further processed with normalization and clustering. Due to the fact that processing-related sample loss occurs differently in different sample treatments, the same amount of total RNA in different samples may not give comparable array results. Thus, an initial data adjustment for different samples is necessary. Two kinds of initial data adjustment methods were used: (1) data adjusted with external control RNAs that were mixed with a fixed amount of total RNA; and (2) data further adjusted with miRNA let-7b (a constitutively expressed miRNA) normalized by 5S RNA (Fig. 7A) or U6 RNA (Fig. 7B) using a Northern blot analysis. After data adjustment, we performed standard data transformation and clustering analyses using Cluster 3.0. The clustering data were then visualized using Java TreeView (Fig. 8). The results showed that the clustering patterns of data adjusted with external and internal controls were different, indicating that data adjustment with different approaches affects array results and interpretation (Fig. 8).
FIGURE 7.
Northern blot analysis of let-7b. Total RNA (10 μg per sample) from mouse spinal cord, brain, liver, and heart were electrophoresed in 15% polyacrylamide gels under denaturing conditions, blotted, and hybridized with 32P-labeled let7b probe. The expression level of let7b relative to 5S rRNA (A) and U6 (B) were used for normalization separately. The numbers corresponding to the tissue with the lowest signal intensity (liver) was set to 100.
FIGURE 8.
Variation of miRNA array clustering introduced by different array normalizations. (A) About 101 detectable miRNAs were hierarchically clustered based on their expression profiles in four different organs by using Gene Cluster 3.0 (complete linkage and Euclidean distance as similarity measure). Data from each miRNA were normalized by different controls and median centered prior to clustering. Samples are in columns and miRNAs in rows. (B) Variation of clustering patterns of highlighted miRNAs. (Let-7/U6) Data were normalized by let-7b level relative to U6 RNA; (Let7/5S) data were normalized by let-7b level relative to 5S rRNA; (External) data were normalized by external controls; Northern validated miRNAs are highlighted in red (e.g., H-miR-124a, H-miR-122a, H-miR-133a, H-miR-486, H-miR-222, H-miR-194). A, B, C, D, and E indicate clustering positions for certain miRNAs or miRNA clusters.
DISCUSSION
Various miRNA array platforms have been developed in the past four years (Krichevsky et al. 2003; Babak et al. 2004; Calin et al. 2004; Liu et al. 2004; Nelson et al. 2004; Sempere et al. 2004; Sioud and Rosok 2004; Jiang et al. 2005; Liang et al. 2005; Lim et al. 2005; Monticelli et al. 2005; Shingara et al. 2005; Castoldi et al. 2006; Grundhoff et al. 2006; Lee et al. 2006; Mattie et al. 2006; Tang et al. 2006; Wang and Wang 2006; Zhao et al. 2006; Wang et al. 2007). One purpose of this study was to reduce the steps of small RNA labeling for array hybridization so that the loss of miRNAs during sample labeling could be minimized. So far, most miRNA array platforms require many steps including adding adapters for the isolated small RNAs, reverse transcription, PCR amplification, and labeling the RNAs with two dyes via RNA transcription. To reduce the pitfalls of certain biases introduced by these complex steps, an RNA-primed, array-based Klenow enzyme (RAKE) miRNA platform was developed to avoid using adapters and the amplification step (Nelson et al. 2004). Other approaches for detecting endogenous miRNAs without amplification have also been developed using splinted ligation (Maroney et al. 2007). We developed a similar strategy to directly label the small RNAs at their 5′ end by introducing radiolabeled phosphates. The radiolabeled small RNAs were hybridized to membranes containing arrayed miRNA probes. This is effective because radioactive signals are highly sensitive and reflect the expression levels of different miRNAs.
This straightforward miRNA array platform has the following advantages: (1) high sensitivity: signals from ∼100 to 300 ng of the isolated small RNAs can reveal the lowest level of endogenous miRNAs at the femtomole range; (2) high specificity: the array can distinguish a single nucleotide difference; (3) repeatability: the array results remain identical for the same samples in repeats and in different experiments of array analysis; (4) cost effectiveness: it costs less than $100 per dual samples; also, the array membrane can be stripped and reprobed many times; (5) simplicity: one can use the array platform without need of special equipment or software for analysis; (6) reliability: the miRNA signal reflects the level of in vivo miRNAs without any signal amplification, thus, the endogenous levels of miRNAs can be estimated using external linear controls; and (7) verifiability: results were in very good agreement with Northern blot analysis because the array is based on a direct hybridization between endogenous miRNAs and the miRNA probes on the membrane, which is virtually a high-throughput technique for Northern blot analysis. In addition, results from our array analysis of mouse tissues revealed new findings about miRNA expressions in these tissues. Like many other array platforms, this array platform has advantages, disadvantages, and unsolved issues as discussed below.
Probe specificity
A challenge in utilizing microarrays or other hybridization-based assays is the problem of potential cross-hybridization. Nonspecific hybridization often results in artifactual expression profiles or increased data variance. To estimate the extent of cross-reaction, we generated a series of 21-mer DNA probes (22 probes, altogether) with a single nucleotide mutation from position 1 to position 21, counted from the 5′ end (Fig. 2A). The mutated probes were printed on the membrane and probed with a synthetic RNA oligo that is complementary to the probes. Results indicate that the array can distinguish a single nucleotide difference at most positions except those mutation positions (∼2 nt) near the 5′ and 3′ ends (Fig. 2B). In other words, small RNAs with 19 consecutive identical bases were not distinguishable by our array system, which was also discussed by Nelson et al. (2004). To minimize cross-hybridization as much as possible, we have designed and synthesized probes that have less than 18 consecutive identical bases for our array platform. The specificity of the probes designed according to these criteria was demonstrated in the mouse tissue array analysis. For example, let-7c and let-7e have only a 3 nt difference, but have less than 18 consecutive identical bases (Table 1). Probes of let-7c and let-7e can distinguish their different expression patterns in mouse tissues (Fig. 4). While let-7c was highly expressed in both spinal cord and brain tissues, let-7e expressed low in spinal cord (Fig. 4A, Row 1 and Column 7) and much higher in brain (Fig. 4B, Row 1 and Column 7). This indicates that probe for let-7c has little, if any, cross-reaction with let-7e. Similar examples can be found for many members of other miRNA families in our study (Fig. 4). Thus, the probes in our array system can distinguish most, but not all, specific members of most miRNA families.
To investigate whether the first and last two nucleotides may drive specificity when probes are shorter, we designed and synthesized four sets of probes of different lengths (17–21 mer) (Supplemental Fig. S2A) that complement, but mismatch at the first and the last nucleotide, with four synthetic RNA oligos (21 nt). These probes were spotted in duplicate on the membrane (Supplemental Fig. S2B) and were hybridized with different amounts (5–40 fmol) of the four radiolabeled synthetic RNA oligos. The results clearly show that the first and the last two nucleotides were not the factors to determine the hybridization specificity (Supplemental Fig. S2B). No hybridization signal was detected when the probes were shorter than 18 mer, which is in agreement with our criteria for probe design.
To further investigate whether variable hybridization conditions, especially above 37°C, could affect the array specificity or cross-reactions, we conducted the miRNA array analysis for mouse brain tissues at 37°C, 42°C, and 50°C, respectively. The results demonstrated that the array specificity has no apparent difference between the conditions of 37°C and 42°C, but array signals were significantly decreased at 50°C (Supplemental Fig. S3), apparently due to a reduced binding ability of the small RNAs to their probes under this temperature condition.
Another important potential source of cross-reaction is pre-miRNAs that may also hybridize with miRNA probes when 15–200 nt small RNAs (for example, small RNAs isolated using the mirVana miRNA Isolation Kit) are used for array analysis, leading to incorrect interpretation of the array results. Our array analysis using RNAs of 60–200 nt revealed that the majority of, except for a few, miRNAs showed no significant cross-reaction between mature miRNAs and their precursors (Fig. 6). In our miRNA array platform, we routinely isolate small RNAs of 15–28 nt for array analysis, effectively avoiding such type of cross-reaction.
Sensitivity of the array platform
The traditional method to titrate the amount of specific miRNAs is quantitative Northern blot analysis (Tang et al. 2003; Tang and Zamore 2004). However, this method is very labor intensive. In our array platform, no amplification was conducted for the endogenous miRNAs and their levels were thus nicely titrated with the external control RNAs, with different sequences and linear ranges among the different control RNAs. In addition, external control RNAs at various concentrations (5–60 fmol) were mixed with 100 μg of total RNAs before gel isolation for small RNAs. The loss of endogenous small RNAs and the input external control RNAs have the same probability during the purification steps and this serves as a titration method for estimating the actual amount of endogenous miRNAs expressed in a certain amount of cells or tissues. This is an important advantage of our microarray platform.
The miRNA-induced silencing complex (miRISC) is a classical Michaelis–Menten RNA–protein complex enzyme (Haley and Zamore 2004) that can bind and cleave the target mRNAs in a multiple turnover manner (Hutvagner and Zamore 2002). miRISC directed post-transcriptional regulation depends, to a certain extent, on the amount of miRISC enzyme. The more miRNA molecules that are in cells, the more miRISCs are formed, and thus, the more target mRNAs are regulated. Indeed, currently well-studied miRNAs are generally the highly expressed miRNAs (Chang et al. 2004; Jopling et al. 2005; Krutzfeldt et al. 2005; Chen et al. 2006; Esau et al. 2006; Wang and Wang 2006). The functions of low-level miRNAs may be in the fine-tuning regulation of their target genes, and thus, may not be easily identified. An estimate of the actual amount of the endogenous miRNA will thus be an important guideline for the study of their specific functions.
Although nonradioactive array platforms are generally preferred, radioactivity still offers one of the most sensitive approaches for the detection of nonamplified miRNAs, as shown in our study. We have included five external control RNAs at the amounts from 5 to 60 fmol (Fig. 4; Table 2, MAC1–5). Most miRNAs expressed in the analyzed mouse organs are under 60 fmol (Fig. 4) and the smallest amount of external control RNA in our array analysis was 5 fmol. From the actual array results, signals that were weaker than the 5 fmol external control RNA were still detectable, indicating that our array can successfully detect amounts under 5 fmol with no amplification of endogenous miRNAs. In addition, small RNAs can be efficiently dephosphorylated and rephosphorylated at their 5′ end using a phosphatase (AP) and a kinase (PNK) (Supplemental Fig. S4), making it an efficient way to directly label the isolated small RNAs for array analysis.
Array reliability and Northern blot validation
The reliability of the miRNA array developed in this study is excellent based on the following observations. First, the most abundant and tissue-associated mouse miRNAs revealed in this study are in agreement with previously published data. For example, our array results showed that miR-124a was most abundant in CNS tissues and that miR-122a is an abundant liver-associated miRNA; these are similar to the previous reports (Jopling et al. 2005; Krutzfeldt et al. 2005; Esau et al. 2006). Second, Northern blot validation results were in agreement with the array results (Fig. 5). Third, clustering analysis of the array results showed that miRNA expression patterns between the brain and spinal cord were conserved, reflecting a conservation of general characteristics of nervous tissues or nerve cells. In addition, our sensitive array platform revealed new tissue-associated miRNAs or new locations of the previously identified miRNAs. For example, miR-486, which is highly expressed in the heart, has not been reported before. We also revealed that the brain cells contained a high level of miR-222, which was identified in the blood (Marsit et al. 2006). It is apparent that our miRNA array platform not only produces results that are consistent with the literature, but also generates new data owing to its high sensitivity.
Northern blot validation is traditionally an essential step for all current miRNA array technologies. In our array analysis, in addition to traditional approaches for data normalization, we also introduced an additional step for array data adjustment by Northern blot analysis of a selected miRNA (let-7b). This additional step may be useful in determining results from ambiguous array data and give accurate data interpretation. Since this step is independent of the miRNA array process and can be done before or after the array analysis, it will not affect the efficiency of the high-throughput array of the samples.
The amount of small RNAs that is isolated from 100 μg of total RNA is about 5–10 times greater than that from the amount (10–20 μg) used for Northern blot validation. Analysis of the external synthetic small RNAs before and after gel purification estimated that about 95% of the small RNAs were recovered in the small RNA isolation step. In our array process, no amplification step was conducted for the isolated small RNAs, and thus they accurately represent the endogenous level of small RNAs. In addition, synthetic miRNA probes (DNA) were printed on the same type of membrane for Northern blot analysis. Therefore, our miRNA array platform is essentially a high-throughput Northern blot analysis. The only difference between our array analysis and Northern blot is that the Northern blot technique transfers the small RNA onto the membrane and DNA probes are labeled for hybridization, while our array analysis uses the opposite approach with DNA printed on the membrane and the RNAs labeled for hybridization. Thus, our array analysis reflects the actual amount of miRNAs from sampled tissue or cells and is 5–10 times more sensitive than Northern blot analysis.
Discrepancies sometimes occur between different array platforms and Northern blot validation for certain miRNAs (unpublished observations).This may be due to using RNA adaptors that were not equally ligated to the miRNAs (Romaniuk et al. 1982; Nelson et al. 2004). Our array platform avoids the use of adapters in miRNA labeling and can considerably minimize the discrepancies between array results and Northern blot validation. Indeed, among the six selected ubiquitous and tissue-associated miRNAs, Northern blot validation results were in extremely high agreement with our array results. However, low-expressed miRNAs that can be detected by our array analysis often cannot be detected by Northern blot analysis (Fig. 5). This is probably due to the higher sensitivity of our array platform over the Northern blot analysis. Nevertheless, when we increased the amount of total RNAs to 100 μg for Northern blot analysis, the sensitivity of Northern blot analysis was not improved much. The exact significance of this phenomenon is not known but may be related to the inability of small amounts of RNA to be bound to the Northern blot membrane or the spreading of signals in Northern blots, possibly due to inaccurate processing of miRNAs in certain tissues.
Array data adjustment, normalization, and controls
Due to their small size, miRNAs are particularly difficult to analyze by a high-throughput array approach and the array data normalization is nontrivial (Davison et al. 2006). In Northern blot analysis, ribosomal RNAs, tRNAs, and U6 are often used as controls for equal loading of total RNAs, data adjustment, and normalization. However, these control RNAs are significantly longer than a miRNA, and it is impossible to probe these control RNAs directly on the same miRNA array membranes. Many previously reported miRNA arrays are based on the input amount of total RNA, assuming that the same amount of total RNA contains comparable amounts of miRNAs, and that small RNAs isolated from the same amount of total RNA are recovered at the same efficiency. Some array platforms use the same amount of isolated small RNAs as array input and for array normalization. In our experience, the small RNAs recovered from different samples may vary dramatically after gel isolation, elution, labeling, and purification. To minimize the discrepancy between different samples, we designed a linear range of external control small RNAs. The probes for these synthetic small RNAs were printed on membranes side by side with the miRNA probes. The same amount of the external control RNAs was mixed to each sample of the same amount before gel isolation so that the fixed amount of external control RNAs could serve as a control for an estimation of the loss of miRNAs during small RNA isolation. In addition, these external controls can serve as a titration for quantification of endogenous miRNAs.
A theoretically better alternative for adjusting array data is to use the above-mentioned Northern blot analysis for a constitutively expressed miRNA normalized by 5S/U6 RNA. For a comparison to the data adjustment by external controls, we selected the miRNA let-7b that is ubiquitously expressed in human and mouse organs (Baskerville and Bartel 2005) as an internal control for Northern blot analysis using either 5S or U6 RNA for equal sample loading normalization. Results showed a difference between the data adjusted by external control RNAs and the data normalized by an internal let-7b control (Fig. 8), indicating an influence on data interpretation by different normalization methods. In addition, internal control normalized by 5S or U6 also produces a different clustering tree. These differences may be attributed to the variation of 5S or U6 RNAs under different conditions (Fig. 7).
The use of Northern blot analysis of one specific miRNA in comparison with U6 or 5S RNA for array data adjustment, rather than only for validation, is a new additional way to normalize data in an array analysis. This normalization is based on the assumption that all the miRNAs from a specific sample under a specific array have a relatively stable ratio between total miRNAs and the proportional loss of miRNAs during purification and labeling steps. This allows for a reasonable adjusting of array signals from different samples with one constitutively expressed miRNA for all the arrayed samples before the actual array comparison. The disadvantage of this normalization is that all of the arrayed samples must be analyzed for one miRNA expression level by Northern blot. However, this step can be done independent of the array process by the sample providers, and thus will not affect the high-throughput manner of array analysis. Current miRNA array platforms use Northern blot analysis only for the validation of array results. Due to improper normalization, many miRNA arrays suffer from discrepancies between the array results and Northern blot analysis (unpublished observation). An extension of Northern blot analysis for the miRNA array data adjustment and normalization steps is helpful for obtaining accurate conclusions.
Although the above technique may be the best possible method for miRNA array normalization to date, it still needs additional refinement. We noticed that U6 and 5S RNA loading controls have different quantitative signals, indicating that U6 or 5S RNA may also change from sample to sample (Figs. 5, 7). U6 and 5S RNAs are frequently used for miRNA arrays and for the validation of results by Northern blot (Krichevsky et al. 2003; Monticelli et al. 2005; Castoldi et al. 2006; Zhao et al. 2006). These two control-adjusted data might result in similar results for some abundant or highly expressed tissue-associated miRNAs, but might give different results for miRNAs with lower expression levels (e.g., see Figs. 5A,D,F, 7), presumably due to different levels of U6 and 5S RNAs in the samples. Further investigation of more reliable controls for miRNA array analysis is needed.
Clustering analysis
Array clustering analysis gives a straightforward comparison of similarities and differences between samples. Our array clustering analyses suggest that the key components of the mouse CNS (brain and spinal cord) have a highly conserved miRNA expression pattern, indicating that neurons in the brain and spinal cord have similar miRNA characteristics (Figs. 4, 5; Table 3). In addition, our analysis showed that the liver and heart are very specialized organs and retain their unique profiling patterns (Fig. 4; Table 3). Array clustering analysis is not only important in finding the differences between different mouse organs, but also in revealing specific miRNA cohorts that may have specialized functions. This may be important in identifying miRNA regulatory networks that are important for specific developmental processes.
Detection of tissue-specific miRNAs and their potential functional relevance
We have identified tissue/organ-associated miRNAs in addition to widely expressed miRNAs. Some tissues express fewer tissue-specific miRNAs than others (Table 3; Supplemental Fig. S1). For example, the liver and spinal cord each only contain one or zero tissue-specific miRNA. In contrast, the brain (12 tissue-specific miRNAs) and the heart (26 tissue-specific miRNAs) contain much more tissue-specific miRNAs. In addition, some miRNAs, though not specifically associated with a particular tissue, are preferentially expressed at high or low levels in certain tissues. The miRNA expression levels presumably play an important role in keeping tissue/organ identity or functions, and misregulation of them may lead to various kinds of diseases (Croce and Calin 2005).
No or little miR-195 is expressed in the normal mouse heart (Fig. 4). It was recently reported that miR-195 is up-regulated during cardiac hypertrophy (van Rooij et al. 2006). The same study showed that cardiac overexpression of miR-195 resulted in pathological cardiac growth and heart failure in transgenic mice, suggesting that misregulation of miR-195 could lead to heart problems. In contrast, muscle-associated miR-1, which counteracts the role of miR-195 and inhibits cardiac growth by suppressing the expression of the basic helix–loop–helix protein Hand2 (Zhao et al. 2005), was specifically expressed in the heart in our array (Fig. 4; Table 3). This indicates that heart-specific miRNA, miR-1, plays an important role in heart functions.
Tissue specificity of miRNA expression is probably less absolute than previously described. MiR-124a, which is thought to be a brain-specific miRNA (Conaco et al. 2006), was indeed detected to be the most abundant miRNA in the mouse brain in our array (Figs. 4B, 5A). In addition, it was also expressed abundantly in the spinal cord (Figs. 4A, 5A) and at a low level in the heart (Fig. 4D). Recently, miR-124a has been shown to be up-regulated in mouse pancreas and target the transcription factor Foxa2 for regulation of pancreas development (Baroukh et al. 2007). Another example is miR-9, which was detected in the mouse brain in this study (Fig. 4; Table 3). However, it also was reported as being expressed in pancreatic insulin-secretion cells and played a negative role in insulin secretion (Plaisance et al. 2006). All of these examples indicate that tissue-specific miRNAs may not be absolutely tissue specific or function specific; rather, they may be detectable in other tissues and have broader roles in gene regulation and function. Extensive array analysis of different tissue or cell types will certainly advance miRNA study by revealing their broader functions.
miRNA biogenesis and miRNA array
The miRNA biogenesis is a complex process of multiple steps and plays an important role in gene regulation by producing mature miRNAs in a temporal and spatial manner. A striking observation is that both pre-miRNAs of miR-690, miR-709, and miR720, and their mature miRNAs were readily detected by our array analyses (Figs. 4, 6A). Northern blot analysis revealed the presence of a large amount of these miRNA precursors constitutively expressed in all of the array mouse tissues. However, their mature miRNAs are difficult to detect by Northern blot (Fig. 6B), although robust signals were detected by the array analysis (Fig. 4). Longer exposed images showed a faint broad band corresponding to each mature miRNA (Fig. 6B). These discrepancies raised several questions regarding the miRNAs to be detected and the reliability of the array technology.
The precursors of miR-690, miR-709, and miR720 were the only three miRNA precursors detected in the array analysis of total RNA (Fig. 6). This indicates that these miRNAs are probably unconventional endogenous small RNAs. First of all, the precursors of these miRNAs accumulated in large amounts in all arrayed tissues (Fig. 6B) and other cell lines (data not shown). In contrast, in the case of most miRNAs, only little or no pre-miRNA accumulates. This suggests that these three miRNAs have a normal processing of primary miRNAs into pre-miRNAs, and an abnormal processing of pre-miRNAs into mature miRNAs. However, whether these accumulated “pre-miRNAs” are the product of a Drosha complex is not clear. Knocking down of Drosha will give insights into the biogenesis of these “precursor” RNAs.
Second, these three miRNAs (miR690, miR-709, and miR-720) were previously identified by extensive sequencing of small RNAs isolated from mouse embryos (Mineno et al. 2006). They appear “unconventional” by having longer loops and more mismatches at the “stem” region. The higher extent of mismatches at the “stem” region may inhibit the cellular Dicer from effective processing. Additional cellular factors may be needed for Dicer to process these miRNA precursors. In certain tissues (e.g., mouse spleen, testis, and lung), mature miR-690, miR-709, and miR720 are better processed and a clearer signal of the mature miRNAs can be detected by Northern blot (Supplemental Fig. S5). It is not clear how these miRNAs are matured from their precursors and why they are not effectively processed. Mutation of the “stem” region of the miRNA precursors for in vitro and in vivo processing will give insights into the biogenesis of these miRNAs.
What are those robust array signals for miR-690, miR-709, and miR720, and why is it difficult to detect them by Northern blotting? Given that the unconventional secondary structures and highly abundant precursors exist to these miRNAs, the processing of the precursor RNAs by the Dicer may not be accurate, resulting in a population of small RNAs with different lengths. This may be the reason why Northern blot detected no sharp band, rather, a faint broad band. In contrast, the array signals came from a dot blot of hybridization between the spotted probes and the enriched small RNAs. This gives a concentrated signal from the small RNAs. Finally, the accumulated precursor RNAs may be subject to degradation, and the degradated products may be enriched in the array hybridization. All of these possibilities contribute to the discrepancy between the array and the Northern blot analyses of these miRNAs.
In summary, we have developed a highly sensitive miRNA array platform and successfully applied it in the analysis of miRNA profiles in different mouse organs. A number of new tissue-associated miRNAs have been revealed in different mouse organs. The array platform has been optimized with various titrations to a linear range. New methods for data adjustment, normalization, and clustering analysis have also been introduced to improve array analysis. This array platform will be widely useful for various kinds of studies.
MATERIALS AND METHODS
Comprehensive miRNA selection for the designed array
A total of 553 miRNA probes, which cover the human and mouse-specific miRNAs, were spotted onto a Hybond N+ membrane by using the Genetix Qpix2 (Advanced Genetic Technologies Center at the University of Kentucky). Each array membrane also contained five quantification and titration probes (MAC1, MAC2, MAC3, MAC4, MAC5; for detailed probe design and experimental justification, see the Results section). After being printed with probes, the membrane was UV cross-linked twice, with 100 μJ/cm2 for half a minute each time, before and after rinsing with 2X SSPE.
Sample preparation: Total RNA isolation, small RNA enrichment, and RNA qualitative and quantitative assessment
Total RNAs were isolated as previously described using conventional methods (Haley et al. 2003; Tang et al. 2003; Tang and Zamore 2004), with some modifications for better quality assurance and standardization. Briefly, 4-month-old FVB mice were anesthetized with an intraperitoneal injection of 0.1 mL Pentobarbital (50 mg/mL, Abbott Laboratories) and transcardically perfused with 0.1 M phosphate buffered saline (PBS) (pH 7.5). Liver, heart, brain, and spinal cord were dissected and snap-frozen in liquid nitrogen. All animal procedures were approved by the university IACUC committee. Tissues/organs were frozen in liquid nitrogen and ground finely with a mortar and pestle. Total RNAs were extracted using Trizol reagent (Invitrogen) with the following modification: total RNAs were precipitated with isopropanol at –20°C overnight for better precipitation of small RNAs. RNA quality was examined prior to miRNA isolation using a Bioanalyzer 2100 (Agilent). An aliquot of total RNA was kept as a control and for future Northern blot validation.
At least 50 μg of total RNAs from each sample were used for small RNA (15–28 nt) isolation using 15% denaturing PAGE. A mixture of synthetic 21-nt control RNAs that were complementary to five MAC probes were incorporated into each sample (see details in the Results section). The gel-purified small RNAs were monitored and quantified by Nanodrop spectrophotometer (Nanodrop Technologies).
Small RNA labeling and hybridization of array membrane
The isolated small RNAs were treated with Antarctic Phosphatase (New England Biolabs) to remove the 5′ phosphates. The phosphatase was heat inactivated and the dephosphorylated small RNAs were then radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). The labeled samples were purified by passing through a G-25 spin column (Roche) and monitored for labeling efficiency by analyzing 1 μL of diluted probes (100 × dilution) on a 15% sequencing gel. Only well-labeled samples were used for further hybridization according to our prior published protocol (Tang et al. 2003; Tang and Zamore 2004).
The arrayed membrane was prehybridized for at least 2 h at 37°C in 50% formamide, 5× SSPE (pH 7.4), 5× Denhardt's reagent, 0.5% SDS, and denatured herring sperm DNA (160 μg/mL). The radiolabeled small RNAs were then added and allowed to continue to hybridize overnight. The overnight hybridized membrane was rinsed and washed three times, 20 min per wash, at 37°C in 2× SSC and 0.1% SDS. The washed membrane was then wrapped and exposed on a PhosphorImager screen.
Array quantification and analysis
Signal detection and documentation
The array was scanned using a PhosphorImager scanner (Typhoon 9400, Molecular Dynamics). Array picture output and quantification was performed using ImageQuant software (Amersham Biosciences) and the intensities of duplicate spots on each array were averaged.
Data normalization
To compare the miRNA expression profiles between different tissues, external and internal controls were used for array data normalization. The external controls were used to remove system-related variations (e.g., amount variations, process-related loss of small RNAs or signals, variations in labeling signal strength, and signal gain differences between scanners), so that biologically relevant variations could be faithfully revealed (see the Results section) (Quackenbush 2001, 2002; Geller et al. 2003). The internal control was a ubiquitously expressed miRNA, normalized by 5S rRNA or U6. The quantification data with various ways of normalization are shown in Supplemental Table S3. The selection of the ubiquitous miRNA was based on a comparison of results from all arrayed samples.
Clustering analysis
Clustering analyses were performed with a hierarchical method using an average linkage and Euclidean distance metric to illustrate relationships among the data from complex array experiments using Cluster 3.0 (de Hoon et al. 2004). The clustering data were visualized using Java TreeView (Saldanha 2004).
Northern blot validation
Northern blot analysis was performed as previously described (Tang et al. 2003; Tang and Zamore 2004). Briefly, 10 μg total RNA isolated from tissues was run on a denaturing 15% Urea–PAGE gel, transferred to membrane, blotted, and probed using DNA oligonucleotides labeled with [γ-32P]ATP. Washed Northern blots were placed on PhosphorImager screens, scanned subsequently using the Typhoon Scanner, and signals were quantified using the ImageQuant software package.
SUPPLEMENTAL DATA
All Supplemental Tables can be found at http://www.uky.edu/∼gtang2/publication/RNA-array-method-suppl-data/TableS1-3.xls). All Supplemental Figures can be found at http://www.uky.edu/∼gtang2/publication/RNA-array-method-suppl-data/Fig-S1-5.pdf.
ACKNOWLEDGMENTS
We thank Dr. Peter Nelson for his critical reading and constructive comments on this manuscript. We thank Ms. Renee Kilty for assistance in mouse tissue collection. This study is partially funded by the Kentucky Tobacco Research and Development Center (KTRDC) (G.T.); the Advanced Genetic Technologies Center (AGTC) of the University of Kentucky (G.T.); USDA-NRI grants 2006-35301-17115 and 2006-35100-17433 (G.T.); NIH/NINDS grant R01NS49126 (H.Z.); and NIH/NCRR Centers of Biomedical Research Excellence grant 1P20RR020171-010005 (H.Z.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.498607.
REFERENCES
- Akao, Y., Nakagawa, Y., Naoe, T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Ambros, V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans . Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Babak, T., Zhang, W., Morris, Q., Blencowe, B.J., Hughes, T.R. Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh, N., Ravier, M.A., Loder, M.K., Hill, E.V., Bounacer, A., Scharfmann, R., Rutter, G.A., Van Obberghen, E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cells lines. J. Biol. Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville, S., Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich, I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Cuppen, E., Plasterk, R.H. Approaches to microRNA discovery. Nat. Genet. 2006;38(Suppl):S2–S7. doi: 10.1038/nr1794. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Liu, C.G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C.D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S.E., Iorio, M.V., Visone, R., Sever, N.I., Fabbri, M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Castoldi, M., Schmidt, S., Benes, V., Noerholm, M., Kulozik, A.E., Hentze, M.W., Muckenthaler, M.U. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J., Nicolas, E., Marks, D., Sander, C., Lerro, A., Buendia, M.A., Xu, C., Mason, W.S., Moloshok, T., Bort, R., et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chen, C.Z., Lodish, H.F. MicroRNAs as regulators of mammalian hematopoiesis. Semin. Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Chen, C.Z., Li, L., Lodish, H.F., Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen, J.F., Mandel, E.M., Thomson, J.M., Wu, Q., Callis, T.E., Hammond, S.M., Conlon, F.L., Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, A.M., Byrom, M.W., Shelton, J., Ford, L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino, A., Calin, G.A., Fabbri, M., Iorio, M.V., Ferracin, M., Shimizu, M., Wojcik, S.E., Aqeilan, R.I., Zupo, S., Dono, M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco, C., Otto, S., Han, J.J., Mandel, G. Reciprocal actions of REST and a micro-RNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean, S., Zanesi, N., Pekarsky, Y., Tili, E., Volinia, S., Heerema, N., Croce, C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(μ)-miR155 transgenic mice. Proc. Natl. Acad. Sci. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, C.M., Calin, G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Cuellar, T.L., McManus, M.T. MicroRNAs and endocrine biology. J. Endocrinol. 2005;187:327–332. doi: 10.1677/joe.1.06426. [DOI] [PubMed] [Google Scholar]
- Cummins, J.M., Velculescu, V.E. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- Davison, T.S., Johnson, C.D., Andruss, B.F. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- de Hoon, M.J.L., Imoto, S., Nolan, J., Miyano, S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Ding, L., Spencer, A., Morita, K., Han, M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans . Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Du, T., Zamore, P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E.G., Ravichandran, L.V., Sun, Y., Koo, S., Perera, R.J., Jain, R., et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Esau, C., Davis, S., Murray, S.F., Yu, X.X., Pandey, S.K., Pear, M., Watts, L., Booten, S.L., Graham, M., McKay, R., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Euling, S., Ambros, V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996;84:667–676. doi: 10.1016/s0092-8674(00)81045-4. [DOI] [PubMed] [Google Scholar]
- Garzon, R., Fabbri, M., Cimmino, A., Calin, G.A., Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gauthier, B.R., Wollheim, C.B. MicroRNAs: “Ribo-regulators” of glucose homeostasis. Nat. Med. 2006;12:36–38. doi: 10.1038/nm0106-36. [DOI] [PubMed] [Google Scholar]
- Geller, S.C., Gregg, J.P., Hagerman, P., Rocke, D.M. Transformation and normalization of oligonucleotide microarray data. Bioinformatics. 2003;19:1817–1823. doi: 10.1093/bioinformatics/btg245. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Grocock, R.J., van Dongen, S., Bateman, A., Enright, A.J. miRBase: MicroRNA sequences, targets, and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gk112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff, A., Sullivan, C.S., Ganem, D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, B., Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Haley, B., Tang, G., Zamore, P.D. In vitro analysis of RNA interference in Drosophila melanogaster . Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., Kim, V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita, Y., Osada, H., Tatematsu, Y., Yamada, H., Yanagisawa, K., Tomida, S., Yatabe, Y., Kawahara, K., Sekido, Y., Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- He, L., Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Helvik, S.A., Snove O., Jr, Saetrom, P. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics. 2007;23:142–149. doi: 10.1093/bioinformatics/btl570. [DOI] [PubMed] [Google Scholar]
- Horvitz, H.R., Sternberg, P.W., Greenwald, I.S., Fixsen, W., Ellis, H.M. Mutations that affect neural cell lineages and cell fates during the development of the nematode Caenorhabditis elegans . Cold Spring Harb. Symp. Quant. Biol. 1983;48:453–463. doi: 10.1101/sqb.1983.048.01.050. [DOI] [PubMed] [Google Scholar]
- Hossain, A., Kuo, M.T., Saunders, G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hwang, H.W., Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio, M.V., Ferracin, M., Liu, C.G., Veronese, A., Spizzo, R., Sabbioni, S., Magri, E., Pedriali, M., Fabbri, M., Campiglio, M., et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Lee, E.J., Gusev, Y., Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Jr, Chang, S., Etchberger, J.F., Ortiz, C.O., Hobert, O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M., Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Jovanovic, M., Hengartner, M.O. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- Kajimoto, K., Naraba, H., Iwai, N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 2006;12:1626–1632. doi: 10.1261/rna.7228806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.K., Lee, Y.S., Sivaprasad, U., Malhotra, A., Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver, J., Kroesen, B.J., Poppema, S., van den Berg, A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., Kosik, K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K.G., Tuschl, T., Manoharan, M., Stoffel, M. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. microRNAs: Runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Kim, H.K., Chung, S., Kim, K.S., Dutta, A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Lee, E.J., Gusev, Y., Jiang, J., Nuovo, G.J., Lerner, M.R., Frankel, W.L., Morgan, D.L., Postier, R.G., Brackett, D.J., Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2006;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li, Y., Wang, F., Lee, J.A., Gao, F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila . Genes & Dev. 2006a;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Zhan, W., Wang, Z., Zhu, B., He, Y., Peng, J., Cai, S., Ma, J. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem. Biophys. Res. Commun. 2006b;348:229–237. doi: 10.1016/j.bbrc.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Liang, R.Q., Li, W., Li, Y., Tan, C.Y., Li, J.X., Jin, Y.X., Ruan, K.C. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni/019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu, C.G., Calin, G.A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, C.D., Shimizu, M., Zupo, S., Dono, M., et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Kirch, S., Ambros, V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., Kutay, U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Chamnongpol, S., Souret, F., Nilsen, T.W. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA. 2007;13:930–936. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit, C.J., Eddy, K., Kelsey, K.T. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Martin, C.M., Astbury, K., O'Leary, J.J. Molecular profiling of cervical neoplasia. Expert Rev. Mol. Diagn. 2006;6:217–229. doi: 10.1586/14737159.6.2.217. [DOI] [PubMed] [Google Scholar]
- Mattie, M.D., Benz, C.C., Bowers, J., Sensinger, K., Wong, L., Scott, G.K., Fedele, V., Ginzinger, D., Getts, R., Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler, M.F., Mattick, J.S. Noncoding RNAs in the nervous system. J. Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C.C., Czech, M.P. Micromanaging insulin secretion. Nat. Med. 2004;10:1297–1298. doi: 10.1038/nm1204-1297. [DOI] [PubMed] [Google Scholar]
- Min, H., Chen, C.Z. Methods for analyzing microRNA expression and function during hematopoietic lineage differentiation. Methods Mol. Biol. 2006;342:209–227. doi: 10.1385/1-59745-123-1:209. [DOI] [PubMed] [Google Scholar]
- Mineno, J., Okamoto, S., Ando, T., Sato, M., Chono, H., Izu, H., Takayama, M., Asada, K., Mirochnitchenko, O., Inouye, M., et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, S., Ansel, K.M., Xiao, C., Socci, N.D., Krichevsky, A.M., Thai, T.H., Rajewsky, N., Marks, D.S., Sander, C., Rajewsky, K., et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, E.G., Lee, R.C., Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Yasuda, T., Saigo, K., Urashima, T., Toyoda, H., Okanoue, T., Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E., Mourelatos, Z. The microRNA world: Small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Kloosterman, W.P., Kauppinen, S., Plasterk, R.H., Mourelatos, Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pallante, P., Visone, R., Ferracin, M., Ferraro, A., Berlingieri, M.T., Troncone, G., Chiappetta, G., Liu, C.G., Santoro, M., Negrini, M., et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- Plaisance, V., Abderrahmani, A., Perret-Menoud, V., Jacquemin, P., Lemaigre, F., Regazzi, R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J. Computational analysis of microarray data. Nat. Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002;32:496–501. doi: 10.1038/ng1032. Suppl. [DOI] [PubMed] [Google Scholar]
- Ramkissoon, S.H., Mainwaring, L.A., Ogasawara, Y., Keyvanfar, K., McCoy J.P., Jr, Sloand, E.M., Kajigaya, S., Young, N.S. Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Romaniuk, E., McLaughlin, L.W., Neilson, T., Romaniuk, P.J. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur. J. Biochem. 1982;125:639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- Rougvie, A.E., Ambros, V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans . Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- Saba, R., Booth, S.A. Target labelling for the detection and profiling of microRNAs expressed in CNS tissue using microarrays. BMC Biotechnol. 2006;6:47. doi: 10.1186/1472-6750-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y., Liang, G., Egger, G., Friedman, J.M., Chuang, J.C., Coetzee, G.A., Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Saldanha, A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingara, J., Keiger, K., Shelton, J., Laosinchai-Wolf, W., Powers, P., Conrad, R., Brown, D., Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveri, L., Tilly, G., Vilotte, J.L., Le Provost, F. MicroRNA involvement in mammary gland development and breast cancer. Reprod. Nutr. Dev. 2006;46:549–556. doi: 10.1051/rnd:2006026. [DOI] [PubMed] [Google Scholar]
- Sioud, M., Rosok, O. Profiling microRNA expression using sensitive cDNA probes and filter arrays. Biotechniques. 2004;37:574–576. 578–580. doi: 10.2144/04374ST01. [DOI] [PubMed] [Google Scholar]
- Slack, F., Ruvkun, G. Temporal pattern formation by heterochronic genes. Annu. Rev. Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- Song, L., Tuan, R.S. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res. C Embryo Today. 2006;78:140–149. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- Tang, G., Zamore, P.D. Biochemical dissection of RNA silencing in plants. Methods Mol. Biol. 2004;257:223–244. doi: 10.1385/1-59259-750-5:223. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes & Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F., Hajkova, P., Barton, S.C., Lao, K., Surani, M.A. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno, B., Cesi, V., Vitali, R., Sesti, F., Giuffrida, M.L., Mancini, C., Calabretta, B., Raschella, G. Silencing of endogenous IGFBP-5 by micro RNA interference affects proliferation, apoptosis and differentiation of neuroblastoma cells. Cell Death Differ. 2005;12:213–223. doi: 10.1038/sj.cdd.4401546. [DOI] [PubMed] [Google Scholar]
- Thompson, B.J., Cohen, S.M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila . Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., Newman, M., Parker, J.S., Morin-Kensicki, E.M., Wright, T., Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij, E., Sutherland, L.B., Liu, N., Williams, A.H., McAnally, J., Gerard, R.D., Richardson, J.A., Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N., Klein, M.E., Varlamova, O., Keller, D.M., Yamamoto, T., Goodman, R.H., Impey, S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia, S., Calin, G.A., Liu, C.G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Ach, R.A., Curry, B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Wang, X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Takeda, A., Mise, K., Okuno, T., Suzuki, T., Minami, N., Imai, H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Weber, F., Teresi, R.E., Broelsch, C.E., Frilling, A., Eng, C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J. Clin. Endocrinol. Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- Weston, M.D., Pierce, M.L., Rocha-Sanchez, S., Beisel, K.W., Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., Kloosterman, W.P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, H.R., Kauppinen, S., Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xie, X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., Kellis, M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., Stephens, R.M., Okamoto, A., Yokota, J., Tanaka, T., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Wang, F., Yang, G.H., Wang, F.L., Ma, Y.N., Du, Z.W., Zhang, J.W. Human microRNA clusters: Genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Samal, E., Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao, J.J., Hua, Y.J., Sun, D.G., Meng, X.X., Xiao, H.S., Ma, X. Genome-wide microRNA profiling in human fetal nervous tissues by oligonucleotide microarray. Childs Nerv. Syst. 2006;22:1419–1425. doi: 10.1007/s00381-006-0173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]