Abstract
The incorporation of unnatural amino acids into proteins is a valuable tool for addition of biophysical probes, bio-orthogonal functionalities, and photoreactive cross-linking agents, although these approaches often require quantities of protein that are difficult to access with chemically aminoacylated tRNAs. THG73 is an amber suppressor tRNA that has been used extensively, incorporating over 100 residues in 20 proteins. In vitro studies have shown that the Escherichia coli Asn amber suppressor (ENAS) suppresses better than THG73. However, we report here that ENAS suppresses with <26% of the efficiency of THG73 in Xenopus oocytes. We then tested the newly developed Tetrahymena thermophila Gln amber suppressor (TQAS) tRNA library, which contains mutations in the second to fourth positions of the acceptor stem. The acceptor stem mutations have no adverse effect on suppression efficiency and, in fact, can increase the suppression efficiency. Combining mutations causes an averaging of suppression efficiency, and increased suppression efficiency does not correlate with increased ΔG of the acceptor stem. We created a T. thermophila opal suppressor, TQOpS′, which shows ∼50% suppression efficiency relative to THG73. The TQAS tRNA library, composed of functional suppressor tRNAs, has been created and will allow for screening in eukaryotic cells, where rapid analysis of large libraries is not feasible.
Keywords: nonsense suppression, frameshift suppression, tRNA, opal suppressor tRNA, unnatural amino acid incorporation
INTRODUCTION
Incorporation of unnatural amino acids (UAAs) site-specifically into proteins is a powerful technique that is seeing increased use. Typically, the UAA is incorporated at a stop codon (nonsense suppression) using an orthogonal tRNA with an anticodon recognizing the stop codon. In higher eukaryotes, nonsense suppression by chemically aminoacylated tRNAs is mostly limited to the Xenopus oocyte, where injection of the mutant mRNA and suppressor tRNA chemically aminoacylated with the UAA is straightforward, and electrophysiology allows for sensitive detection of UAA incorporation (Dougherty 2000; Beene et al. 2003). Previously, only a single UAA could be incorporated into a given protein expressed in Xenopus oocytes, but frameshift suppression allows for the simultaneous incorporation of three UAAs, using the amber stop codon (UAG) and the quadruplet codons, CGGG and GGGU (Rodriguez et al. 2006).
In developing optimal procedures for nonsense suppression, two key issues must be addressed. The first is “orthogonality”; the suppressor tRNA must not be recognized by any of the endogenous aminoacyl-tRNA synthetases (aaRSs) of the expression system, as this would lead to competitive incorporation of natural amino acids at the mutation site. In our companion paper, we developed and evaluated the orthogonality of a library of suppressor tRNAs, showing several new mutations increase the orthogonality of amber suppressor tRNAs (Rodriguez et al. 2007, companion paper, this issue). The second issue is suppression efficiency. In order to produce adequate quantities of protein, optimal use of the chemically aminoacylated tRNA is essential, as this stoichiometric reagent is often not available in large quantities. This is especially true when considering incorporation of biophysical probes for fluorescence or cross-linking strategies that often require more protein than the much-used electrophysiological approaches.
THG73, an amber suppressor tRNA from Tetrahymena thermophila with a G73 mutation, has been used extensively to incorporate >100 residues in 20 different proteins in Xenopus oocytes (Dougherty 2000; Beene et al. 2003). In the present work, we evaluate a number of tRNAs for their suppression efficiency in Xenopus oocytes compared to THG73. We find that an Escherichia coli Asn amber suppressor (ENAS) tRNA that was shown to incorporate UAAs better than THG73 in vitro (Cload et al. 1996) is in fact significantly less efficient than THG73 in Xenopus oocytes.
We also evaluate several other tRNAs that contain mutations in the second to fourth positions of the acceptor stem on THG73. Our study of tRNA orthogonality showed such mutations can have interesting consequences on aminoacylation in vivo (Rodriguez et al. 2007). A number of studies have shown that acceptor stem, anticodon stem, D stem, and T stem structure can influence suppression efficiency, often finding that replacement of non-Watson–Crick base pairs with canonical pairs increases efficiency (Hou and Schimmel 1992; Buttcher et al. 1994; Ohtsuki et al. 2005). We, therefore, created a library of T. thermophila Gln amber suppressor (TQAS) mutants that strengthened the acceptor stem with C–G/G–C pairs and replaced the non-Watson–Crick pair U4-G69 with C4-G69. Many of the mutant tRNAs had increased suppression efficiency over THG73, but there was a lack of correlation with the ΔG of the acceptor stem in Xenopus oocytes. Intriguingly, a mutant tRNA with U2-C71 and mutations in the third and fourth positions was found to suppress UAG more efficiently than THG73 in Xenopus oocytes. In contrast, when creating T. thermophila Gln opal suppressor (TQOpS) tRNAs, the U2-C71 mutation had an adverse effect on suppression efficiency compared to C2-G71 tRNA (TQOpS′). Thus, in vivo nonsense suppression efficiencies of both UAG and UGA stop codons are affected by mutations in the acceptor stem of tRNAs. Overall, we have created a TQAS tRNA library that is functional in Xenopus oocytes and an opal suppressor tRNA for the incorporation of UAAs by chemical aminoacylation.
RESULTS
Electrophysiology assay
All experiments were performed by suppression in the nicotinic acetylcholine receptor (nAChR), which is a pentameric ion channel composed of α, β, γ, and δ subunits in the ratio of 2:1:1:1, respectively. For comparison of the suppression efficiency and the incorporation of UAAs, the well-characterized αW149 site was utilized. This site can only function with the incorporation of aromatic amino acids or aromatic UAAs, because it makes a cation-π interaction with ACh (Zhong et al. 1998). In order to compare suppression efficiencies across different batches of oocytes, we normalized the average maximal current for each tRNA to the average maximal current for THG73, obtained concurrently. This ratio allows for comparison of suppression efficiencies even if protein expression varies between batches of oocytes.
During the course of this research, we noticed variations in aminoacylation depending on whether the oocytes were obtained from frogs purchased from Xenopus Express or Nasco (Rodriguez et al. 2007, companion paper, this issue). Xenopus laevis frogs from Xenopus Express are caught in Africa, while Nasco frogs are bred in a laboratory and are from a more restricted gene pool (L. Northey, pers. comm.). We, therefore, tested the suppression efficiency of tRNAs to see if there was any difference between Xenopus Express and Nasco oocytes. When different suppliers are used in experiments, they are explicitly labeled with each figure.
ENAS and ENAS A71 suppression efficiency
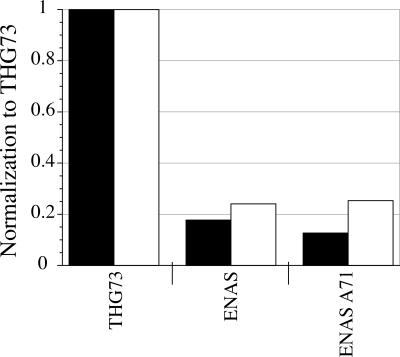
The ENAS tRNA has been shown previously to have a greater suppression efficiency than THG73 in an E. coli in vitro translation system (Cload et al. 1996). ENAS has been used extensively to incorporate UAAs in vitro and can also tolerate substitution to the anticodon to suppress quadruplet codons for the incorporation of multiple UAAs (Murakami et al. 2003, 2006). Therefore, ENAS may be a valuable alternative to THG73 for the creation of amber and/or frameshift suppressor tRNAs in vivo. ENAS contains the insertion G1-C72 to allow for optimal T7 RNA polymerase transcription and thus has an 8 base-pair (bp) acceptor stem (Cload et al. 1996). THG73 and the yeast Phe frameshift suppressor tRNAs (YFFSCCCG and YFaFSACCC) are derived from eukaryotic cells and have a 7 bp acceptor stem (Rodriguez et al. 2006). When analyzing the structure of ENAS, we noticed that the second position of the acceptor stem contains the non-Watson–Crick base pair U2-C71, and thus we created the variant ENAS A71 to form the canonical pair U2-A71 (Fig. 1) that is present in the wild-type E. coli Asn tRNA (Kleina et al. 1990). Suppression of α149UAG with either ENAS-W or ENAS A71-W resulted in substantially diminished suppression efficiency (Fig. 2), with the best case being only 26% relative to THG73-W. Overall, ENAS is not a viable alternative to THG73 for the incorporation of UAAs in Xenopus oocytes.
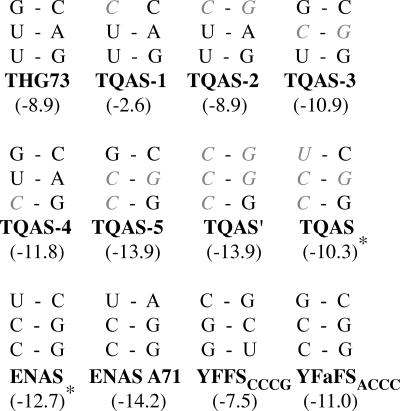
FIGURE 1.
THG73 mutations and tRNAs studied. The second to fourth positions of the acceptor stem are shown for all tRNAs, with mutations in gray italics. ENAS and TQAS contain the same nucleotides at these positions. Below each tRNA is the ΔG (kcal/mol) of the entire acceptor stem calculated using Mfold (Zuker 2003). TQOpS′ and TQOpS have the same ΔG as TQAS′ and TQAS, respectively. * ΔG calculated as written in Materials and Methods.
FIGURE 2.
ENAS-W and ENAS A71-W suppression at α149UAG. tRNA-W (21 ng per oocyte) average current was normalized by THG73-W average current and bars represent this average ratio (Total oocytes tested are 40, 17>n>11). Black and white bars correspond to Xenopus Express and Nasco oocytes, respectively. ENAS-W and ENAS A71-W are <26% of the THG73-W current when suppressing at α149UAG. Therefore, neither ENAS nor ENAS A71 offer improved suppression over THG73 in Xenopus oocytes.
Another option would be to create frameshift suppressors from ENAS. However, previous work has shown that frameshift suppressors derived from amber suppressor tRNAs are less efficient in rabbit reticulocyte lysate (Taira et al. 2005) and in Xenopus oocytes (Rodriguez et al. 2006), so we did not screen frameshift suppressor tRNAs derived from ENAS.
Testing THG73 acceptor stem mutations suppression efficiency
We then tested the TQAS tRNA library (shown in Fig. 1) developed recently (Rodriguez et al. 2007, companion paper, this issue) for suppression efficiency at α149UAG. The mutation G2C on THG73 (TQAS-1) results in the placement of C2 C71 at the second position of the acceptor stem. According to a free energy calculation by Mfold (Zuker 2003), this causes a distortion in the acceptor stem whereby the following base pairs are formed, C2-G73 and G1-C74, while the CA extension is reduced by two nucleotide lengths. Not surprisingly, TQAS-1-W shows only 1% of the suppression efficiency of THG73-W in both Xenopus Express and Nasco oocytes (Fig. 3), a value only slightly greater than α149UAG mRNA.
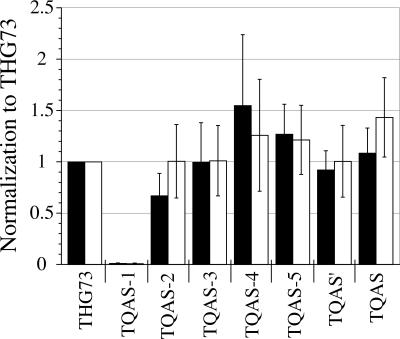
FIGURE 3.
THG73 acceptor stem mutations suppressing at α149UAG. tRNA-W (9 ng per oocyte) average currents were normalized to THG73-W average current, the ratios were averaged, and error bars represent the standard error of ≥3 normalization experiments (17>n>8 oocytes per experiment, for a total of 709 oocytes tested). Bar colors are the same as in Fig. 2. TQAS-1 is nonfunctional because it represents <1% of THG73-W. All other tRNAs show significant current and acceptance by the translational machinery. TQAS-4-W, TQAS-5-W, and TQAS-W all have greater average suppression efficiency than THG73-W at α149UAG.
The single helix pair mutations C2-G71 and C3-G70 (TQAS-2 and TQAS-3, respectively) show similar suppression efficiency to THG73-W in both Xenopus Express and Nasco oocytes (Fig. 3). The single mutation U4C (TQAS-4) removes the non-Watson–Crick pair at the fourth position and increases suppression efficiency by 55% and 26% in Xenopus Express and Nasco oocytes, respectively (Fig. 3).
Combining the C3-G70 and C4 mutations (TQAS-5) shows an increase in suppression efficiency of 26% and 21% in Xenopus Express and Nasco oocytes, respectively (Fig. 3). Combining all of the functional single helix pair mutations created TQAS′, which suppresses equivalently to THG73-W even though there are five mutations in the acceptor stem (Fig. 3). Placement of the ENAS second to fourth helix pairs on THG73 created TQAS (Fig. 1). TQAS-W is more efficient in Nasco oocytes with 43% increase in suppression efficiency, compared to 9% increase in suppression efficiency in Xenopus Express oocytes (Fig. 3). The structures of Figure 1 constitute a library of amber suppressor tRNAs containing various acceptor stem mutations, which function in vivo comparably or superior to the parent tRNA, THG73.
Combining mutations causes averaging of the suppression efficiency
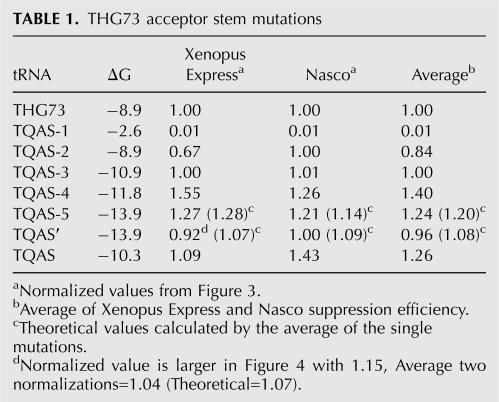
While analyzing suppression efficiency, we noticed a trend where combining the acceptor stem mutations caused an averaging of the single mutations. Table 1 lists values from Figure 3 and also shows the average of the two normalization experiments for each tRNA-W. TQAS-5 contains the mutations from both TQAS-3 and TQAS-4 and has a suppression efficiency of 1.27 and 1.21, relative to THG73-W, in Xenopus Express and Nasco oocytes, respectively. The average of TQAS-3 and TQAS-4 is 1.28 and 1.14 in Xenopus Express and Nasco oocytes, respectively (Table 1). TQAS′ contains the mutations from TQAS-2, TQAS-3, and TQAS-4 and has suppression efficiencies of 0.92 and 1.00 in Xenopus Express and Nasco oocytes, respectively. The average of TQAS-2, TQAS-3, and TQAS-4 is 1.07 and 1.09 in Xenopus Express and Nasco oocytes, respectively (Table 1).
TABLE 1.
THG73 acceptor stem mutations
Suppression efficiency of the acceptor stem mutations does not correlate with ΔG
Previous work has shown that non-Watson–Crick mutations within tRNAs have an adverse effect on suppression efficiency (Hou and Schimmel 1992; Buttcher et al. 1994; Ohtsuki et al. 2005), but these mutations would also reduce the free energy (ΔG) of the stem regions. Therefore, we calculated the ΔG of the acceptor stem using Mfold (Zuker 2003). Mfold does not recognize the U–C pair, and therefore we calculated the ΔG as described in Materials and Methods. Plotting the TQAS library suppression efficiency as a function of ΔG showed no correlation in Xenopus Express or Nasco ooctyes. TQAS-5 and TQAS′ share the highest ΔG values of the library, but suppression efficiency is different due to averaging of single mutations (Table 1). Therefore, ΔG of the acceptor stem is not a reliable predictor for tRNA suppression efficiency under the conditions currently used.
Testing amber, opal, and frameshift suppression efficiency
Incorporating multiple UAAs simultaneously requires the use of a unique stop or quadruplet codon for each UAA. Previously, we have incorporated three UAAs simultaneously using THG73, YFFSCCCG, and YFaFSACCC suppressor tRNAs at the corresponding suppression sites, UAG, CGGG, and GGGU (Rodriguez et al. 2006). Suppression efficiency of the opal (UGA) codon has been shown to be comparable to the amber (UAG) codon in mammalian cells when using suppressor tRNAs that are aminoacylated by endogenous aaRSs or by the import of exogenous E. coli aaRSs (Capone et al. 1986; Kohrer et al. 2004). The opal codon has also been utilized to incorporate an UAA in mammalian cells using a tRNA/synthetase pair (Zhang et al. 2004). However, when using a chemically aminoacylated tRNA, an opal suppressor that efficiently suppresses the opal codon and is adequately orthogonal is currently lacking. Sisido and colleagues tested a yeast Phe opal suppressor in rabbit reticulocyte lysate, but the suppression efficiency was only 15% (compared to 65% for the yeast Phe amber suppressor) (Taira et al. 2005). An opal suppressor created by changing the anticodon of THG73 to UCA resulted in large amounts of aminoacylation in vitro (Cload et al. 1996). We replaced the anticodons of TQAS′ and TQAS with UCA to create TQOpS′ and TQOpS, respectively. Both TQOpS′ and TQOpS show reduced aminoacylation when compared to THG73 in Xenopus oocytes (Rodriguez et al. 2007, companion paper, this issue).
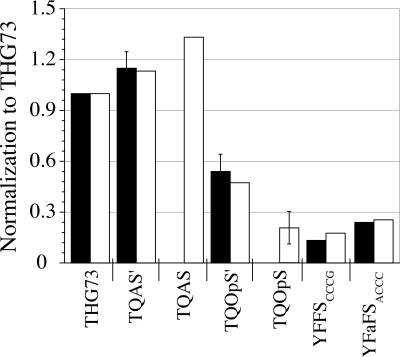
Suppression efficiencies of THG73-W, TQAS′-W, TQAS-W, TQOpS′-W, TQOpS-W, YFFSCCCG-W, and YFaFSACCC-W were evaluated at the α149 suppression site (Fig. 4). All mRNA and tRNAs were normalized to allow for identical conditions. Amber suppression is the most efficient, and the suppression efficiency follows the order of TQAS-W>TQAS′-W>THG73-W in Nasco oocytes (Fig. 4). Opal suppression with TQOpS′-W and TQOpS-W has a suppression efficiency of 48% and 21%, respectively, relative to THG73-W in Nasco oocytes (Fig. 4). TQAS-W and TQOpS-W were not tested in Xenopus Express oocytes because TQAS was not originally selected as an orthogonal tRNA until screening in Nasco oocytes (Rodriguez et al. 2007, companion paper, this issue), but all other tRNAs show comparable suppression efficiency in both Xenopus Express and Nasco oocytes (Figs. 3, 4). Overall, TQOpS′-W shows the greatest opal suppression efficiency with 54% and 48% in Xenopus Express and Nasco oocytes, respectively, and is a better suppressor tRNA than either frameshift suppressor (Fig. 4). The suppression efficiency trend is thus TQAS-W>TQAS′-W>THG73-W>TQOpS′-W>YFaFSACCC-W≈TQOpS-W>YFFSCCCG-W.
FIGURE 4.
Amber, opal, and frameshift suppressor tRNAs suppression at α149. All tRNA-W (7.5 ng per oocyte) average current was normalized to THG73-W average current. TQAS′-W, TQOpS′-W, and TQOpS-W were performed twice and error bars represent the standard error. Total oocytes tested are 161 oocytes, where 16>n>5 for each experiment. Bar colors are the same as in Fig. 2. Amber suppression (THG73-W, TQAS′-W, or TQAS-W) is the most efficient. Opal suppression is variable, with TQOpS′-W and TQOpS-W suppressing 48% and 21%, respectively, of THG73-W in Nasco oocytes. YFFSCCCG-W suppresses less than YFaFSACCC-W, as previously seen in Xenopus oocytes (Rodriguez et al. 2006). TQAS′-W and TQAS-W show increased suppression compared to THG73-W.
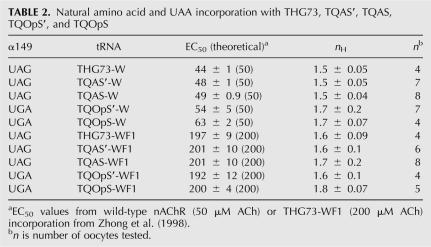
Natural amino acid and UAA incorporation with selected suppressor tRNAs
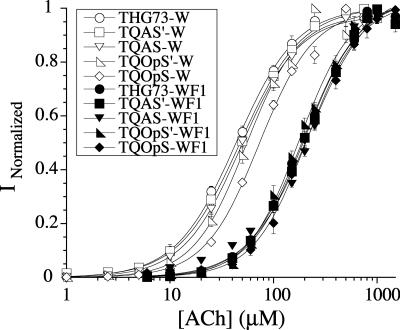
To evaluate incorporation of a natural amino acid and an UAA using TQAS′, TQAS, TQOpS′, and TQOpS, we chose to suppress the well-characterized, nonpromiscuous site α149. The Trp at α149 makes a cation-π interaction with ACh, and the incorporation of the UAA, 5-F-Tryptophan (WF1), results in an approximately fourfold increase in the EC50 for ACh. Incorporation of Trp at the α149UAG/UGA site is a wild-type recovery experiment because it places the natural amino acid at the suppression site. All tRNA-W were injected along with mRNA containing the appropriate codon at site α149, and the EC50 was determined by fits to the Hill equation (Fig. 5). All tRNA-W showed the correct EC50 except for TQOpS-W, which showed a slightly higher EC50 than the wild-type nAChR (Fig. 5; Table 2). Injection of α149UGA with TQOpS (74 mer) resulted in only 3% of the current relative to the injection of α149UGA with TQOpS-W and an EC50 for the aminoacylation product could not be determined. The natural amino acid must be aromatic for functional receptors when suppressing at the α149 site and cannot be Trp (EC50 would have been wild-type) and, therefore, is most likely either Tyr or Phe, which are expected to produce substantially higher EC50 values than Trp (Zhong et al. 1998). However, TQOpS-W weakly suppresses the α149UGA site relative to TQOpS′ (Fig. 4), which is the better suppressor tRNA in both Xenopus Express and Nasco oocytes.
FIGURE 5.
Fits to the Hill equation for wild-type recovery and UAA incorporation at α149. Suppression of tRNA-W (9 ng per oocyte) at α149(UAG or UGA) places the natural amino acid and results in wild-type EC50 (∼50 μM ACh) for all tRNAs tested except TQOpS-W (white diamond), which gave the same EC50 in two experiments (see Table 2). Incorporation of WF1 at α149 results in a fourfold increase in EC50 (200 μM ACh) (Zhong et al. 1998). All tRNA-WF1 suppressing at α149(UAG or UGA) give similar EC50s and show that all tRNAs are able to incorporate an UAA. All experiments were done in Nasco oocytes and give the same EC50s as W or WF1 incorporation in Xenopus Express oocytes (Rodriguez et al. 2006). EC50s, n H, and n are listed in Table 2.
TABLE 2.
Natural amino acid and UAA incorporation with THG73, TQAS′, TQAS, TQOpS′, and TQOpS
We then incorporated the UAA, WF1 at the α149 suppression site with the injection of tRNA-WF1 and determined the EC50s (Fig. 5; Table 2). All tRNAs are able to incorporate WF1 at the α149 suppression and show the correct EC50s (Fig. 5; Table 2). All experiments were performed in Nasco oocytes and give the same EC50s as W or WF1 incorporation at α149UAG in Xenopus Express oocytes (Rodriguez et al. 2006). TQAS′ and TQAS are both viable suppressor tRNAs for the incorporation of natural amino acids and UAAs, which suggests that the entire T. thermophila Gln amber suppressor (TQAS) tRNA library would also be able to incorporate natural amino acids and UAAs.
DISCUSSION
THG73 is an amber suppressor previously shown to suppress UAAs better than a modified yeast Phe amber suppressor (MN3) in Xenopus oocytes (Saks et al. 1996). An E. coli in vitro translation has shown that ENAS suppresses better than THG73 (Cload et al. 1996). We show that in Xenopus oocytes both ENAS and ENAS A71 suppress <26% relative to THG73 (Fig. 2). Less suppression efficiency may result from less acceptance of E. coli-derived amber suppressor tRNAs and/or the extra base pair in the acceptor stem by the eukaryotic translational machinery. Therefore, neither ENAS nor ENAS A71 offer an improvement over THG73. As a result, frameshift suppressor tRNAs derived from ENAS were not screened.
Interestingly, many accepter stem mutations show no adverse effects on function, and there is no statistically significant difference in suppression efficiency in Xenopus Express and Nasco oocytes (Fig. 3), which was seen for aminoacylation (Rodriguez et al. 2007). Most in vivo assays employ suppressor tRNAs that are aminoacylated in vivo by endogenous or exogenous aaRS(s) (Capone et al. 1986; Kleina et al. 1990; Jahn et al. 1991; Hou and Schimmel 1992; Buttcher et al. 1994; McClain et al. 1998; Kohrer et al. 2004). By chemically aminoacylating mutant suppressor tRNAs, we avoid any bias that may result from suppressor tRNAs being differentially aminoacylated. Therefore, we are able to look at mutant tRNA-W interactions with the translational machinery after aminoacylation by evaluating protein production, or suppression efficiency. All mutations to the acceptor stem of THG73, excluding the disruptive C2 C71 of TQAS-1, result in functional suppressor tRNAs, and many have improved suppression efficiency over THG73 (Fig. 3). TQAS-4, TQAS-5, and TQAS all have increased suppression efficiency (Fig. 3; Table 1) and contain the U4C mutation, which removes a non-Watson–Crick U–G pair at the fourth position of the acceptor stem.
Previous work has established that removing non-Watson–Crick base pairs increases suppression efficiency in vitro and in vivo (Hou and Schimmel 1992; Buttcher et al. 1994; Ohtsuki et al. 2005). Intriguingly, TQAS contains U2-C71 and results in a functional suppressor tRNA with increased suppression efficiency relative to THG73 (Fig. 3; Table 1). C–U/U–C pairs have been identified in the acceptor stem of eukaryotic tRNAs (Goodenbour and Pan 2006). However, selection schemes for orthogonal tRNA/synthetase pairs have not identified such mismatches in E. coli tRNAs, even though ∼98.4% of the randomized library would contain a mismatch (Anderson and Schultz 2003). Therefore, non-Watson–Crick base pairs are tolerated by the eukaryotic translational machinery and may serve some special function in higher eukaryotes, as previously suggested (Goodenbour and Pan 2006).
Combining the acceptor stem mutations resulted in an averaging of the suppression efficiency (Table 1). This result is intriguing because it reflects the interactions of the chemically aminoacylated mutant tRNA-W with proteins such as EF-1α and the ribosome, but not with aaRSs. All mutations are in the acceptor stem (Fig. 1) and maintain the rest of the tRNA body constant. TQAS-4 contains the single mutation U4C, which replaces the non-Watson–Crick base pair U–G, and shows the highest average suppression efficiency (Table 1). This mutation most likely increases interactions with EF-1α, the ribosome, and/or another protein. However, this mutation, combined with other mutations, results in reduced suppression efficiency in TQAS-5, TQAS′, and TQAS (Table 1). This clearly illustrates that the mutations are not additive and no single mutation dominates in influencing the suppression efficiency. This suggests there is no dominant protein interaction that strictly influences the suppression efficiency, but rather the ensemble of all protein interactions equally affects the overall suppression efficiency.
The acceptor stem ΔG does not correlate with suppression efficiency. Removal of non-Watson–Crick base pairs has been shown to increase suppression efficiency in various regions of tRNAs (Hou and Schimmel 1992; Buttcher et al. 1994; Ohtsuki et al. 2005), which would also increase the ΔG of the tRNA in helix-forming regions. Using our TQAS library, we related suppression efficiency as a function of ΔG of the acceptor stem and found no overall trend. The replacement of the non-Watson–Crick base pair U4-G69 with C4-G69 (TQAS-4) resulted in increased average suppression efficiency, but was not significantly different from the TQAS library (Fig. 3). TQAS-5 and TQAS′ have the same ΔG (Fig. 1), but different suppression efficiencies (Fig. 3). This difference is caused by various combinations of single mutations that result in an averaging in suppression efficiency (Table 1) due to interactions with proteins in the translational machinery. Therefore, it is difficult to predict the tRNA with the best suppression efficiency just by ΔG of the acceptor stem under the conditions used in these experiments.
An efficient opal suppressor tRNA is highly desirable for the incorporation of multiple UAAs in eukaryotic cells. By replacing the anticodon of TQAS′ and TQAS with UCA, we created the opal suppressers TQOpS′ and TQOpS, respectively. TQOpS′-W shows suppression efficiency of ∼50% relative to THG73-W, while TQOpS-W has a suppression efficiency of 21% (Fig. 4). Intriguingly, the amber suppressor TQAS-W has greater suppression efficiency than TQAS′-W (Figs. 3, 4), but the opal suppressors show the reverse trend (Fig. 4). Therefore, UAG and UGA suppression shows a differential preference for acceptor stem mutations. TQOpS′-W suppresses better than either frameshift suppressor, YFFSCCCG-W and YFaFSACCC-W (Fig. 4). TQOpS′ incorporates W and the UAA, WF1, at α149UGA (Fig. 5; Table 2) and, therefore, is a viable suppressor tRNA for the incorporation of multiple UAAs in vivo.
TQAS′ and TQAS can incorporate W and the UAA, WF1, at α149UAG (Fig. 5; Table 2). This suggests that the entire TQAS tRNA library, excluding TQAS-1, will be able to incorporate UAAs in vivo. TQAS′ and TQAS were specifically screened because they showed the least amount of aminoacylation in Xenopus Express and Nasco oocytes, respectively (Rodriguez et al. 2007). Therefore, both orthogonal tRNAs, in their respective genetic background, are able to incorporate UAAs. The TQAS tRNA library, therefore, offers a diverse range of acceptor stem mutations for the selection of orthogonal tRNAs (Rodriguez et al. 2007). The TQAS library will be valuable for screening in other eukaryotic cells, where screening of randomized libraries may not be feasible.
MATERIALS AND METHODS
Materials
All oligonucleotides were synthesized by Integrated DNA Technologies. NotI was from Roche Applied Science. BamHI, EcoRI, FokI, T4 DNA ligase, and T4 RNA ligase were from NEB. Kinase Max, T7 MEGAshortscript, and T7 mMessage mMachine kits were from Ambion. ACh chloride and yeast inorganic pyrophosphatase were purchased from Sigma-Aldrich. 6-nitroveratryloxycarbonyl protected dCA-W and dCA-WF1 were prepared as reported by Zhong et al. (1998).
tRNA gene preparation and tRNA transcription
THG73, YFFSCCCG, and YFaFSACCC subcloned in the pUC19 vector were previously made (Saks et al. 1996; Rodriguez et al. 2006). Genes for ENAS (sequence from Cload et al. 1996) with flanking EcoRI and BamHI overhangs were phosphorylated using Kinase Max kit, annealed, and ligated with T4 DNA ligase into EcoRI and BamHI linearized pUC19 vector as described (Nowak et al. 1998). ENAS A71 (original sequence from Kleina et al. 1990 and containing the G1-C72 insertion for T7 polymerase transcription) was created by QuikChange mutagenesis on ENAS in pUC19. Acceptor stem mutations on THG73 were created by QuikChange mutagenesis and are shown in Figure 1 (gray italics). Replacing the anticodon of TQAS′ and TQAS from CUA to UCA by QuikChange mutagenesis created TQOpS′ and TQOpS, respectively. All mutations were verified by DNA sequencing (California Institute of Technology Sequencing/Structure Analysis Facility). Template DNA for tRNA lacking the 3′CA was prepared by FokI digestion, and tRNA was transcribed using the T7 MEGAshortscript kit with 0.5 μL of yeast inorganic pyrophosphatase (40 U/mL in 75 mM Tris, 10 mM MgCl2, and pH 7.0). tRNA was desalted using CHROMA SPIN-30 DEPC-H2O columns (BD Biosciences), and the concentration was determined by absorption at 260 nm.
nAChR gene preparation and mRNA transcription
The masked α, β, γ, and δ subunits of the nAChR subcloned in the pAMV vector were previously prepared (Rodriguez et al. 2006). All four subunits terminate with the opal (UGA) stop codon and each UGA was mutated to the ochre (UAA) codon to avoid possible suppression by TQOpS′ and TQOpS. α149(UAG, CGGG, and GGGU) were previously prepared on the masked constructs (Rodriguez et al. 2006). α149UGA was prepared by QuikChange mutagenesis. Mutations were verified by DNA sequencing (California Institute of Technology Sequencing/Structure Analysis Facility). DNA was linearized with NotI, and mRNA was prepared with the T7 mMessage mMachine kit with 0.5 μL of yeast inorganic pyrophosphatase. mRNA was purified by using the RNeasy Mini kit (Qiagen) and quantified by absorption at 260 nm.
ΔG calculations of each tRNA acceptor stem
The entire acceptor stem for each tRNA was used rather than the entire tRNA because only secondary interactions are determined by Mfold (Zuker 2003) and the correct clover-leaf structure was not obtained for each tRNA. The entire acceptor stem, seven helix pairs for THG73, TQAS library, YFFSCCCG, and YFaFSACCC and eight helix pairs for ENAS and ENAS A71 due to the insertion of G1-C72 for T7 RNA polymerase transcription, with a U8 linker was utilized, and default parameters (37°C) of Mfold were used to calculate ΔGs (Zuker 2003) (Fig. 1, values listed for each tRNA acceptor stem). For TQAS and ENAS, a U–C pair is present in the acceptor stem and this pair is not recognized by Mfold (Zuker 2003), even though the U–C pair can form hydrogen bonding interactions in RNA helixes (Holbrook et al. 1991; Kierzek et al. 1999; Tanaka et al. 2000). Previously, non-Watson–Crick base-pair thermodynamics (37°C) were obtained by determining the stabilizing/destabilizing effect upon placement in a RNA helix (seven base pairs total with mismatch) and relating those parameters to a reference RNA helix (six base pairs) (Kierzek et al. 1999). The U–C pair was removed to create a reference RNA helix and the ΔG of TQAS(ΔU2-C71) and ENAS(ΔU2-C71) with a U8 linker was determined using Mfold; 0.26 kcal/mol was added to the determined ΔG value to obtain the number listed in Figure 1 for TQAS and ENAS; 0.26 kcal/mol is the average value of two separate measurements of C–U/U–C pairs in RNA helixes and was used because the exact nucleotide sequence was not measured (Kierzek et al. 1999). The ΔGs for TQOpS′ and TQOpS are the same for TQAS′ and TQAS, respectively.
dCA-W and dCA-WF1 ligation to suppressor tRNAs
Seventy-five micromolars (instead of 300 μM, because there was no change in ligation efficiency) of 6-nitroveratryloxycarbonyl protected dCA-W or dCA-WF1 were coupled to suppressor tRNAs by using T4 RNA ligase for 30 min as previously reported (Nowak et al. 1998; Petersson et al. 2002), desalted using CHROMA SPIN-30 DEPC-H2O columns, and quantified by absorption at 260 nm. tRNA ligation efficiency was qualitatively determined by MALDI mass spectrometry (Petersson et al. 2002), and all tRNA ligations were identical within each prepared group and >80%.
In vivo suppression experiments
Prior to in vivo suppression experiments, all tRNAs and mRNAs were simultaneously made and normalized by UV and densiometric analysis using AlphaEaseFC Stand Alone (Alpha Innotech). Stage VI oocytes of Xenopus laevis were prepared as described (Quick and Lester 1994). All tRNAs were refolded at 65°C for 2 min and 6-nitroveratryloxycarbonyl protected dCA-W or dCA-WF1 was deprotected for 5 min by UV irradiation before injection (Saks et al. 1996). Oocytes were injected with 50 nL of mRNA alone or with tRNA and incubated at 18°C for 44–52 h; 20 ng of mRNA in a subunit ratio of 10:1:1:1 for α149UAG:β:γ:δ were injected (Fig. 2). Figures 3 and 5 used the same ratio, but 40 ng and 60 ng of mRNA, respectively, were injected per oocyte. In Figure 4, 40 ng of mRNA in a subunit ratio of 10:1:1:1 for α149(UAG, UGA, CGGG, or GGGU):β:γ:δ. Amounts of tRNA injected are listed with the legend of each figure.
Electrophysiology
Recordings employed two-electrode voltage clamps on the OpusXpress 6000A (Molecular Devices). ACh was stored at −20°C as a 1 M stock, diluted in Ca2+-free ND96, and delivered to oocytes by a computer-controlled perfusion system. For all experiments, the holding potential was −60 mV. Dose–response data were obtained from at least 11 ACh concentrations, and all tRNA suppression experiments were tested with a single 1 mM ACh dose. Dose–response relations were fit to the Hill equation to determine the EC50 and Hill coefficient (n H). All reported values are represented as a mean±SE of the tested oocytes (number of oocytes [n] is listed with each figure).
ACKNOWLEDGMENTS
E.A.R. is a National Science Foundation Predoctoral Fellow. This work was supported by the NIH (NS 34407 and NS 11756).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.667607.
REFERENCES
- Anderson, J.C., Schultz, P.G. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- Beene, D.L., Dougherty, D.A., Lester, H.A. Unnatural amino acid mutagenesis in mapping ion channel function. Curr. Opin. Neurobiol. 2003;13:264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- Buttcher, V., Senger, B., Schumacher, S., Reinbolt, J., Fasiolo, F. Modulation of the suppression efficiency and amino acid identity of an artificial yeast amber isoleucine transfer RNA in Escherichia coli by a G–U pair in the anticodon stem. Biochem. Biophys. Res. Commun. 1994;200:370–377. doi: 10.1006/bbrc.1994.1458. [DOI] [PubMed] [Google Scholar]
- Capone, J.P., Sedivy, J.M., Sharp, P.A., RajBhandary, U.L. Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: Use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol. Cell. Biol. 1986;6:3059–3067. doi: 10.1128/mcb.6.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cload, S.T., Liu, D.R., Froland, W.A., Schultz, P.G. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem. Biol. 1996;3:1033–1038. doi: 10.1016/s1074-5521(96)90169-6. [DOI] [PubMed] [Google Scholar]
- Dougherty, D.A. Unnatural amino acids as probes of protein structure and function. Curr. Opin. Biotechnol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- Goodenbour, J.M., Pan, T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, S.R., Cheong, C., Tinoco I., Jr, Kim, S.H. Crystal structure of an RNA double helix incorporating a track of non-Watson–Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Schimmel, P. Novel transfer RNAs that are active in Escherichia coli . Biochemistry. 1992;31:4157–4160. doi: 10.1021/bi00132a001. [DOI] [PubMed] [Google Scholar]
- Jahn, M., Rogers, M.J., Soll, D. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kierzek, R., Burkard, M.E., Turner, D.H. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- Kleina, L.G., Masson, J.M., Normanly, J., Abelson, J., Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J. Mol. Biol. 1990;213:705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Kohrer, C., Sullivan, E.L., RajBhandary, U.L. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: Concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004;32:6200–6211. doi: 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H., Schneider, J., Bhattacharya, S., Gabriel, K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Natl. Acad. Sci. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, H., Kourouklis, D., Suga, H. Using a solid-phase ribozyme aminoacylation system to reprogram the genetic code. Chem. Biol. 2003;10:1077–1084. doi: 10.1016/j.chembiol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Murakami, H., Ohta, A., Ashigai, H., Suga, H. A highly flexible tRNA acylation method for nonnatural polypeptide synthesis. Nat. Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- Nowak, M.W., Gallivan, J.P., Silverman, S.K., Labarca, C.G., Dougherty, D.A., Lester, H.A. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- Ohtsuki, T., Manabe, T., Sisido, M. Multiple incorporation of nonnatural amino acids into a single protein using tRNAs with nonstandard structures. FEBS Lett. 2005;579:6769–6774. doi: 10.1016/j.febslet.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Petersson, E.J., Shahgholi, M., Lester, H.A., Dougherty, D.A. MALDI-TOF mass spectrometry methods for evaluation of in vitro aminoacyl tRNA production. RNA. 2002;8:542–547. doi: 10.1017/s1355838202026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick, M.W., Lester, H.A. Methods for expression of excitability proteins in Xenopus oocytes. In: Narahashi T., editor. Ion channels of excitable cells. Academic Press; San Diego, CA: 1994. pp. 261–279. [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D.A. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc. Natl. Acad. Sci. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, E.A., Lester, H.A., Dougherty, D.A. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: Minimizing misacylation. RNA. 2007 doi: 10.1261/rna.666807. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks, M.E., Sampson, J.R., Nowak, M.W., Kearney, P.C., Du, F.Y., Abelson, J.N., Lester, H.A., Dougherty, D.A. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. J. Biol. Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- Taira, H., Fukushima, M., Hohsaka, T., Sisido, M. Four-base codon-mediated incorporation of nonnatural amino acids into proteins in a eukaryotic cell-free translation system. J. Biosci. Bioeng. 2005;99:473–476. doi: 10.1263/jbb.99.473. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Kojima, C., Yamazaki, T., Kodama, T.S., Yasuno, K., Miyashita, S., Ono, A., Ono, A., Kainosho, M., Kyogoku, Y. Solution structure of an RNA duplex including a C-U base pair. Biochemistry. 2000;39:7074–7080. doi: 10.1021/bi000018m. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Alfonta, L., Tian, F., Bursulaya, B., Uryu, S., King, D.S., Schultz, P.G. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, W., Gallivan, J.P., Zhang, Y., Li, L., Lester, H.A., Dougherty, D.A. From ab initio quantum mechanics to molecular neurobiology: A cation-π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]