Abstract
Multiple steps in mRNA processing and transcription are coupled. Notably, the processing of mRNA 3′ ends is linked to transcription termination by RNA polymerase II. Previously, we found that the yeast hnRNP protein Npl3 can negatively regulate 3′ end mRNA formation and termination at the GAL1 gene. Here we show that overexpression of the Hrp1 or Rna14 subunits of the CF IA polyadenylation factor increases recognition of a weakened polyadenylation site. Genetic interactions of mutant alleles of NPL3 or HRP1 with RNA15 also indicate antagonism between these factors. Npl3 competes with Rna15 for binding to a polyadenylation precursor and inhibits cleavage and polyadenylation in vitro. These results suggest that an important function of hnRNP proteins is to ensure the fidelity of mRNA processing. Our results support a model in which balanced competition of Npl3 with mRNA processing factors may promote recognition of proper polyadenylation sites while suppressing cryptic sites.
Keywords: mRNA 3′ end processing, transcription termination, cleavage, polyadenylation
INTRODUCTION
Processing of eukaryotic mRNA 3′ ends is coupled to transcription termination by RNA polymerase II (RNAPII). The same transcribed sequences that signal polyadenylation also contribute to release of polymerase from the template. Processing of mRNA 3′ ends occurs in two steps: endonucleolytic cleavage and addition of a poly(A) tail (Colgan and Manley 1997; Zhao et al. 1999a). In yeast, the multisubunit complex responsible for cleavage and polyadenylation is composed of a cleavage factor (CF) I and cleavage/polyadenylation factor (CPF). This machinery is highly conserved, with most of the subunits having orthologs in mammalian cells (for reviews, see McCracken et al. 1997; Zhao et al. 1999a; Hirose and Manley 2000; Sadowski et al. 2003; Rosonina et al. 2006).
In Saccharomyces cerevisiae, multiple sequences at the 3′ end of the gene determine where cleavage will occur (Guo and Sherman 1996; Greger and Proudfoot 1998). Two particularly important sequences usually located upstream of the poly(A) site are the UA-repeat and an A-rich element, although the adherence to these consensus motifs can vary greatly (Guo et al. 1995; Graber et al. 1999, 2002). These elements are recognized by CF I, which can be separated into two components: CF IA is a complex of Rna14, Rna15, Clp1, and Pcf11 (Minvielle-Sebastia et al. 1994; Kessler et al. 1996; Amrani et al. 1997; Takagaki and Manley 1997); CF IB consists of the RNA binding protein Hrp1 (Kessler et al. 1997; Gross and Moore 2001a). The UA-repeat is the likely docking site for Hrp1 (Kessler et al. 1997; Chen and Hyman 1998; Valentini et al. 1999), while the A-rich element is recognized by Rna15 with the help of Rna14 (Gross and Moore 2001b). Rna15 alone also has affinity for U-rich regions (Takagaki and Manley 1997; Gross and Moore 2001b). Interactions between Hrp1 and the Rna14/15 complex further enhance recognition of polyadenylation sites (Gross and Moore 2001b).
In addition to their role in cleavage, the poly(A) signals and many of the components of the cleavage/polyadenylation machinery are required for transcription termination downstream of the poly(A) site (for reviews, see Proudfoot 2000, 2004; Proudfoot et al. 2002; Buratowski 2005). For production of functional mRNA, recognition of the UA-repeat and A-rich elements is necessary to identify authentic poly(A)/termination signals. However, the lack of highly conserved polyadenylation signals in yeast and the general A/U-rich nature of the yeast genome suggests that additional mechanisms may be in place to suppress the use of cryptic sites.
We previously presented evidence that the hnRNP protein Npl3 might play such a role. An allele of NPL3 (npl3-120) was isolated in a screen for mutations that improved 3′ end processing and termination at a gene with a partial deletion of the 3′end processing signal (Bucheli and Buratowski 2005). This allele also improved termination at several wild-type poly(A) sites and increased recruitment of Rna15 to the 3′ ends of genes. Temperature-sensitive alleles of NPL3 can be suppressed by mutant alleles of genes encoding the 3′ end processing proteins Hrp1 and Rna15, suggesting a genetic balance between the functions of these proteins (Chen and Moore 1992; Henry et al. 1996; Kessler et al. 1996). All of these results are consistent with a model where cotranscriptional binding of Npl3 to the nascent mRNA transcript antagonizes recognition of 3′ end sequences by the poly(A)/termination machinery. Two nonexclusive mechanisms could lead to this antagonism: Npl3 could increase the elongation rate of RNAPII to narrow the window in which poly(A) factors can act, or Npl3 could compete with poly(A) factors for binding to the nascent RNA. Npl3 might bind and mask many weak or cryptic poly(A) sites, while a “real” poly(A) signal with higher affinity for Rna14/15 and Hrp1 could shift the balance to favor 3′end processing. Here we provide direct evidence for the competition model.
We show that altering the balance of Npl3 and 3′end processing factors influences the efficiency of 3′end processing and, in turn, that of termination. Overexpression of the processing factors Hrp1 and Rna14 can enhance termination downstream of a defective poly(A) site. Furthermore, Npl3 competes with Rna15 for binding to a polyadenylation precursor and inhibits cleavage and polyadenylation in vitro. These results support the conclusion that Npl3 is an important modulator of mRNA 3′end formation.
RESULTS
Overexpression of Hrp1 or Rna14 suppresses defective transcription termination
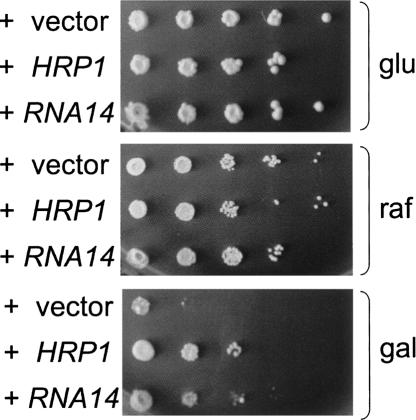
A small deletion of the poly(A) signal of the GAL10 gene (gal10Δ56) leads to a yeast strain that cannot properly metabolize galactose because of read-through transcription into the GAL7 promoter. The lack of Gal7 protein results in toxicity of galactose, even when the alternative carbon source raffinose is present (Greger and Proudfoot 1998; Bucheli and Buratowski 2005; Kaplan et al. 2005). This phenotype can be suppressed by mutations that enhance RNAPII termination (Greger and Proudfoot 1998; Bucheli and Buratowski 2005; Kaplan et al. 2005). Therefore, overexpression of proteins that contribute to recognition of poly(A) sites might also be expected to allow growth of a gal10Δ56 strain on galactose. Accordingly, the gal10Δ56 strain was transformed with a multicopy plasmid library, and transformants were screened for growth on media containing galactose and raffinose, resulting in the isolation of several genomic plasmids. Sequencing of the suppressing plasmids revealed that they carried either the HRP1 or RNA14 genes. Subcloning showed that overexpression of only HRP1 or RNA14 was sufficient for suppression, although the suppression by RNA14 was weaker than that by HRP1 (Fig. 1).
FIGURE 1.
Overexpression of Hrp1 or Rna14 suppresses the sensitivity of gal10Δ56 to galactose. Serial 10-fold dilutions of a gal10Δ56 strain (CKY265), previously transformed with 2-μm plasmids containing either HRP1 or RNA14 as indicated, were spotted on SC-leu media supplemented with 2% glucose (glu), 2% raffinose (raf), or 2% raffinose/1% galactose (gal). Plates were incubated for 3 d (glu), 5 d (raf), or 7 d (gal) at 30°C. The gal10Δ56 strain transformed with a 2-μm vector is shown as control.
Hrp1 and Rna14 are both components of CF I. Furthermore, Rna14 increases the affinity of Rna15 for RNA (Noble et al. 2004). Accordingly, we tested whether overexpression of Rna15, another CF I subunit, can rescue the Gal− phenotype of gal10Δ56. However, transformation with a 2-μm plasmid containing RNA15 did not allow growth in the presence of galactose (data not shown). Therefore, either Rna15 is not limiting for polyadenylation/termination or the 2-μm plasmid does not express high enough levels of Rna15 for suppression.
Rna15 and Hrp1 are the two subunits within the CF I complex that have RNA recognition motif (RRM) domains, and both have been shown to contact RNA (Gross and Moore 2001a). The finding that gal10Δ56 can be suppressed by either the overexpression of HRP1 or RNA14 or by certain mutant alleles of NPL3 suggested that CF I could be competing with Npl3 for RNA binding. A prediction of this model is that suppression of gal10Δ56 by NPL3 mutants (which compete less well with wild-type CF I) would be reduced by a second mutation in HRP1 or RNA14/15 that reduced CF I activity. To test this idea, several alleles of HRP1 (hrp1-1, hrp1-3, hrp1-5, hrp1-6) (Henry et al. 1996), rna15 (rna15-2, rna15-58) (Amrani et al. 1997; Hammell et al. 2002), or rna14 (rna14-49, rna14-64) (Hammell et al. 2002) were crossed into the gal10Δ56 and npl3-120 gal10Δ56 backgrounds (Fig. 2). The rna15-2 allele, which displays defocused polyadenylation activity as seen by utilization of cryptic poly(A) sites (Sparks and Dieckmann 1998), was unable to sporulate in combination with the npl3-120 allele (data not shown). Furthermore, multiple mutant alleles of hrp1 (hrp1-5 and hrp1-6), rna15 (rna15-58), and rna14 (rna14-49) reversed the Gal+ phenotype of the npl3-120, gal10Δ56 strain (data not shown; Fig. 2). Therefore, the phenotype of Npl3-120 is not due to bypass suppression of normal polyadenylation, since it requires functional Hrp1 and Rna15. When mutant Npl3 is combined with mutant Hrp1, Rna14, or Rna15, the balance is restored and the original Gal- phenotype is restored.
FIGURE 2.
HRP1 and RNA15 functions are essential for the npl3-mediated suppression of gal10Δ56. Serial 10-fold dilutions of strains with single or double mutations (hrp1-5, rna15-58, and npl3-120) in the gal10Δ56 background were spotted onto YPD or YPGal plates and grown for 2 d at 30°C. Alleles of hrp1-5 or rna15-58 with wild-type GAL10 are shown as controls for growth on YPGal.
Npl3 competes with Rna15 for RNA binding
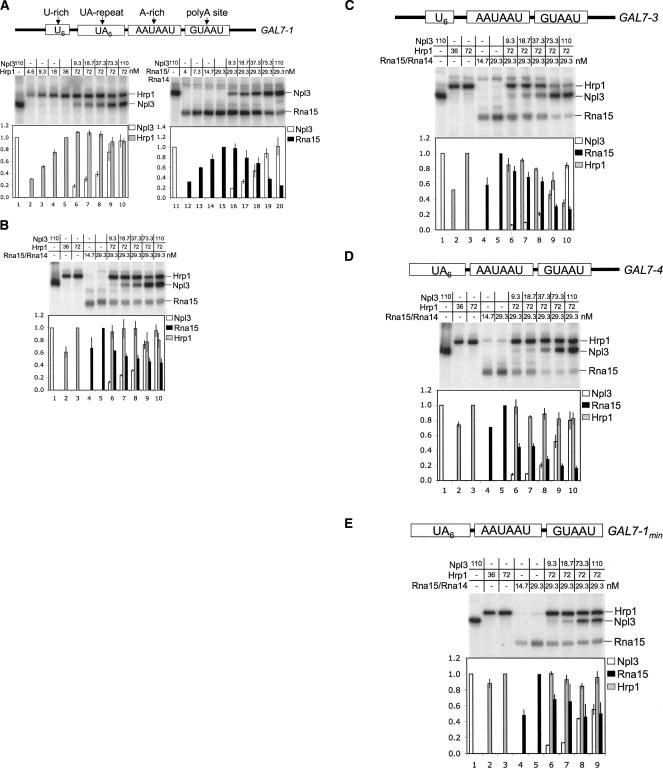
The genetic interactions between Npl3 and Hrp1 or Rna15 could be explained by direct competition for the same RNA binding sites. Gel shift experiments (Supplemental Fig. 1) were used to find protein concentrations in the linear range for binding to a GAL7-1 RNA that carries the upstream U-rich sequence, the UA-repeat and A-rich polyadenylation signals, and the poly(A) site itself (Fig. 3A; Gross and Moore 2001b). To directly visualize possible competition of Npl3 with Hrp1 or the Rna14/Rna15 complex, UV cross-linking assays were performed with recombinant proteins and radioactively labeled GAL7-1 RNA. Binding was quantitated as a fraction of the maximal binding (Fig. 3A, lanes 1, 11 for Npl3, lane 5 for Hrp1, or lane 15 for Rna15/Rna14). Increasing Npl3 led to a clear decrease in the cross-linking signal for Rna15 alone (see Supplemental Fig. 2A) or within the complex with Rna14 (Fig. 3A, right panel). In contrast, the interaction of Hrp1 with RNA was not noticeably altered upon the addition of Npl3, with only a slight reduction at the two highest concentrations of Npl3 used (73 and 110 nM). Therefore, Npl3 appears to compete directly with Rna15 but not Hrp1.
FIGURE 3.
Npl3 competes with Rna15 for binding to the GAL7 3′UTR transcript. Recombinant Npl3, Hrp1, or Rna15/Rna14 complex were incubated with radioactively labeled RNA, UV cross-linked, and resolved in denaturing 10% SDS-PAGE gels. (A) Schematic diagram of the long transcript GAL7-1, including a U-rich, UA-repeat, and A-rich sequences upstream of the poly(A) site is at top. Representative UV-cross-link experiments are shown where increasing Npl3 is added to reactions containing Hrp1 or Rna15/Rna14. The graph below each gel shows quantitation for the average of three experiments, with error bars showing the standard deviation. Binding levels were calculated as a fraction relative to a reaction containing the highest concentration of the individual RNA-bound protein. (B) A UV cross-linking experiment similar to A was done where increasing levels of Npl3 were added to a reaction containing both Hrp1 and Rna15/Rna14 bound to the GAL7-1 RNA. (C) The same combination of proteins as that shown in B is shown with the GAL7-3 RNA. This template contains a deletion of the UA-repeat. (D) Same as B except for a truncated template (GAL7-4) where sequence upstream of the UA-repeat is deleted. (E) Same as B but with a minimal GAL7 (GAL7-1 min) sequence, which has an additional deletion that spans the region downstream of the poly(A) site.
The UV cross-linking experiment was repeated with all four proteins; here, the amount of Hrp1 and Rna15/Rna14 remained constant while that of Npl3 was increased (Fig. 3B). Again, the cross-linking for Rna15 but not Hrp1 decreased upon the addition of increasing concentrations of Npl3. A similar experiment using Rna15 in the absence of Rna14 gave comparable results (see Supplemental Fig. 2B), although Rna15 cross-linking was even more sensitive to increasing Npl3, as expected due to Rna14 stabilization of the RNA–Rna15 interaction (Gross and Moore 2001b; Noble et al. 2004).
It has been shown previously that the strong interaction of Hrp1 with poly(A) signals is through the UA-repeat (Gross and Moore 2001b). In agreement, the Hrp1 signal is diminished upon the addition of Npl3 if we use an RNA (GAL7-3) that lacks the UA-sequence (Fig. 3C; Supplemental Fig. 2C). In this case, it is likely that the Hrp1 association is primarily through nonspecific interactions with the RNA or interactions with Rna14/Rna15.
Cross-linking to an RNA substrate (GAL7-4) in which the U-rich region upstream of the UA-repeat has been deleted (Fig. 3D) shows that Rna15 cross-linking is more sensitive to the addition of Npl3, while that of Hrp1 is unchanged (Fig. 3D). Similar results for GAL7-4 are observed when Npl3 is competed with Rna15 only (Supplemental Fig. 2D). These results are consistent with a preference of Rna15 for U-rich stretches (Takagaki and Manley 1997).
Finally, we tested an RNA substrate containing only the three core processing signals (UA-repeat, A-rich sequence, and cleavage/poly[A] site, but no further downstream sequences) (Fig. 3E). Interestingly, UV cross-linking of Npl3 to this minimal RNA (GAL7-1 min) was reduced by ∼50% in the presence of Hrp1 and Rna15/Rna14. As before, binding of Hrp1 was unaffected by the addition of Npl3, while Rna15 was competed, albeit less effectively. The presence of Rna14 strengthens the interaction of Hrp1 and Rna15 to the RNA, but not that of Npl3 (cf. Fig. 3E and Supplemental Fig. 2E). These results suggest that Npl3 may make additional contacts with RNA outside of the minimal poly(A) sequences such as the upstream poly(U) sequence or sequences 3′ to the cleavage/poly(A) site. However, in total, the results from these UV cross-linking experiments support the hypothesis that Npl3 directly competes with Rna15 for binding to the RNA, and can have an indirect effect on Hrp1.
Npl3 binds to specific sites in the GAL7 3′ untranslated regions
As a consensus RNA recognition sequence for Npl3 binding has not yet been identified, we sought to determine whether there are preferred RNA binding sites for Npl3 on the GAL7 3′ untranslated regions (UTRs). Recombinant Npl3 was UV cross-linked to the GAL7 3′ UTR and the RNA was then subjected to primer extension (PE) with reverse transcriptase. Protein–RNA cross-links block extension by reverse transcriptase, and their approximate positions can be determined by the length of the DNA products. Without RNA, only the 22-nucleotide (nt) primer band is seen. Several PE stops are seen on the RNA in the absence of protein, presumably due to spontaneous or UV-induced RNA damage. Addition of wild-type Npl3 leads to cross-links throughout the probe, but with clear site preferences (Fig. 4A,B). In addition to strong stops at or downstream from the poly(A) site, other PE stops (marked by bars) indicate that Npl3 also interacts with the UA-repeat and A-rich elements. There is also a set of strong stops ∼60–90 nt upstream of the UA-repeat (Fig. 4B, see the expanded region). Interestingly, many of these regions contain UA or UG motifs, and structural experiments are underway to determine if these sequences are part of an Npl3 recognition sequence (P. Deka, M. Bucheli, C. Moore, S. Buratowski, and G. Varani, in prep.).
FIGURE 4.
Wild-type and mutant Npl3 have distinct RNA cross-linking patterns. Npl3 and Npl3-120 were UV cross-linked to GAL7-1 RNA. Primer extension was then performed using a radiolabeled primer downstream of the poly(A) site; cross-links result in a block for reverse transcriptase and therefore give a band near the site of protein interaction. The RNA sequence, extrapolated from size markers, is shown next to the gels. (B) Same as A except a region (marked with an asterisk) further upstream of the poly(A) signal was expanded for analysis. Solid and dotted lines indicate PE stops for Npl3 and Npl3-120, respectively. (C) Coomassie staining showing the amount of each protein used in the cross-linking/primer extension analysis.
We previously identified a mutant NPL3 allele (npl3-120) that suppresses the Gal- phenotype of the gal10Δ56 strain (Bucheli and Buratowski 2005). Interestingly, the pattern of stops for the mutant protein shows a decrease in some of the strong PE stops observed for the wild-type protein, along with more frequent PE stops in other regions (Fig. 4A,B). These results suggest that Npl3 is not a completely nonspecific RNA binding protein and that the mutant Npl3-120 has altered specificity but not abolished RNA binding. The altered specificity perhaps leads to reduced competition with the polyadenylation machinery for RNA binding.
Npl3 inhibits mRNA 3′ end processing in vitro
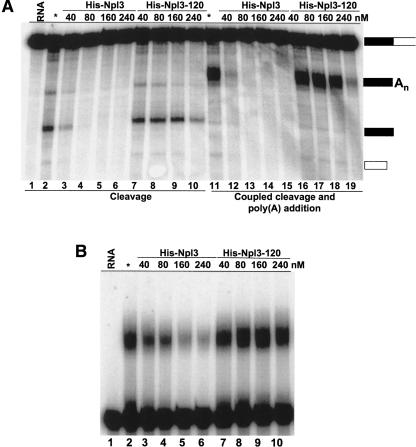
A prediction of the competition model is that high levels of Npl3 should negatively affect mRNA 3′end processing. We examined the effects of adding recombinant Npl3 to an in vitro mRNA processing system consisting of whole-cell extract (WCE) and radioactively labeled GAL7-1 precursor RNA (Fig. 5A). The addition of ATP allows both cleavage and poly(A) addition (Fig. 5A, lanes 11–19), while substituting dATP for ATP blocks poly(A) synthesis but permits cleavage (Fig. 5A, lanes 1–10). The addition of recombinant Npl3 at a concentration of 40 nM strongly inhibits cleavage and the coupled cleavage/polyadenylation reaction (Fig. 5A, cf. lanes 2,3 and lanes 11,12). Increasing the amount of Npl3 completely shuts down processing (Fig. 5A, lanes 4–6, 12–14). The mutant Npl3-120 is much less effective at inhibiting processing and does so at only the highest concentrations (Fig. 5A, lanes 7–10, 16–19). In a reaction for polyadenylation using a transcript that ends at the poly(A) site, Npl3 caused a reduction in the accumulation of product, although inhibition of polyadenylation is not as dramatic as that of cleavage (Fig. 5B, lanes 3–6). The mutant Npl3-120 did not affect this step (Fig. 5B, lanes 7–10). These results show that binding of Npl3 to the RNA substrate can inhibit processing of the transcript.
FIGURE 5.
Npl3 inhibits in vitro mRNA 3′ end processing. (A) In vitro mRNA 3′ end cleavage and polyadenylation assays. Yeast wild-type whole-cell extracts (30 μg; lanes 2–19) were incubated with radioactively labeled GAL7-1 RNA. Reactions were initiated with either dATP (for activation of cleavage but not polyadenylation, lanes 1–10) or ATP (allowing both cleavage and polyadenylation, lanes 11–19) for 20 min at 30°C. Whole-cell extract was supplemented with increasing concentrations of recombinant His6-Npl3 (lanes 3–6,7–10) or its mutant His6-Npl3–120 (lanes 7–10,16–19) as indicated before initiating the reaction. Products were resolved on a denaturing 5% polyacrylamide gel and visualized with a PhosphorImager. The position of the precursor and product RNAs are marked schematically on the right. Unreacted precursor RNA is shown in lane 1, and lanes 2 and 11 show RNA incubated with whole-cell extract but no additional Npl3. (B) Poly(A) addition assays. The assays were performed as in A except that precleaved GAL7-9 RNA (lane 1) was used instead as the RNA precursor.
Detection of Npl3 in the WCEs by immunoblotting shows that the amounts of rNpl3 added to the processing reactions are in large excess to that of endogenous Npl3 and Rna15 (data not shown). Therefore, we have forcefully shifted the competition by adding Npl3 in excess to 3′end processing factors, resulting in clear inhibition of cleavage and polyadenylation.
DISCUSSION
The interplay between different RNA binding proteins and their target RNA sequences is critical for proper mRNA 3′ end formation. Genetic interactions between NPL3 and RNA15/HRP1 have been previously observed (Henry et al. 1996). Here we present both genetic and biochemical data that Npl3 directly competes with CF I for RNA binding at the 3′ end of GAL7. Our results demonstrate that the direct effect of Npl3 competition is to prevent the binding of the CF I subunit Rna15 to the RNA.
Npl3, Hrp1, and Rna15 each have short and/or degenerate recognition sequences. Similarly, yeast poly(A) sequences appear to be quite degenerate and redundant. Recognition of poly(A) sites is likely to consist of multiple weak interactions rather than a single high-affinity one. Because of this low complexity, it is likely that cryptic sequences resembling poly(A) sites would appear in the genome (unless they were actively selected against). Therefore, it may be necessary to suppress promiscuous processing at weak or cryptic poly(A) signals. Chromatin immunoprecipitation experiments suggest that Npl3 binds throughout the mRNA (Lei and Silver 2002; Bucheli and Buratowski 2005), and we believe this binding may antagonize cryptic polyadenylation and termination by blocking binding of CF IA to all but the authentic, strongest poly(A) signals. Similarly, Hrp1/CF IB is found across RNAPII-transcribed genes (Komarnitsky et al. 2000), and clustered Hrp1 sites may focus CF IA binding to the poly(A) site (Gross and Moore 2001b). In agreement with this idea, Minvielle-Sebastia et al. (1998) have shown that reconstitution of the 3′ end processing reaction without Hrp1 causes variability in the exact site of cleavage.
Genetic results provide further support for the competition model. A mutated poly(A) site in GAL10 can be rescued by either increasing the levels of Hrp1 or Rna14 in the cell, or by a defect in Npl3. The mutant Npl3-120 has reduced in vitro inhibition of cleavage and polyadenylation compared with wild-type Npl3 and is less effective at competing with Rna15 for RNA binding (Supplemental Fig. 3). It is important to remember that npl3-120 cannot be a complete loss of function allele since NPL3 is an essential gene. Npl3-120 still binds RNA but shows a distinctly different pattern of RNA binding, as assayed by our cross-linking/PE analysis. The Npl3-120 mutant allele carries a single amino acid replacement (L225S) that maps within the second RRM domain. This particular point mutant was isolated multiple times in the gal10Δ56 suppression screen, perhaps suggesting that binding is subtly changed so as not to completely lose normal Npl3 function (Bucheli and Buratowski 2005). Further structural studies will address how this particular mutant affects RNA binding (P. Deka, M. Bucheli, C. Moore, S. Buratowski, and G. Varani, in prep.).
Selection of the proper 3′end processing site has important downstream implications, including defining the window for transcription termination and determining the extent of 3′ UTRs that may contribute to the stability of the mRNA. Because of this significance, proper poly(A) site selection is likely subject to intense regulation. In the female germline of flies, polyadenylation site “switching” has been shown to occur as a result of competition between RNA binding proteins for their target sequences on the template (Gawande et al. 2006). In this instance, the master sex-switch protein SXL competes with CstF-64, the ortholog of Rna15, for binding to the enhancer GU-rich element on the enhancer of rudimentary (e[r]) mRNA. Thus, a distal polyadenylation site can be selected, thereby altering the gene expression of e(r) (Gawande et al. 2006). In mammalian cells, binding of hnRNPs such as F, H, HU, and PTB to sequences in the vicinity of poly(A) sites can inhibit 3′ end processing, and in some cases, this has been attributed to blocking CstF-64 association (Veraldi et al. 2001; Castelo-Branco et al. 2004; Alkan et al. 2006; Wilusz and Beemon 2006; Zhu et al. 2007). While these are examples of regulated use of normal poly(A) sites, it is possible that these higher eukaryotic RNA binding proteins also function like Npl3 in suppressing weak or cryptic sites within the precursor RNA.
The competition of RNA binding proteins for 3′ processing sites might be further regulated by post-translational modifications of these proteins. The C-terminal domain of Npl3 contains an SR-like domain that has been shown to undergo phosphorylation and methylation, modifications that affect shuttling of this protein into and out of the nucleus, respectively (Shen et al. 1998; Gilbert et al. 2001). Phosphorylation of Npl3 has also been shown to affect RNA binding, but it is not yet clear whether this effect is due to reduced affinity of this protein for the RNA or increased specificity. Thus the outcome of the competition for RNA binding might be mainly determined by the strength of the poly(A) signals but may also be influenced by modification of Npl3.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used in this study were FY23 (Mat a ura3–52 leu2Δ1 trp1Δ63; F. Winston, Harvard Medical School) and CKY265 (ura3–52 his3Δ200 leu2Δ1 trp1Δ63 met15Δ0 lys2–128 gal10Δ56; this allele has also been referred to as gal10ΔpolyA). For pRS425/HRP1, the BamHI-HindIII fragment of pG48 (a genomic clone containing HRP1) containing the sequence from position −642 relative to the Start codon through position +196 relative to the Stop codon was cloned into the BamHI-HindIII–digested pRS425. For pRS425/RNA14, the HindIII-StuI fragment from genomic plasmid pG73 containing 3504 bp from position −1033 relative to the Start codon/StuI site through position +445 relative to the Stop codon/HindIII site was cloned into the HindIII-SmaI–digested pRS425 vector. The RNA15 2-μm plasmid was kindly provided by F. Lacroute (CNRS, Gif-sur-Yvette, France) (Minvielle-Sebastia et al. 1991).
Multicopy suppressor screen
To identify high-copy suppressors that would allow a strain containing the gal10Δ56 allele (described previously in Bucheli and Buratowski 2005; Kaplan et al. 2005) to grow in the presence of galactose, CKY265 was transformed with a high-copy yeast genomic DNA library (constructed in the 2-μm LEU2 plasmid YEp13) kindly provided by J. Kaplan (University of Utah, Salt Lake City, Utah). Approximately 9000 Leu+ transformants were replica-plated onto Synthetic Complete media lacking leucine (SC-leu) and then incubated at 30°C (Rose et al. 1990). Plates were supplemented with 2% raffinose and 1% galactose as carbon sources as well as Antimycin A (1 μg/mL). Plasmids from colonies that were Gal-resistant were isolated (pG48 and pG73) and retransformed to determine plasmid linkage (Robzyk and Kassir 1992). These library plasmids were then sequenced, revealing insertions containing the HRP1 and RNA14 genes, respectively. The HRP1 and RNA14 genes were subsequently cloned into pRS425 vectors (pRS425/HRP1 and pRS425/RNA14). Plasmids were retransformed into the original strain, and these genes were confirmed as multicopy suppressors of the galactose sensitivity conferred by gal10Δ56.
Purification of recombinant proteins
Escherichia coli strain BL21(DE3) was transformed with pSBEThis7-NPL3 or pSBEThis7-npl3-120, and recombinant Npl3 was purified as described previously (Bucheli and Buratowski 2005). Recombinant Hrp1 and Rna15 were expressed and purified as described previously (Gross and Moore 2001a). The Rna15/Rna14 complex was expressed and purified as described by Noble et al. (2004).
Gel mobility shift assays
For gel electrophoretic mobility shift assays (EMSA) shown in Supplemental Figure 1, GAL7 RNAs were synthesized by in vitro transcription from linearized plasmid pJCGAL7–1 (Gross and Moore 2001a). Heat denatured [32P] GAL7-1 RNA (∼10,000 cpm) was mixed with recombinant Npl3, Hrp1, Rna15, or Rna15/Rna14 complex in binding buffer (1 mM magnesium acetate, 50 mM potassium acetate, 10% glycerol, 20 mM HEPES at pH 7.2, 1 mM DTT, 33 μg/mL tRNA). All reactions were incubated for 20 min at room temperature. Glycerol was added to 25%, and reactions were loaded on 5% polyacrylamide gels (30:0.8 acrylamide:bis) in 0.5× TBE buffer, dried, and exposed to a PhosphorImager screen for quantitation. All experiments were done in triplicate.
UV cross-linking assays
UV cross-linking experiments were performed as described previously (Kessler et al. 1997). GAL7-1(wild-type) or GAL7-3 (deletion of UA-repeat), GAL7-4 (poly(A) upstream region deleted) RNAs were synthesized by in vitro transcription from linearized plasmids pJCGAL7-1, pJCGAL7-3, or pJCGAL7-4 (Gross and Moore 2001a). For GAL7 min, a region spanning the T3 promoter to immediately downstream from the GAL7 cleavage/poly(A) site (primers: 5′-AATTAACCCTCACTAAAGG-3′; 5′-CGCTCGAGATAACTAGAATAGCATTAAGAATCAGA-3′) was amplified using the pJCGAL7–4 plasmid as a template, and the PCR product generated was used for RNA synthesis by in vitro transcription. Denatured [32P] GAL7-1 GAL7-3, GAL7-4, or GAL7-1 min RNA (∼100,000 cpm) was mixed with recombinant Npl3, Hrp1, Rna15, or Rna15/Rna14 complex in binding buffer (1 mM magnesium acetate, 50 mM potassium acetate, 10% glycerol, 20 mM HEPES at pH 7.2, 1 mM DTT, 33 μg/mL tRNA). All reactions were incubated for 20 min at room temperature, and then UV-irradiated in an Ultra-Lum/UVC-515 ultraviolet multilinker set at 1800 μJ (×100). The samples were digested with RNase A (Sigma, 0.9 mg/mL) for 30 min at 37°C. Loading buffer was added, and reactions were resolved by 10% SDS–polyacrylamide gel (30:0.8 acrylamide:bis), dried, and exposed to the PhosphorImager screen.
RNA processing assays with WCEs
Yeast WCEs were prepared as described previously (Zhao et al. 1999b). 32P-Labeled full-length GAL7 RNA and precleaved GAL7 RNA were synthesized by in vitro transcription from linearized plasmids pJCGAL7-1 and pJCGAL7-9. In vitro 3′ end processing assays were carried out as described elsewhere (Gross and Moore 2001a). Briefly, reactions were performed in a volume of 15 μL containing 1 mM ATP, 10 mM creatine phosphate, 1 mM magnesium acetate, 75 mM potassium acetate, 2% polyethylene glycol 8000 (PEG 8000), 1 mM dithiothreitol, 0.1 mg/mL of bovine serum albumin (New England Biolabs), 0.4U of RNasin (Promega), 30 μg of yeast WCE, and a radioactively labeled RNA precursor (∼300,000 cpm). Reactions were assembled on ice and incubated for 20 min at 30°C. For supplementation with recombinant proteins, indicated amounts of His6-Npl3 or His6-Npl3-120 were added to extracts and incubated for 15 min at 25°C before initiating the reaction by addition of 32P-labeled GAL7 RNA. Reactions were stopped by the addition of proteinase K (33 μg/mL) and incubated for 15 min at 30°C. The reaction mixture was diluted with 30 μL TE buffer (10 mM Tris-Cl, 1 mM EDTA at pH 7.5) and extracted once with phenol-chloroform-isoamyl alcohol. One-tenth of the reaction was resolved on a 5% acrylamide–8.3 M urea gel and then visualized by a PhosphorImager.
Detection of protein–RNA cross-links by PE
Unlabeled RNA was synthesized by in vitro transcription from linearized plasmids pJCGAL7-1 (Gross and Moore 2001a). Unlabeled GAL7-1 RNA (28–56 ng/μL) was mixed with recombinant His6-Npl3 (130 ng) or His6-Npl3–120 (210 ng) in binding buffer (1 mM magnesium acetate, 50 mM potassium acetate, 10% glycerol, 20 mM HEPES at pH 7.2, 1 mM DTT) as described above, final volume 5 μL (Kessler et al. 1997). Reactions were incubated for 20 min at room temperature, and then UV irradiated in a UV Cross-linker/FB-UV-1000 (Fisher Biotech) set at 1800 μJ. For labeled primers, 10 pmol of DNA oligomer 5′-GGAAAGGACCACATTACATAAC-3′ was labeled at the 5′end using γ-[32P]-ATP and T4 DNA kinase as described previously (Sambrook and Russell 2001). PE was performed using M-MLV reverse transcriptase (Invitrogen) as described previously (Sambrook and Russell 2001). The samples were then treated with Pronase (2 mg/mL; Roche Applied Science), plus 0.5% SDS for 30 min at 50°C and phenol:chloroform extracted. Loading buffer was added, and reactions were electrophoresed on 8% polyacrylamide denaturing gels (19:1 acrylamide:bis, 7 M urea, 1× TBE), dried, and exposed to the PhosphorImager screen. Run in parallel were size markers. A 10-base pair (bp) DNA ladder was labeled with 5′end using γ-[32P]-ATP and T4 DNA kinase and denatured as described (Invitrogen).
SUPPLEMENTAL DATA
All Supplemental Figures can be found at http://tfiib.med.harvard.edu/pubs/BucheliSupFigs.pdf.
ACKNOWLEDGMENTS
We thank L. Vasilieva for technical advice and helpful discussions and A. Bohm for help with expression and purification or the Rna14/15 complex. M.E.B. was supported by fellowships from the American Cancer Society (PF-03-224-01-GMC) and National Institutes of Health (1K01CA115515-01A1). C.D.K. was supported by a fellowship from the Helen Hay Whitney Foundation. This work was supported by grant GM56663 from the National Institutes of Health to S.B. and GM68887 to C.L.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.607207.
REFERENCES
- Alkan, S.A., Martincic, K., Milcarek, C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Wyers, F., Dufour, M.E., Aggerbeck, L.P., Lacroute, F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli, M.E., Buratowski, S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski, S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., Proudfoot, N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Hyman, L.E. A specific RNA–protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 1998;26:4965–4974. doi: 10.1093/nar/26.21.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Moore, C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan, D.F., Manley, J.L. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Gawande, B., Robida, M.D., Rahn, A., Singh, R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006;25:1263–1272. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W., Siebel, C.W., Guthrie, C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, J.H., Cantor, C.R., Mohr, S.C., Smith, T.F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, J.H., McAllister, G.D., Smith, T.F. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger, I.H., Proudfoot, N.J. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae . EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S., Moore, C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. 2001a;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S., Moore, C.L. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 2001b;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Sherman, F. 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- Guo, Z., Russo, P., Yun, D.F., Butler, J.S., Sherman, F. Redundant 3′ end-forming signals for the yeast CYC1 mRNA. Proc. Natl. Acad. Sci. 1995;92:4211–4214. doi: 10.1073/pnas.92.10.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell, C.M., Gross, S., Zenklusen, D., Heath, C.V., Stutz, F., Moore, C., Cole, C.N. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, M., Borland, C.Z., Bossie, M., Silver, P.A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, Y., Manley, J.L. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Kaplan, C.D., Holland, M.J., Winston, F. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- Kessler, M.M., Zhao, J., Moore, C.L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J. Biol. Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- Kessler, M.M., Henry, M.F., Shen, E., Zhao, J., Gross, S., Silver, P.A., Moore, C.L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky, P., Cho, E.J., Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P., Silver, P.A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., Bentley, D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Winsor, B., Bonneaud, N., Lacroute, F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Preker, P.J., Keller, W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Beyer, K., Krecic, A.M., Hector, R.E., Swanson, M.S., Keller, W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, C.G., Walker, P.A., Calder, L.J., Taylor, I.A. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004;32:3364–3375. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., Dye, M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Robzyk, K., Kassir, Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., Hieter, P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Rosonina, E., Kaneko, S., Manley, J.L. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Sadowski, M., Dichtl, B., Hubner, W., Keller, W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shen, E.C., Henry, M.F., Weiss, V.H., Valentini, S.R., Silver, P.A., Lee, M.S. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes & Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks, K.A., Dieckmann, C.L. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki, Y., Manley, J.L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini, S.R., Weiss, V.H., Silver, P.A. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA. 1999;5:272–280. doi: 10.1017/s1355838299981633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi, K.L., Arhin, G.K., Martincic, K., Chung-Ganster, L.H., Wilusz, J., Milcarek, C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, J.E., Beemon, K.L. The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J. Virol. 2006;80:9634–9640. doi: 10.1128/JVI.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., Moore, C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999a;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Kessler, M., Helmling, S., O'Connor, J.P., Moore, C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 1999b;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Zhou, H.L., Hasman, R.A., Lou, H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]