Abstract
Induction of gene silencing using intracellularly expressed silencing triggers has been explored for large-scale loss-of-function screening, creation of knockdown cell lines or knockdown animals, and disease intervention. In all of these applications, the use of highly potent silencing constructs can maximize the possibility of obtaining target knockdown and thereby is intrinsically important for the chance of success. Several attempts have been made to improve the potency of a silencing construct. Results published in high profile journals such as Nature Biotechnology and Nature Genetics suggest that shRNAs with a 29-nucleotide (nt) stem is much more potent than shRNAs with a 19-nt stem, and miR30-based silencing constructs are much more potent than shRNA-based constructs. In this study, we systematically investigated several parameters, including the use of shRNA- or miR30-based scaffolds, the length of shRNA, and the selection of shRNA sequences for their impact on the knockdown efficiency of a silencing construct. Our studies revealed that the optimal configurations for a potent silencing trigger could be an shRNA with a 19-nt stem and a 9-nt loop. By comparing properties that favor the functional shRNAs and siRNAs using a set of 190 shRNAs against 19 targets and 360 siRNAs against four targets, we found that the functional shRNAs and siRNAs displayed similar but not identical nucleotide preferences. Based on the characteristic nucleotide preferences in the functional versus the nonfunctional shRNAs, we developed a computer program that outperforms an advanced siRNA selection algorithm for the enrichment of highly functional shRNAs.
Keywords: RNAi, miR30, microRNA, shRNA, siRNA
INTRODUCTION
RNA interference (RNAi) is a process of gene silencing using double-stranded RNA (dsRNA) (Fire et al. 1998; Montgomery et al. 1998; Elbashir et al. 2001a,b). Because of its ease of use and the perceived specificity, RNAi is becoming the method of choice to study gene function in mammalian cells and animals (Dorsett and Tuschl 2004; Hannon and Rossi 2004). In addition to using chemically synthesized short interfering RNA (siRNA) for gene silencing, induction of gene silencing using intracellularly expressed silencing triggers is also widely used for many applications. For example, large libraries of DNA vector-based RNAi reagents are being used for various loss-of-function screens; cell lines or transgenic animals that express silencing triggers against the targets of interests are being created, and therapeutic approaches based on vector-expressed silencing triggers are being investigated (Brummelkamp et al. 2002; Miyagishi and Taira 2002; Paddison et al. 2002; Sui et al. 2002; Xia et al. 2002; Brummelkamp et al. 2003; Carmell et al. 2003; Rubinson et al. 2003; Berns et al. 2004; Paddison et al. 2004; Xia et al. 2004; Li et al. 2005a,b; Stegmeier et al. 2005; Grimm et al. 2006). In all of these applications, the use of highly potent silencing constructs maximizes the possibility of obtaining target knockdown and thereby is intrinsically important for the chance of success. Several attempts have been made to improve the potency of a silencing construct. Results published in high profile journals such as Nature Biotechnology and Nature Genetics suggest that shRNAs with a 29-nucleotide (nt) stem is much more potent than shRNAs with a 19-nt stem, and the miR30-based silencing construct is much more potent than the shRNA-based construct (Silva et al. 2005; Siolas et al. 2005; Stegmeier et al. 2005). In the publications that reported the substantial improvement of knockdown efficiency using the miR30 scaffold or using shRNAs with 29-nt stems, the shRNAs used for comparison contain a 4-nt loop. However, shRNAs with longer loops are more commonly used and, in our hands, are more efficient for target knockdown. Therefore, the broad applicability of the reported findings remains to be determined. In addition to the attempts of improving knockdown efficiency using the miR30 scaffold or using longer shRNAs, the potency of a hairpin-based silencing trigger can also be improved by selecting the best targeting sequence. Although it is expected that the hairpin-based silencing triggers and siRNAs will share some properties because siRNAs are the final product of hairpin-based silencing triggers, the hairpin-based silencing triggers may also possess unique features to ensure their efficient processing in cells. Thus far, no systematic studies have been carried out to investigate the rules that govern the selection of potent shRNAs. In this study, we systematically investigated the importance of the scaffold (shRNA or miR30), the length of shRNA, and the selection of target sequences, for their impact on the potency of target knockdown. These studies revealed an optimal configuration for the hairpin-based silencing trigger and identified properties that define a potent shRNA sequence.
RESULTS
U6-expressed shRNAs are more efficient than miR30-based embedded siRNAs for target knockdown
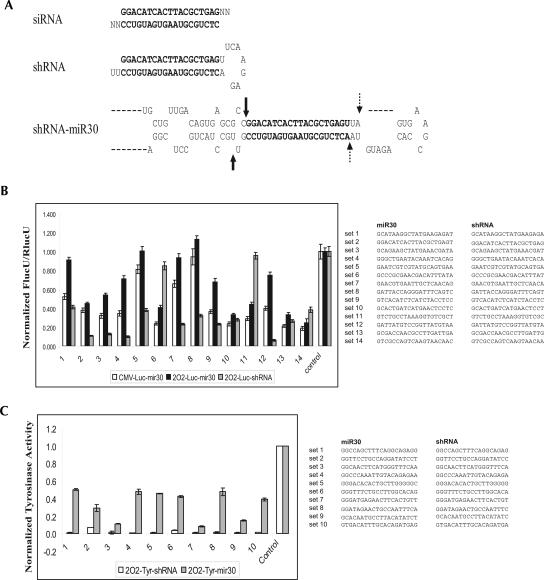
DNA-based intracellular expression of siRNAs is an alternative to synthetic siRNAs for induction of gene-specific silencing in mammalian cells. Several different types of constructs have been utilized for DNA-based siRNA expression, including vectors that use RNA polymerase III to express short hairpin RNAs and vectors that express long transcripts mimicking human microRNAs (Dickins et al. 2005; Silva et al. 2005; Stegmeier et al. 2005; Zeng et al. 2005a). It has recently been reported that constructs using the miR30 scaffold substantially outperform those using the shRNA scaffold for target knockdown when shRNAs with a 29-nt stem and a 4-nt loop were used for comparison (Silva et al. 2005; Stegmeier et al. 2005). Because shRNAs with a 19-nt stem and a longer loop (UUCAAGAGA) are more commonly used (Brummelkamp et al. 2002), we decided to systematically compare the overall knockdown efficiency of the miR30-scaffold versus shRNAs with a 19-nt stem and 9-nt loop. Toward this end, we created a panel of pairwise shRNA- and miR30-based knockdown constructs against luciferase according to the standard design for each type of construct (Fig. 1A). Using these designs, the shRNA-based construct is expected to produce a 19-nt siRNA after processing, and the miR30-based construct is expected to express a >250-nt transcript, which will be processed into a mixture of several 19-nt and 20-nt siRNAs. According to the literature-described processing sites for the miR30-based transcripts, the major product of the miR30-based construct is a 20-nt siRNA with nucleotides 1–19 identical to the siRNA processed from the corresponding shRNA-based construct (Fig. 1B, right panel; Cullen 2004; Silva et al. 2005; Zeng et al. 2005b). This design theoretically will enable us to evaluate the impact of the scaffold on overall construct efficacy. In the first set of constructs, a previously described U6 promoter variant, 2O2, was used to drive the expression of both the shRNA- and miR30-based silencing triggers (Lin et al. 2004), and the abilities of these constructs to knockdown luciferase were determined based on their abilities to inhibit the luciferase activity in a cotransfection assay. It was found that, in the majority of the pairs tested (11 out of 14), the shRNA-based constructs induced a better knockdown of luciferase compared with their miR30-based counterparts. In contrast, only in three out of the 14 pairs, did the miR30-based constructs induce a better knockdown of luciferase compared with the corresponding shRNA-based constructs (Fig. 1B, 2O2-Luc-shRNA versus 2O2-Luc-miR30, sets 6,11,14).
FIGURE 1.
shRNA-based constructs outperform miR30-based constructs in target knockdown. (A) The schematic depiction of a siRNA and the corresponding shRNA or miR30-based silencing triggers. The bold characters represent the double-stranded part of a siRNA sequence that each set of silencing triggers is designed to produce in cells. The solid and broken arrows represent the main cleavage sites in miR30-based silencing triggers by Drosha and Dicer, respectively. (B) H1299 cells were transfected with 0.05 μg of each shRNA- or miR30-based construct, 0.15 μg of the firefly luciferase reporter, pGL3-control, and 0.015 μg of the renilla luciferase reporter, pRL-TK. The luciferase activities in transfected cells were determined, and the results were normalized to the luciferase activities in cells that were transfected with pGL3-control, pRL-TK, and a control vector (control). The Y-axis represents the normalized luciferase activities of each sample. The right panel listed the siRNAs against luciferase that were expected to be produced from the miR30- or shRNA-based constructs. (C) H1299 cells were transfected with 1 μg of shRNA- or miR30-based constructs and 1 μg of an expression plasmid encoding mouse tyrosinase. The tyrosinase activities in each transfected cells were determined, and the results were normalized to the tyrosinase activities in cells that were transfected with the tyrosinase expression plasmid and a control plasmid (Control). The Y-axis represents the normalized tyrosinase activities from each sample. The right panel lists the siRNAs against mouse tyrosinase that were expected to be produced from the miR30- or shRNA-based constructs.
Unlike the shRNA-based constructs, which require the use of RNA polymerase III-dependent promoters to obtain a robust target knockdown, an efficient target knockdown with the miR30-based constructs can be obtained using both the RNA polymerase III- or RNA polymerase II-dependent promoters (Dickins et al. 2005; Silva et al. 2005; Stegmeier et al. 2005; Zeng et al. 2005a). To examine whether using an RNA polymerase II-dependent strong promoter such as the CMV promoter will lead to a better performance of the miR30-based constructs, we engineered another set of constructs that utilize the CMV promoter to drive the expression of the same set of miR30-based silencing triggers against luciferase. Although the CMV-driven miR30-based constructs produced slightly better knockdowns of the luciferase activity compared to their 2O2-driven counterparts (Fig. 1B, CMV-Luc-mir30 versus 2O2-Luc-mir30), the shRNA-based constructs still significantly outperformed the CMV-driven miR30-based constructs in 10 out of 14 pairs tested (Fig. 1B, 2O2-Luc-shRNA versus CMV-Luc-miR30). More importantly, in all of the 14 sets of constructs that were tested, the four constructs that produced the best knockdown of luciferase were all based on shRNAs (Fig. 1B, 2O2-Luc-shRNA in sets 2,3,4,12).
To further investigate whether the shRNA-based constructs are also better than the miR30-based constructs in knocking down targets other than luciferase, we engineered another panel of shRNA- and miR30-based constructs that were designed against mouse tyrosinase. Interestingly, in this panel of constructs, all of the shRNA-based constructs were found to substantially outperform the miR30-based constructs in knocking down mouse tyrosinase (Fig. 1C). Collectively, these results suggest that, although it is possible to obtain a relatively potent knockdown construct using the miR30-based design, the simple shRNA-based construct will likely produce a better target knockdown. Therefore, we focused on the shRNA-based construct for further study.
shRNAs with a 19-nt stem are superior to shRNAs with a 29-nt stem for target knockdown in the context of a 9-nt loop
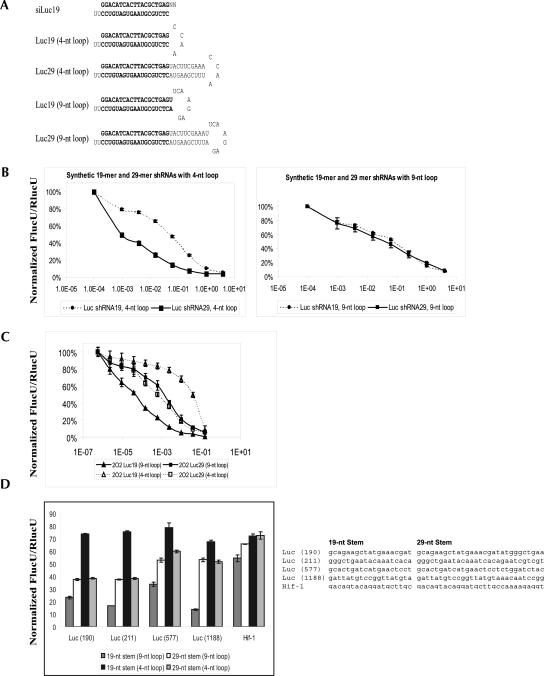
For shRNA-based knockdown constructs, the length of shRNA is one of the factors that could affect the knockdown efficiency. It has been shown that, in the context of a 4-nt loop, synthetic shRNAs with a 29-nt stem are much more potent than shRNAs with a 19-nt stem (Siolas et al. 2005). To address whether the observed improvement on knockdown efficiency using longer shRNAs in the context of a 4-nt loop is applicable for shRNAs with a 9-nt loop, we obtained synthetic shRNAs that contain a 29-nt or 19-nt stem and a 9-nt or 4-nt loop. In this set of shRNAs, all of them are designed to produce the same siRNA against luciferase after intracellular processing (Fig. 2A). Consistent with what was described in the literature, the shRNA with a 29-nt stem was much more potent than the shRNA with a 19-nt stem in the context of a 4-nt loop (Fig. 2B, left panel). In contrast, the shRNAs with a 19-nt or 29-nt stem in the context of a 9-nt loop were found to have similar potencies in knocking down luciferase (Fig. 2B, right panel). These results suggest that in the context of a 9-nt loop, increasing the length of the stem of an shRNA does not appear to improve its potency in target knockdown.
FIGURE 2.
shRNAs with a 19-nt stem outperform shRNAs with a 29-nt stem in the context of a 9-nt loop. (A) The schematic depiction of a siRNA and the corresponding shRNAs with a 19-nt or 29-nt stem and 4-nt or 9-nt loop. The bold characters represent the double-stranded part of a siRNA against luciferase. (B) Different amounts of synthetic shRNAs with a 19-nt or 29-nt stem and a 4-nt or 9-nt loop were cotransfected with the luciferase reporters pGL3-control and pRL-TK into H1299 cells. (C) Different amounts of vectors that express the luciferase-targeting shRNAs with a 19-nt or 29-nt stem and a 4-nt or 9-nt loop were cotransfected with the luciferase reporter pGL3-control and pRL-TK into H1299 cells. (D) Vectors that express the luciferease or HIF-1α-targeting shRNAs with a 19-nt or 29-nt stem and a 4-nt (CCAA) or 9-nt (UUCAAGAGA) loop were cotransfected with the pGL3-control (Fluc) or HIF-1 reporter (pHRE) together with the pRL-TK (Rluc) reporter into H1299 cells. In B, C, and D the luciferase activities in transfected cells were determined using the Dual-Glo assay and normalized to the luciferase activities in cells that were transfected with the luciferase reporters and a control plasmid (control). The Y-axis represents the normalized luciferase activities (Fluc/Rluc) in each transfected cells. The X-axis in B and C represents the doses (Log(μg)) of the shRNA expression constructs.
To further determine whether the results obtained using synthetic shRNAs are applicable to shRNAs expressed in cells, we created and tested the target knockdown triggered by four DNA constructs that express shRNAs of different stems and loops, meanwhile producing the same siRNA against luciferase (Fig. 2A). When a 4-nt loop was used, increasing the length of the stem from 19-nt to 29-nt indeed substantially improved the knockdown efficiency of the shRNA construct (Fig. 2C, 2O2-Luc19 [4-nt loop] versus 2O2-Luc29 [4-nt loop]). However, in the context of a 9-nt loop, increasing the length of the stem from 19 nt to 29 nt resulted in a less potent construct (Fig. 2C, 2O2-Luc19 [9-nt loop] versus 2O2-Luc29 [9-nt loop]). Overall, the DNA construct that was designed to produce the shRNA with a 19-nt stem and a 9-nt loop induced the best knockdown of luciferase (Fig. 2C, 2O2-Luc19 [9-nt loop] versus others). To further determine whether similar results can be obtained using shRNAs targeting different sites, we created additional sets of shRNAs with different combinations of the stems and loops (19-stem/4-loop, 19-stem/9-loop, 29-stem/4-loop, and 29-stem/9-loop). Among these, four sets of shRNAs were designed to target four different sites of the luciferase gene (Luc 190, 211, 577, 1188) and one set of shRNAs was designed to target HIF-1α. In each set of shRNAs, the shRNA with a 19-nt stem and a 9-nt loop (UUCAAGAGA) was found to exhibit the most robust target knockdown, confirming that shRNA with a 19-nt stem and a 9-nt loop is the optimal configuration for a knockdown construct (Fig. 2D).
Functional shRNAs and siRNAs exhibit similar but not identical sequence preferences
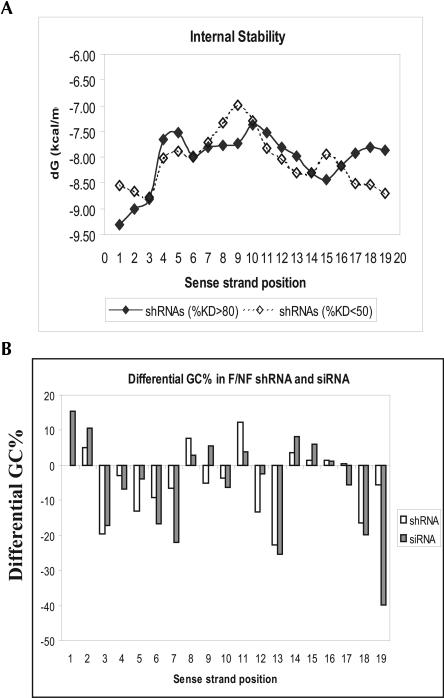
Upon deciding on the optimal configuration of an shRNA, the selection of shRNA sequences becomes the major factor that will affect the potency of a knockdown construct. Because siRNAs are the final product of shRNAs, potent shRNAs are expected to follow a similar set of rules as potent siRNAs. However, because of the additional processing steps required for shRNAs to function, potent shRNA may also possess unique features to ensure their efficient processing in cells. Moreover, because Dicer-dependent shRNA processing will likely lead to more efficient RISC loading, it is possible that some siRNA features that are critical for RISC entry might be alleviated in shRNAs. Over the last several years, we have created and tested 190 shRNAs against 19 targets. An initial analysis of these shRNAs indicated that the functional shRNAs exhibited lower thermodynamic stability at the 3′ end of the sense strand and higher thermodynamic stability at the 5′ end of the sense, which is consistent with the reported thermodynamic property of the functional siRNAs (Fig. 3A; Khvorova et al. 2003).
FIGURE 3.
The functional shRNAs exhibit similar but not identical sequence preference compared with the functional siRNAs. (A) The internal thermodynamic stability of the functional (>80% knockdown) vs. the nonfunctional (<50% knockdown) shRNAs at each position of the shRNA duplex (sense strand) was calculated as described in the literature (Khvorova et al. 2003). (B) The differential %GC in the functional vs. nonfunctional shRNA or siRNAs at each position of the siRNA duplex was calculated using the formula: %GC of the functional shRNAs (or siRNAs) − %GC of the nonfunctional shRNAs (or siRNAs). One hundred and fifty shRNAs against 14 targets were used for the analysis. shRNAs that induced >75% silencing were considered functional (n = 75), and shRNAs that induced <50% silencing were considered nonfunctional (n = 40). shRNAs that mediated 50%–75% knockdown were excluded from analysis. A proprietary data set of 360 siRNAs for four genes (Luc, Cyclo, GADPH, and DBI) from Dharmacon was analyzed in parallel. siRNAs that induced >80% silencing were considered functional (n = 136), and siRNAs that induced <50% knockdown were considered nonfunctional (n = 118). siRNAs that mediated 50%–80% target knockdown were excluded from analysis. The Y-axis represents the differential %GC of the functional shRNAs (or siRNAs) vs. the nonfunctional shRNAs (or siRNAs). The higher the value, the stronger preference of GC is in the functional shRNAs or siRNAs. Negative values indicate a preference for AU at a particular position. The numbers 1–19 on the X-axis correspond to the nucleotide positions in the sense strand of the shRNA or siRNA duplex.
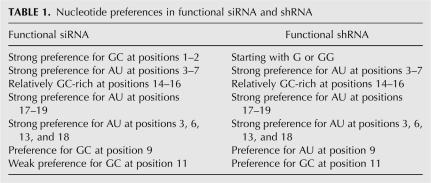
To evaluate the differences and similarities between functional shRNAs and siRNAs, we calculated ISP, GC content profile, and positional nucleotide preferences to identify parameters that separate functional siRNAs and shRNAs from their nonfunctional counterparts. One way to visualize the general duplex properties that favor the functional shRNAs or siRNAs is to examine the differential GC distribution in the functional versus nonfunctional shRNAs or siRNAs at each position of the duplex. A training data set of 150 shRNAs against 14 targets was first analyzed. From these analyses, it is clear that siRNA and shRNA profiles are very similar with both the functional shRNAs and siRNAs having lower GC content at the 3′ end of the sense strand compared to the nonfunctional shRNAs or siRNAs. These results suggest that thermodynamic bias for RISC entry is an important event in the context of Dicer-dependent RISC loading. The relative importance of the 5′ end stability (sense strand) in the context of shRNAs cannot be analyzed because all shRNAs were designed to start with G for optimal expression using the U6 promoter. In addition to the similarities at both ends of the duplex, the functional shRNAs and siRNAs also displayed a similar GC profile along the duplex (Fig. 3B). For example, both the functional shRNAs and siRNAs were found to contain a highly AU-rich region from positions 3–7 and a relatively GC-rich region from positions 14–16. Furthermore, the functional siRNAs and shRNAs both displayed a very strong preference for AU at positions 3, 6, 13, and 18, which highlights the importance of these positions in determining the siRNA or shRNA functionality (Fig. 3B). The similar GC profiles and the matched GC preference at the positions 3, 6, 13, and 18 in the functional siRNAs and shRNAs suggest that, using our shRNA configuration, the processing of shRNAs may indeed happen predominantly at the predicted sites, and the common features found in the functional shRNAs and siRNAs likely represent essential properties that define a potent siRNA duplex. Although the functional shRNAs and siRNAs displayed remarkable similarities in GC preference along the duplex, there were also noticeable differences between the two. For example, the functional shRNAs had a strong preference for GC at position 11 and for AU at positions 9 and 12, while these preferences were much less prominent or reversed in the functional siRNAs (Fig. 3B; Table 1).
TABLE 1.
Nucleotide preferences in functional siRNA and shRNA
A standard regression model based on the shRNA data set predicts the shRNA functionality more accurately than the regression models built on the siRNA data set
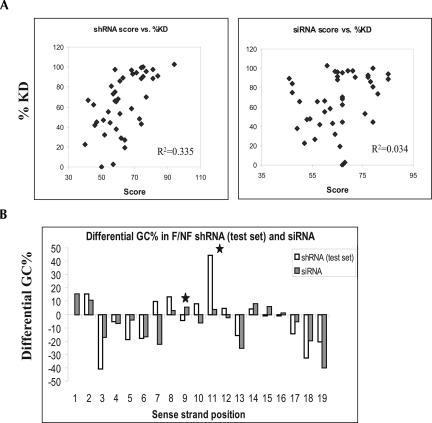
The characteristic GC preference at various positions of the functional shRNA duplexes suggests that descriptive algorithms might be derived to predict the functionality of a given shRNA similar to the algorithms developed for siRNA functionality prediction. As an attempt to develop an algorithm to facilitate the selection of potent shRNA sequences, we performed regression analyses on various characteristics of the 150 shRNAs in the training data set for their association with the shRNA functionality. Using these characteristics as parameters, a correlation coefficient of 0.55 was obtained between the cumulative regression value and experimental functionality for the shRNA training set (data not shown). In this context, a higher regression score implies that the shRNA will be more potent in knocking down the target. To further test how the scoring scheme will perform on an independent data set, we analyzed 40 additional shRNAs that were designed against five more targets. Because these shRNAs were not included in the previous GC distribution study that was used to derive the shRNA prediction algorithm, the 40 shRNAs represented an independent data set for evaluating the performance of the shRNA prediction program. A positive correlation was found between the shRNA scores and the degrees of target knockdown using this independent testing data set (Fig. 4A, left panel, R2 = 0.335). Among shRNAs with a score of 70 or more (13 out of 40 shRNAs), 78% (10/13) of the shRNAs were highly potent (>85% knockdown of the target), and 16% (2/13) of the shRNAs were nonfunctional (<50% knockdown), whereas, in the entire population, 38% (15/40) of the shRNAs were nonfunctional (<50% knockdown) and 38% (15/40) of the shRNAs were highly potent (>85% knockdown). These results suggest that potent shRNAs can be enriched using our shRNA-scoring algorithm.
FIGURE 4.
A computer program enriches the functional shRNAs. (A) The shRNA prediction program (left panel) or an advanced siRNA prediction program (right panel) was used to generate prediction scores on a set of 40 shRNAs against five targets. The prediction scores generated from each program were plotted against the degrees of target knockdown produced by these shRNAs. (B) The differential %GC of the functional vs. nonfunctional shRNA from the 40 shRNAs in the testing data set was analyzed using the same method as in Fig. 3B. The differential %GC in functional vs. nonfunctional siRNAs was generated using the same set of 360 siRNAs as in Fig. 3B.
Because siRNAs and shRNAs displayed very similar GC profiles along the duplex, we next tested whether the siRNA selection algorithm has the same predictive power as the shRNA algorithm. The eight-component rules that were described in the literature for siRNA selection were first used to calculate prediction scores for a subset of 80 shRNAs from the training data set of 150 shRNAs (Reynolds et al. 2004). To our surprise, scores produced using the eight-component rules failed to exhibit any correlation with the degrees of target knockdown induced by these shRNAs (R2 = 0.0001), indicating that the eight rules that have been successfully used to enrich the functional siRNAs may not be sufficient to predict the shRNA functionality. We next examined the performance of an advanced siRNA selection algorithm for the prediction of the shRNA functionality. This siRNA selection program was derived from thousands of siRNA data points collected at Dharmacon and is currently being used as the basis for the selection of the SmartPool siRNAs. Interestingly, the scores generated using this advanced siRNA prediction program appeared to predict the functionality of the 40 shRNAs in our testing data set with less accuracy compared with the shRNA algorithm (Fig. 4A, R2 = 0.034 using the siRNA algorithm versus R2 = 0.335 using the shRNA algorithm). Considering that this advanced siRNA selection algorithm has been used to predict the functionality of thousands of siRNAs accurately (data not shown), the reduced prediction power of this algorithm for shRNAs suggests that some of the subtle differences in nucleotide preference between the functional siRNAs and shRNAs such as the differences in nucleotide preference at positions 9, 11, and 12, might be important components for the prediction of shRNA activity.
In order to identify which of these observed differences might contribute to the better prediction power of the shRNA algorithm over the siRNA algorithm, we examined the differential GC distribution in the functional versus the nonfunctional shRNAs from the testing data set. It is expected that the observed GC preference at a particular position in the training data set should be preserved in the testing data set if the GC preference at this position truly contributes to the functionality of shRNAs. Consistent with what was observed in the training data set, the functional shRNAs from the test data set shared a number of common features with the functional siRNAs, including the preference for GC at 5′ of the sense strand, the preference for AU at 3′ of the sense strand, the presence of an AU-rich stretch from positions 3 to 6, and the preference for AU at positions 3, 6, 13, and 18 (Fig. 4B). Interestingly, the functional shRNAs in the testing data set displayed a very strong preference for GC at position 11 while the functional siRNAs only exhibited a weak preference for GC at this position. In addition, the functional shRNAs prefer AU at position 9, while the functional siRNAs favor GC at this position. Considering that these differences in nucleotide preference between the functional siRNAs and shRNAs in the testing data set were also observed in the training data set, the strong GC preference at position 11 and the preference for AU but not GC at position 9 might represent some of the “shRNA-specific components” that are not fully taken into account in the advanced siRNA prediction algorithm.
DISCUSSION
To facilitate the creation of potent silencing constructs, we systematically investigated several parameters including the use of shRNA- or miR30-based design, the length of shRNA, and the selection of shRNA sequences for their impact on the knockdown efficiency of a silencing construct. Our studies revealed that only when a particular 4-nt loop was used for shRNA are silencing constructs using the 29-nt stem shRNAs or the miR30-based scaffold more potent than constructs using the conventional 19-nt stem shRNAs. However, when a 9-nt loop (UUCAAGAGA) was used for shRNA, the conventional shRNA-based scaffold clearly outperformed the miR30-based scaffold for target knockdown. In addition, using the same 9-nt loop, shRNA with a 19-nt stem outperformed shRNA with a 29-nt stem. Therefore, one of the optimal configurations for a potent silencing trigger could be an shRNA with a 19-nt stem and a 9-nt loop. The discrepancy between our results and what was reported in the literature may arise from the use of different loops for shRNAs. In the publications that reported the substantial improvement of knockdown potency using the miR30-based constructs or using shRNAs with a 29-nt stem, shRNAs with a 4-nt loop were used for comparison. However, shRNAs with a 4-nt loop are typically less active compared with shRNAs with a commonly used 9-nt loop (data not shown; Figs. 2B, 3C). Therefore, the much-hyped improvement using the miR30-based design or using shRNAs with 29-nt stems represents an improvement over the less optimal performance of shRNAs with a 4-nt loop rather than a general advancement in the design of potent hairpin-based silencing triggers. The inferior performance of shRNAs with a 4-nt loop could result from an inefficient processing of this type of shRNAs by Dicer. In fact, it has been shown that the processing of synthetic shRNAs with 19-nt stems in the context of a 4-nt loop is independent of Dicer (Siolas et al. 2005). In contrast, shRNAs with 19-nt stems and a longer loop may still serve as good substrates for Dicer. Therefore, employing strategies that aim to improve Dicer-dependent processing of silencing triggers, such as using miR30-based design or longer shRNAs, is not expected to offer much benefit compared with standard shRNAs with a 19-nt stem and a 9-nt loop. To test this hypothesis, we examined the ability of recombinant Dicer to process synthetic 19-nt hairpin RNAs with a 4-nt or 9-nt loop. As we suspected, the hairpin RNA with a 9-nt loop was processed by Dicer. In contrast, the hairpin RNA with a 4-nt loop was not processed (data not shown).
Beyond the configuration of hairpin-based silencing triggers, the selection of a promoter can also affect the knockdown efficiency of a silencing construct. In addition to the human U6 promoter used in this study, the human H1 promoter is also widely used for the expression of shRNA. The U6 and H1 promoters require different leading nucleotide sequences to obtain optimal expression. For example, the U6 promoter requires the shRNA to start with a G or GG, and the H1 promoter requires the shRNA to start with a G or A for optimal expression. We have compared the potency of knockdown constructs that use the U6 or H1 promoter to drive the expression of an shRNA that starts with a G. Under our experimental conditions, the U6-driven shRNA construct produced much better knockdown compared to the H1-driven shRNA construct (data not shown). The CMV promoter is not suitable for the expression of shRNAs in our hands. Although there are reports in the literature indicating that functional shRNAs can be expressed from the CMV promoter, we found that the CMV-driven shRNA constructs are often several orders of magnitude less efficient in knocking down the target compared with the U6-driven shRNA constructs (data not shown).
Much effort has been devoted to identifying rules for the prediction of siRNA functionality, and several computer programs have been reported to be very successful in selecting potent siRNA sequences (Reynolds et al. 2004; Huesken et al. 2005). However, the algorithms for shRNA sequence selection have not been reported. Upon comparing a limited set of siRNAs and shRNAs for their GC profiles and their abilities for target knockdown, we found that functional siRNAs and shRNAs share a number of common features, including the preference for AU at the 3′ end of the sense strand, the preference for GC at the 5′ end of the sense strand, the presence of an AU-rich stretch at positions 3–6, and the strong preference for AU at positions 3, 6, 13, and 18. We suspect that these common features likely represent the essential properties of a potent siRNA duplex. Among these common features, the strong preference for AU at the 3′ end of the sense strand is consistent with the reported low internal stability at this region, which is thought to promote the entry of the guide strand into RISC (Khvorova et al. 2003). Along the same line, the preference for GC in the functional siRNAs or shRNAs at the 5′ end of the sense strand could further ensure the asymmetric internal stability of a siRNA duplex to prevent the loading of the sense strand into RISC. The presence of an AU-rich stretch at positions 3–6 and the strong preference of AU at positions 3, 6, 13, and 18 in both the functional shRNAs and siRNAs are intriguing. However, a better understanding of the RNAi mechanism will be required to obtain a mechanistic insight of how these features could affect the siRNA or shRNA potency. The most prominent differences between a functional siRNA and shRNA are the nucleotide preferences at positions 9 and 11. Compared with the functional siRNAs, the functional shRNAs exhibit a much stronger preference for GC at position 11 and a preference for AU but not GC at position 9. Because our shRNA data set is relatively small, we cannot completely exclude the possibility that these differences between the functional siRNAs and shRNAs may result from normal variations of the shRNA data. However, because these differences are preserved in the functional shRNAs from both the training data set and the testing data set, we suspect that the strong preference for GC at position 11 and the preference for GC but not AU at position 9 may represent properties that are important for shRNA function (the nucleotide preferences of functional shRNAs and siRNAs are summarized in Table 1). Based on the characteristic nucleotide preferences in the functional versus the nonfunctional shRNAs, we developed a computer program for the selection of shRNA sequences. Although the size of our shRNA data set is quite small, the computer program performs surprisingly well to enrich the highly functional shRNAs. These results suggest that, as for siRNAs, descriptive functions can be developed to facilitate the selection of the functional shRNA sequences. With the collection of a larger shRNA data set, it is expected that similar analysis could lead to the development of a more accurate algorithm to facilitate the selection of the functional shRNAs.
MATERIALS AND METHODS
Selection of shRNA sequences
All shRNA sequences were selected using the following rules: (1) starting with G or GG for efficient expression using the U6 promoter; (2) GC percentage ranging from 30% to 80%; and (3) more than 75 base pairs (bp) downstream from the translation initiation site in the target mRNA.
Creation of DNA vectors that express shRNA or miR30-based silencing triggers
A modified U6 promoter, 2O2, was used to drive the expression of all shRNAs (Lin et al. 2004). The 2O2 promoter was developed as a tetracycline-responsive U6 promoter variant in the presence of the tet repressor, tetR. In our experimental setting, the 2O2 promoter was used as a constitutively active promoter because it was used in cells that do not express tetR. The shRNA expression constructs were created using PCR-based methods. Briefly, a 5′ primer that hybridizes to the 2O2 promoter and a 3′ primer that contains an shRNA sequence and a sequence complementary to the 3′ of the 2O2 promoter were used to amplify a DNA fragment that contains both the 2O2 promoter and the shRNA sequences. The PCR fragment was then cloned into pBluescript II(SK+) to create the shRNA expression construct. The DNA vectors that express the miR30-based silencing triggers were also created using PCR-based methods. The partial sequence, including the promoter and the miR30-like transcript, of a representative 2O2-driven miR30-based construct that targets luciferase is presented here: ggcaaaacgcaccacgtgacggagcgtgaccgcgcgccgagcgcgcgccaaggtcgggcaggaagagggcctatttcccatgattccatcatatttgcatatacgatacaaggctgttagagagataattagaattaatttgactgtaaacacaaagatattagtataaaatacgtgacgtagaaagtaataatttcttgggtagtttgcagttttaaaattatgttttaaaatggactatcatatgcttaccgtaacttgaaactccctatcagtgatagagattatatatctccctatcagtgatagagaccgtcgactagggataacagggtaattgtttgaatgaggcttcagtactttacagaatcgttgcctgcacatcttggaaacacttgctgggattacttcttcaggttaacccaacagaaggctcgagaaggtatattgctgttgacagtgagcgccGGACATCACTTACGCTGAGTtagtgaagccacagatgtaACTCAGCGTAAGTGATGTCCgttgcctactgcctcggaattcaaggggctactttaggagcaattatcttgtttactaaaactgaataccttgctatctctttgatacatttttacaaagctgaattaaaatggtataaattaaatcacttttttggtacccaattcgccctatagtgagt. The underscored sequences represent the 2O2 promoter. The capital letters represent the duplex sequence of a luciferase siRNA that is embedded in the miR30-like transcript. For all PCR-based cloning, the PCR fragment was verified by sequencing.
Target knockdown induced by shRNA or miR30-based constructs
To determine the degree of target knockdown induced by an shRNA- or miR30-based construct against luciferase, the shRNA- or miR30-based construct was cotransfected into H1299 cells with the pGL3-control (Fluc) and pRL-TK (Rluc) reporters (Promega) using the previously described protocol (Lin et al. 2004). The luciferase activities in transfected cells were determined 48 h after transfection using the Dual-Glo luciferase assay system (Promega).
To determine the degree of target knockdown induced by an shRNA- or miR30-based construct against mouse tyrosinase, the shRNA- or miR30-based construct was cotransfected into H1299 cells with a plasmid encoding the mouse tyrosinase. The tyrosinase activities in transfected cells were determined 48 h after transfection. The degree of target knockdown triggered by a knockdown construct against human HIF-1α was estimated by cotransfecting the knockdown construct with pHRE, a HIF-1 reporter containing the hypoxia responsible element (HRE) from the enolase promoter, together with the pRL-TK reporter (Promega). Twenty-four hours after transfection, cells were subjected to hypoxia treatment for 16 h as previously described (Li et al. 2005b), and the luciferase activity was determined using the Dual-Glo luciferase assay system. To determine the degree of target knockdown triggered by shRNAs against other targets, the cDNAs that contain the coding sequences of these targets were cloned in-frame into the pcDNA3.1/V5-His vector (Invitrogen) to create expression vectors that produce the V5-His tagged proteins. The shRNA expression construct was cotransfected with the expression vector of each target into the H1299 cells. The cells were lyzed 48 h after transfection, and the protein levels of each target in transfected cells were determined by Western blotting using the anti-V5 antibody (Sigma) and densitometry scanning of the X-ray film.
Tyrosinase assay
Cells that were cotransfected with an shRNA- or miR30-based knockdown construct and the tyrosinase-expression plasmid were lysed 48 h after transfection using the CelLytic-M lysis buffer (Sigma). After centrifugation, the supernatant of the cell lysate was mixed with 0.2% 3,4-Dihydroxy-L-phenylalanine in 0.1 M PBS at pH 6.0 using a 1:2 ratio. The mixtures were incubated at 37°C in a moisture chamber, and the O.D. 570 nm was determined after colors developed.
Target knockdown induced by siRNAs
siRNAs were designed, synthesized, and tested for target knockdown as previously described (Khvorova et al. 2003; Reynolds et al. 2004). Briefly, silencing of the target was determined 24 h after siRNA transfection by measuring mRNA levels using the branched DNA assay (Panomics) or using the luciferase assay in the cases of luciferase siRNAs; 100 nM of each siRNA was used for transfection, and the transfection efficiency was maintained at over 95%.
Calculation of internal stability profile (ISP) and base-pair preferences for siRNA and shRNAs
The set of 150 shRNAs and 390 siRNAs was used for initial analysis. The siRNA sequences used are a collection of siRNAs targeting fLUC, PPIB, DBI, and GAPDH. These siRNAs were designed, synthesized, and tested for target knockdown as previously described (Khvorova et al. 2003; Reynolds et al. 2004). shRNAs that induced >75% target knockdown were considered functional (n = 75), shRNAs that induced <50% target knockdown were considered nonfunctional (n = 40), and shRNAs that mediated 50%–75% target knockdown were excluded from the analysis. siRNAs that induced >80% silencing of the target were considered functional (n = 136), siRNAs that induced <40% silencing of the target were considered nonfunctional (n = 118), and siRNAs that mediated 40%–80% target knockdown were excluded from the analysis. Expanding the analysis to different functionality cutoffs for both siRNAs and shRNAs produced similar data (data not shown). The ISP was calculated as previously described (Khvorova et al. 2003). The differential positional preference was calculated as the difference between the percentages of a nucleotide in a particular position in functional versus nonfunctional subsets.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.599107.
REFERENCES
- Berns, K., Hijmans, E.M., Mullenders, J., Brummelkamp, T.R., Velds, A., Heimerikx, M., Kerkhoven, R.M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Nijman, S.M., Dirac, A.M., Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Zhang, L., Conklin, D.S., Hannon, G.J., Rosenquist, T.A. Germline transmission of RNAi in mice. Nat. Struct. Biol. 2003;10:91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Dickins, R.A., Hemann, M.T., Zilfou, J.T., Simpson, D.R., Ibarra, I., Hannon, G.J., Lowe, S.W. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Dorsett, Y., Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grimm, D., Streetz, K.L., Jopling, C.L., Storm, T.A., Pandey, K., Davis, C.R., Marion, P., Salazar, F., Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J., Rossi, J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Huesken, D., Lange, J., Mickanin, C., Weiler, J., Asselbergs, F., Warner, J., Meloon, B., Engel, S., Rosenberg, A., Cohen, D., et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Li, M.J., Kim, J., Li, S., Zaia, J., Yee, J.K., Anderson, J., Akkina, R., Rossi, J.J. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005a;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Li, L., Lin, X., Staver, M., Shoemaker, A., Semizarov, D., Fesik, S.W., Shen, Y. Evaluating hypoxia-inducible factor-1α as a cancer therapeutic target via inducible RNA interference in vivo. Cancer Res. 2005b;65:7249–7258. doi: 10.1158/0008-5472.CAN-04-4426. [DOI] [PubMed] [Google Scholar]
- Lin, X., Yang, J., Chen, J., Gunasekera, A., Fesik, S.W., Shen, Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004;577:376–380. doi: 10.1016/j.febslet.2004.10.033. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M., Taira, K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Montgomery, M.K., Xu, S., Fire, A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans . Proc. Natl. Acad. Sci. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison, P.J., Silva, J.M., Conklin, D.S., Schlabach, M., Li, M., Aruleba, S., Balija, V., O'Shaughnessy, A., Gnoj, L., Scobie, K., et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S., Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rubinson, D.A., Dillon, C.P., Kwiatkowski, A.V., Sievers, C., Yang, L., Kopinja, J., Rooney, D.L., Zhang, M., Ihrig, M.M., McManus, M.T., et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Silva, J.M., Li, M.Z., Chang, K., Ge, W., Golding, M.C., Rickles, R.J., Siolas, D., Hu, G., Paddison, P.J., Schlabach, M.R., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P.S., Paddison, P.J., Hannon, G.J., Cleary, M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., Hu, G., Rickles, R.J., Hannon, G.J., Elledge, S.J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y., Forrester, W.C. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Mao, Q., Paulson, H.L., Davidson, B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Xia, H., Mao, Q., Eliason, S.L., Harper, S.Q., Martins, I.H., Orr, H.T., Paulson, H.L., Yang, L., Kotin, R.M., Davidson, B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Cai, X., Cullen, B.R. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005a;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., Cullen, B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005b;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]