Abstract
microRNAs (miRNAs) are noncoding small RNAs that regulate gene expression at the translational level by mainly interacting with 3′ UTRs of their target mRNAs. Archived formalin-fixed paraffin-embedded (FFPE) specimens represent excellent resources for biomarker discovery. Currently there is a lack of systematic analysis on the stability of miRNAs and optimized conditions for expression analysis using FFPE samples. In this study, the expression of miRNAs from FFPE samples was analyzed using high-throughput locked nucleic acid-based miRNA arrays. The effect of formalin fixation on the stability of miRNAs was also investigated using miRNA real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. The stability of miRNAs of archived colorectal cancer FFPE specimens was characterized with samples dating back up to 10 yr. Our results showed that the expression profiles of miRNAs were in good correlation between 1 μg of fresh frozen and 1–5 μg of FFPE samples (correlation coefficient R 2 = 0.86–0.89). Different formalin fixation times did not change the stability of miRNAs based on real-time qRT-PCR analysis. There are no significant differences of representative miRNA expression among 40 colorectal cancer FFPE specimens. This study provides a foundation for miRNA investigation using FFPE samples in cancer and other types of diseases.
Keywords: microRNA, FFPE, expression, microarray
INTRODUCTION
miRNAs are naturally occurring small, single-stranded, noncoding RNAs that mediate gene expression at the post-transcriptional and translational levels in both plants and animals (Ambros 2004; Bartel 2004). The total numbers of identified miRNAs and our understanding of miRNA biology are still increasing (Ruby et al. 2006).
The first miRNA, lin-4, was initially discovered over a decade ago in Caenorhabditis elegans and controls the timing and progression of the nematode life cycle (Lee et al. 1993; Feinbaum and Ambros 1999; Reinhart et al. 2000). However, the importance of this research was not appreciated until recently with the discoveries of hundreds of miRNAs in worm, fly, and mammalian genomes (Lagos-Quintana et al. 2001, 2003; Brennecke et al. 2003; Berezikov et al. 2005). Many miRNAs are evolutionarily conserved, further indicating that these miRNAs are involved with essential biological processes such as development (Sempere et al. 2003; Baskerville and Bartel 2005).
miRNAs are derived from endogenous genes that are initially transcribed as large RNA transcripts that are capped and polyadenylated. RNA polymerase II has been shown to be involved in their transcription in the nucleus to form large pre-miRNA transcripts (Basyuk et al. 2003; Cai et al. 2004). miRNAs are then further processed to become mature miRNAs through a complicated process. This involves a large protein complex, which includes RNase III Drosha and its cofactor Pasha (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004). Large miRNA transcripts are processed by Drosha into 70 nucleotide (nt) hairpin pre-miRNAs that are transported by exportin-5 to the cytoplasm (Bohnsack et al. 2004). Mature miRNAs 18–27 nt in length are processed from pre-miRNAs by another RNaseIII nuclease Dicer (Lee et al. 2002).
In the past, most studies have focused on protein coding genes and their regulation at the transcriptional level. Over the past few years, however, post-transcriptional and translational controls regulated by naturally occurring noncoding RNAs have emerged as an interesting field of research. Translational control mediated by miRNAs provides the cell with a more precise, immediate, and energy-efficient way of controlling the expression of proteins, since it can induce rapid changes in protein synthesis without the need for transcriptional activation and subsequent mRNA processing steps. In addition, translational control provides the cell with greater flexibility in responding to various cytotoxic stresses. The main function of miRNAs is to repress gene expression at the translational level by binding to the 3′ UTR of the messenger RNA. Although the exact function of most of the newly discovered miRNAs is just emerging, their ability to regulate cell proliferation and cell death has been recently shown (Chan et al. 2005). We, and others, have also shown that miRNAs may be good biomarkers for cancer diagnosis and prognosis (Calin et al. 2005; Cummins et al. 2006; Nakajima et al. 2006; Xi et al. 2006a,b). Our group first demonstrated that some miRNAs are key components of the p53 tumor suppressor gene function (Xi et al. 2006b), and over 46% of putative promoters of miRNAs contain p53 binding sequences; this was further confirmed independently by several groups in recent reports (Chang et al. 2007; He et al. 2007; Tarasov et al. 2007).
However, we realized that several issues still remain regarding the quality and the expression profiles of miRNAs in formalin-fixed paraffin-embedded (FFPE) specimens. One is the effect of formalin fixation on miRNA, and the second is the quality and degree of correlation of the miRNA global expression profile between FFPE and fresh frozen samples. To a large extent, these issues have not been systematically investigated.
The goal of this study was to systematically investigate the miRNA expression profiles between FFPE samples and fresh frozen samples using miRNA array analysis. In archived clinical FFPE specimens, the length of formalin fixation varies widely among different hospitals. Therefore, the effect of formalin fixation on miRNA expression was also investigated. Several miRNA expression analysis approaches have been developed in recent years, including Northern blot (Cummins et al. 2006), polymerase chain reaction (PCR)-based (Chen et al. 2005), serial analysis of gene expression (SAGE)-based (Cummins et al. 2006), and microarray-based (Babak et al. 2004; Calin et al. 2004; Nelson et al. 2004; Castoldi et al. 2006; Wang et al. 2007) analyses. miRNA expression analysis on FFPE samples has been reported previously using different platforms (Nelson et al. 2004; Wang et al. 2007). We chose the locked nucleic acid (LNA)-based miRNA array for this study mainly because, at the time, this was one of the most sensitive miRNA array platforms for miRNA expression analysis and it was available to us (Castoldi et al. 2006). Unlike several reported miRNA array expression studies using regular DNA capture oligo probes, it does not require RNA fractionation or enrichment of the input RNA as a starting material. LNA-modified capture probes provide Tm-normalized hybridization conditions for all miRNAs and also increase the hybridization specificity with single nucleotide resolution. Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis was also applied to access the stability issues of miRNAs in archived colon cancer FFPE specimens.
RESULTS
Effect of formalin fixation on miRNA expression
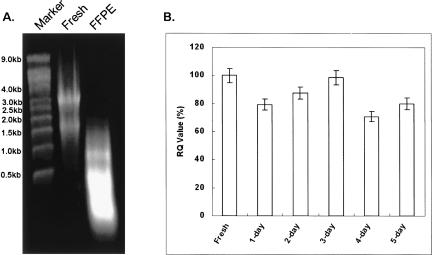
It is well known that after surgery, clinical samples are fixed in formalin for various amounts of time in different hospitals. The preserved tissue specimens are perfectly fine for traditional histopathological analysis. To study the potential impact of formalin fixation on miRNA stability, fresh mouse liver tissue was subjected to buffered formalin fixation from 1 d up to 5 d. Figure 1 represents a schematic flow diagram of sample processing and miRNA analysis from mouse liver. A representative RNA image is shown in Figure 2A (lane 1, RNA ladder; lane 2, fresh frozen sample; lane 3, FFPE sample). RNAs isolated from different days have similar gel image appearances. In general, total RNA yield from one tissue slice (2 × 4 cm2, 10 μm thick) is at least 10 μg, and it is sufficient for both real-time qRT-PCR and LNA-based miRNA array analysis. Our results showed that the expression of miRNA was relatively stable and not affected by formalin fixation based on real-time qRT-PCR analysis (Fig. 2B).
FIGURE 1.
Schematic illustrations of sample processing and miRNA expression analysis from mouse liver tissue.
FIGURE 2.
Effect of formalin fixation on miRNA expression. (A) Representative RNA agarose gel image (lane 1, RNA ladder; lane 2, fresh frozen sample; lane 3, FFPE sample). (B) The expression of hsa-miR-16 and RUN6B was analyzed using real-time QRT-PCR analysis from formalin-fixed mouse liver samples.
Correlation of miRNA expression between fresh frozen and FFPE samples
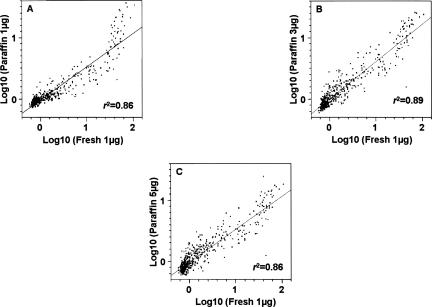
To determine the degree of correlation of miRNA expression profiles of FFPE samples with fresh frozen samples, the miRNA expression profiles of FFPE samples were compared to that of fresh frozen samples. In order to determine the optimal concentration of total RNAs from FFPE samples for miRNA expression analysis using the LNA-based array, 1–5 μg of total RNAs from FFPE samples were used for comparison purposes. In general, our results showed a good correlation of miRNA expression profiles of the FFPE samples compared to the fresh frozen samples (Fig. 3), with correlation coefficients ranging from R 2 = 0.86 to 0.89. This result was further strengthened by analyzing the miRNA expression profiles among different amounts of FFPE samples with R 2 = 0.90–0.96 (Fig. 4).
FIGURE 3.
Correlation analysis of the global miRNAs expression between fresh frozen and FFPE samples. (A–C) The comparisons of different input RNA amounts (1, 3, and 5 μg) of FFPE samples versus 1 μg fresh frozen sample.
FIGURE 4.
(A–C) Correlation analysis of the global miRNAs expression between different amounts of total RNA input (1, 3, and 5 μg) from FFPE samples.
Correlation of mRNA transcripts expression between fresh frozen and FFPE samples
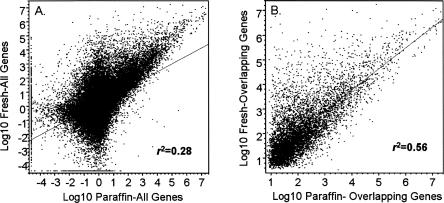
To determine the degree of correlation of mRNA expression profiles of FFPE samples to that of fresh frozen samples, in parallel, we also investigated the mRNA expression profiles between FFPE samples versus fresh frozen samples. Results in Figure 5 show a poor correlation of mRNA profiles of FFPE samples to that of fresh frozen samples with a correlation coefficient of R 2 = 0.28 (Fig. 5A; all genes) and R 2 = 0.56 (Fig. 5B; overlapping genes).
FIGURE 5.
Correlation analysis of the messenger RNA expression between fresh frozen and FFPE samples. (A) Global mRNAs expression comparison. (B) Overlapping mRNAs expression comparison.
Stability of miRNAs from FFPE clinical samples
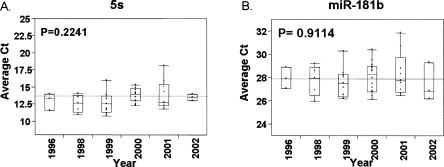
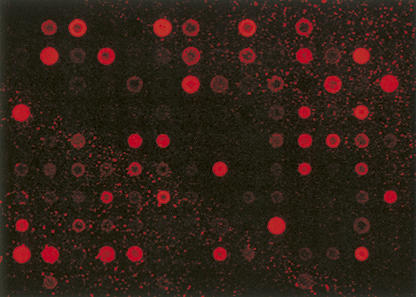
To further investigate the stability of miRNAs in FFPE samples over time, RNA samples were isolated from colon cancer FFPE samples up to 10 yr old. Due to the lack of paired fresh frozen clinical specimens for a global correlation study, instead real-time miRNA qRT-PCR analysis was used to quantify the expression levels of miRNAs. Based on the real-time qRT-PCR analysis, the expression of miRNAs was quite stable (Fig. 6). We also performed the miRNA array analysis on this sample, and Figure 7 shows a representative image with good quality signals of miRNA expression from a 1996 colorectal cancer FFPE sample.
FIGURE 6.
Quantitative real-time PCR expression analysis of 5S (A) and miR-181b (B) from archived clinical colon cancer specimens. The line represents the trend of median expression levels of miRNAs in FFPE samples from different years. P-value was calculated by Wilcoxon's signed-rank sum test.
FIGURE 7.
Representative miRNA array image from a 10-yr-old FFPE colon cancer sample.
DISCUSSION
miRNAs hold great promise as potential biomarkers for cancer diagnosis and prognosis as well as novel targets. To realize such potential, expression analysis of miRNA expression using archived FFPE samples is one of the key components. The effect of formalin fixation and paraffin embedding on miRNA stability and the expression level in clinical samples has not been systematically investigated. In this study, the expression profiles of miRNAs were compared using fresh frozen and corresponding FFPE samples using one of the currently available LNA-based miRNA arrays. The advantages of the LNA-based miRNA array are its high specificity and sensitivity toward miRNAs and that it does not require any sample enrichment or fractionation (Castoldi et al. 2006). This was achieved by the use of mixed DNA/LNA-modified capture probes (Nielsen et al. 2003). The elevated thermal duplex stability allows a normalized melting temperature of the capture probes by modifying LNA content and oligonucleotide length.
The effect of formalin fixation on miRNA expression was first evaluated using real-time qRT-PCR analysis. Based on our results, the expressions of miRNAs were not significantly affected by formalin fixation for up to 5 d (Fig. 2B). This demonstrates the clear advantage of using miRNAs for biomarker discovery compared to mRNAs. Although the exact reason for this better stability of miRNAs is not clear, we speculate that the small size of the miRNAs, as well as the secondary structure, may contribute to the stability.
We further demonstrated a high correlation of global miRNA expression profiles between FFPE samples and corresponding fresh frozen samples using LNA-based miRNA array analysis (Figs. 3, 4). Our data further confirmed the results of Nelson et al. (2004) in which FFPE samples can be used for miRNA profiling analysis. These results provide a solid foundation for using miRNAs as biomarkers and using FFPE samples in target discovery studies.
In parallel, we also compared the expression profiles of mRNA transcripts using a Codelink oligo array in which we used both fresh frozen and FFPE samples. In contrast to the good correlation of miRNA results, we showed that the correlation between these two types of samples was only R 2 = 0.56 in the best situation, suggesting that a large portion of mRNA transcripts were not able to generate a good signal due to degradation (Fig. 5). These results, taken together, suggest that miRNAs may be a better choice for expression profiling analysis than mRNA when using FFPE samples.
Furthermore, the stability of the miRNAs from archived colorectal cancer specimens was characterized using real-time qRT-PCR analysis. Our results indicate that the expression of miRNA is quite stable over a 10-yr period (Fig. 6). Because of the lack of paired clinical archived fresh frozen specimens, we could not perform a direct comparison of miRNAs expression profiles between fresh frozen and corresponding FFPE clinical specimens. Nonetheless, we could still address this critical issue based on this and data from paired mouse liver samples. It is still convincing that miRNAs are quite stable over time. This notion was further supported by analyzing the miRNA expression using 10-yr-old FFPE colon cancer samples with an LNA-based miRNA array, and good quality signals were detected in these samples (Fig. 7).
In summary, in this study, we systematically investigated the miRNA expression profiles of FFPE samples with LNA-based miRNA array anlysis. We concluded that the expression of miRNAs from FFPE samples was in good correlation with fresh frozen samples and that formalin fixation did not significantly alter the stability of the miRNAs. Profiling miRNAs using archived FFPE specimens is feasible, and it may hold great promise for biomarker and novel target discovery for cancer research and other diseases.
MATERIALS AND METHODS
Sample preparation
One adult mouse was sacrificed by cervical separation. The entire liver was quickly removed and rinsed with ice-cold 1× PBS three times (Fig. 1). One sixth of the liver tissue was snap-frozen in liquid nitrogen. The rest was split into five pieces and then fixed in 10% buffered formalin (Fisher Scientific) from 1 d to 5 d. A portion of the 3-d formalin-fixed liver tissue was followed by the routine clinical sample preparation protocol of dehydration and paraffin embedding. Forty colorectal cancer FFPE specimens were obtained from the Department of Surgery, Institute of Gastroenterology, Tokyo Women's Medical University, Japan. All patients signed detailed consent forms before the study was conducted. The experimental samples were pre-made on standard slides with 10 μm thick FFPE tissues using the standard method by the Department of Pathology, Tokyo Women's Medical University, Japan.
RNA isolation
For snap-frozen and formalin-fixed samples, a tissue homogenizer (OMNI International) was utilized to disrupt cells in TRIzol reagent (Invitrogen). Total RNA was isolated from 500 μL aliquots. For FFPE samples, the blocks were dissected into 10 μm thick pieces, and one tissue slice was placed in 1.5 mL nuclease-free microcentrifuge tubes. One milliliter of Xylene (EMD) was added for deparaffinization with vortexing for 5 min at room temperature. Samples were left at 60°C for 3 min and then centrifuged at 14,000 rpm for 7 min at room temperature. Supernatants were removed, and 1 mL of 100% ethanol was added with vortexing for 7 min. This ethanol washing step was repeated twice. Samples were air dried, and 360 μL of digestion buffer containing 30 mM Tris-HCl, 20 mM EDTA, 1% SDS (all from Sigma), and nuclease-free water were added to the tubes. Samples were homogenized and 40 μL of Proteinase K (Qiagen, Inc.) solution was added. Samples were incubated at 56°C until the tissue was completely digested (≤3 h). Subsequently, 2 volumes of TRIzol LS reagent (Invitrogen) were added and vortexed for 5 min at room temperature.
One hundred microliters of 1-bromo-3-chloropropane (BCP) solution (Molecular Research Center, Inc.) were added into the samples. After 2 min vortexing, the samples were incubated for 3 min at room temperature. The upper aqueous phase containing the RNA was transferred to a new tube after centrifuging at 14,000 rpm for 7 min. An equal volume (100 μl) of 100% isopropanol (Sigma) was added, and precipitation was performed at −80°C for 60 min. After the sample was centrifuged at 14,000 rpm for 7 min at 4°C, the supernatant was removed and 1 mL of 75% ethanol was added for washing. We repeated centrifuging and removing supernatant. The RNA pellet was air dried and eluted in 30 μL of nuclease-free water.
miRNA reverse transcription and TaqMan-based qRT-PCR analysis for formalin-fixed samples
Primer sets for specific reverse transcription including hsa-miR-16 and endogenous control RUN6B and the high-capacity cDNA Archive Kit were utilized following the manufacturer's protocol. All reagents and instruments were from Applied Biosystems Inc., except where otherwise indicated. Briefly, the reaction master mix containing 10× RT Buffer, 5× RT Primers, MultiScribe Reverse Transcriptase, RNase Inhibitor, and nuclease-free water was mixed with 20 ng of each total RNA sample. Mixtures were incubated for 30 min at 16°C, 30 min at 42°C, then 5 min at 85°C. qRT-PCR was carried out using Applied Biosystems 7500 Real-Time PCR System. The PCR master mix containing TaqMan 2× Universal PCR Master Mix (No Amperase UNG), 10× TaqMan assay, and RT products in 20 μL volume were processed as follows: 95°C for 10 min and then 40 cycles of 95°C for 15 sec and 60°C for 60 sec (n = 3). The signal was collected at the endpoint of every cycle. The fresh sample also was processed at the same time.
miRNAs expression analysis using LNA miRNA array
One microgram of total RNA from a snap-frozen sample and 1 μg, 3 μg, and 5 μg total RNA from FFPE samples were labeled using the miRCURY LNA microRNA Array labeling kit (Exiqon Inc.). All reagents used for this part were from Exiqon unless specifically mentioned. In brief, 10 μL 2.5× labeling buffer, 2 μL Hy5 fluorescent label, 5 μL labeling enzyme, and 1 μL loading control were incubated with different amounts of total RNA samples in a 25 μL volume at 0°C for 60 min in the dark and then at 65°C for 15 min to stop the labeling reaction. The labeled samples were mixed with equal volumes of 2× hybridization buffer at 95°C for 3–5 min and cooled down in seconds. The samples were loaded on the miRCURY LNA microRNA Array (Exiqon Inc.), which were precovered with 50 μL volume plastic chambers (Grace Bio-Labs). After the loading port were sealed, the slides were placed in prehumidified hybridization slide chambers and incubated at 60°C for 16 h. After we removed the plastic chambers, the slides were washed in three steps. The first step was to immediately soak the slides in prewarmed 2× salt buffer, containing 0.2% detergent solution, for 2 min. The second step was to rinse the slides with 1× salt buffer for 10 sec and then wash for 2 min in fresh 1× salt buffer. The third step was to use 0.2× salt buffer to wash the slides for 2 min. After they were washed, the slides were quick dried by centrifuging at 1000 rpm for 5 min. The scanning was performed by an Axon GenePix Professional 4200A microarray scanner (Molecular Devices Corp.). The images were gridded and analyzed by ImaGene 7.0 (BioDiscovery Inc.). Per chip normalization was performed by dividing each gene's measurement by the 50th percentile of all measurements in the single array. All normalized data were exported for further analysis. One 1996 clinical FFPE sample was also processed for the LNA miRNA array with the above procedure.
mRNA transcript expression analysis using mRNA microarray
CodeLink Mouse Whole Genome Bioarray (GE Healthcare/Amersham Biosciences), containing ∼36,000 gene probes, was used to generate gene expression profiles of mRNAs from snap-frozen and FFPE samples. The RNA samples were treated with DNase to avoid potential DNA contamination. Fifteen microliters of DNase treatment mix (2 μL RQ1 RNase-Free DNase, 11 μL RQ1 DNase 10× reaction buffer, and 2 μL RNasin Plus RNase inhibitor; Promega) were incubated with each sample in 100 μL volume at 37°C for 30 min. We then added 115 μL ultrapure phenol:chloroform:isoamyl alcohol (25:24:1, Invitrogen Inc.). After 2 min of vortexing, samples were centrifuged at 14,000 rpm for 7 min. The upper aqueous phase was carefully taken out and mixed with 2 volumes of 100% ethanol and 1/10 volume of 3M sodium acetate (pH 5.5) (Ambion Inc.). Samples were kept at room temperature for 15–30 min, then centrifuged at 14,000 rpm for 7 min. After we removed the supernatant, 1 mL of 75% ethanol was added for washing. We repeated the centrifuging and then air dried the pellets. The samples were dissolved in nuclease-free water. A NanoDrop ND-100 spectrophotometer (NanoDrop Technologies) was used to determine the concentration for all samples.
Double-stranded cDNAs were generated using 2 μg total RNA from both fresh frozen and FFPE samples. All reagents and protocols were provided by GE Healthcare/Amersham Biosciences unless otherwise indicated. After purification, the double-stranded cDNAs were used as templates to generate cRNA via an in vitro transcription reaction using T7 RNA polymerase and biotin-11-UTP (Perkin-Elmer). Biotin-labeled cRNA (10 μg) was fragmented and hybridized to the Mouse Whole Genome Bioarray. The arrays were washed and stained with Cy5-streptavadin. After they were washed, the dried slides were scanned by an Axon GenePix Professional 4200A microarray scanner under Genepix Pro 5.1 software (Molecular Devices Corp.). The images were gridded by Codelink 4.1 software (GE Healthcare/Amersham Biosciences) and exported to GeneSpring Software 7.2 (Agilent). Per chip normalization was done by dividing each gene's measurement by the 50th percentile of all measurements in the single array. Normalized data were exported for subsequent analysis.
miRNA reverse transcription and SYBR Green-based qRT-PCR analysis for clinical FFPE samples
mirVana qRT-PCR Primer Sets (Ambion Inc.) for miRNA specific reverse transcription including hsa-miR-181b endogenous control 5S rRNA were utilized according to the manufacturer's protocol. Briefly, the reaction master mix containing mirVana 5× RT buffer, 1× mirVana RT primer, ArrayScript Enzyme Mix, and nuclease-free water were mixed with 20 ng of each total RNA sample. These mixtures were incubated for 30 min at 37°C, then 10 min at 95°C. qRT-PCR was carried out using the Applied Biosystems 7500 Real-Time PCR System and mirVana qRT-PCR miRNA Detection Kit (Ambion Inc.). The PCR master mix containing mirVana 5× PCR buffer (with SYBR Green I), 50× ROX, SuperTaq Polymerase, mirVana PCR primers, and RT products were processed as follows: 95°C for 3 min and then 40 cycles of 95°C for 15 sec and 60°C for 35 sec (n = 3). The signal was collected at the endpoint of every cycle.
Statistical analysis
Statistical analysis was performed using the JMP 6.0 software (SAS Institute Inc.). All normalized data from snap-frozen and FFPE samples were analyzed to determine the correlation coefficient of expression. Wilcoxon's signed-rank sum test was employed to analyze the miRNA expression difference among clinical archived samples from different years. P-values of <0.05 were interpreted as statistically significant.
ACKNOWLEDGMENTS
We appreciated the critical reviews of our manuscript from Dr. Mikkel Noerholm of Exiqon Inc. This study was supported in part by NIH1R21CA114043 (J.J.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.642907.
REFERENCES
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Babak, T., Zhang, W., Morris, Q., Blencowe, B.J., Hughes, T.R. Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville, S., Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuk, E., Suavet, F., Doglio, A., Bordonne, R., Bertrand, E. Human let-7 stem–loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–6597. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov, E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H.A., Cuppen, E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Bohnsack, M.T., Czaplinski, K., Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Cai, X.Z., Hagedorn, C.H., Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Liu, C.G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C.D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Ferracin, M., Cimmino, A., Di Leva, G., Shimizu, M., Wojcik, S.E., Iorio, M.V., Visone, R., Sever, N.I., Fabbri, M., et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Castoldi, M., Schmidt, S., Benes, V., Noerholm, M., Kulozik, A.E., Hentze, M.W., Muckenthaler, M.U. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.A., Krichevsky, A.M., Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang, T.C., Wentzel, E.A., Kent, O.A., Ramachandran, K., Mullendore, M., Lee, K.H., Feldmann, G., Yamakuchi, M., Ferlito, M., Lowenstein, C.J., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Ridzon, D.A., Broomer, A.J., Zhou, Z., Lee, D.H., Nguyen, J.T., Barbisin, M., Xu, N.L., Mahuvakar, V.R., Andersen, M.R., et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins, J.M., He, Y., Leary, R.J., Pagliarini, R., Diaz L.A., Jr, Sjoblom, T., Barad, O., Bentwich, Z., Szafranska, A.E., Labourier, E., et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B.J., Plasterk, R.H.A., Ketting, R.F., Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Feinbaum, R., Ambros, V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans . Dev. Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- Gregory, R.I., Yan, K.P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- He, L., He, X., Lim, L.P., de Stanchina, E., Xuan, Z., Liang, Y., Xue, W., Zender, L., Magnus, J., Ridzon, D., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., Tuschl, T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J.J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Nakajima, G., Hayashi, K., Xi, Y., Kudo, K., Uchida, K., Takasaki, K., Ju, J. Noncoding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics & Proteomics. 2006;3:311–324. [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Nielsen, J.T., Stein, P.C., Petersen, M. NMR structure of an α-L-LNA:RNA hybrid: Structural implications for RNase H recognition. Nucleic Acids Res. 2003;31:5858–5867. doi: 10.1093/nar/gkg800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., Sokol, N.S., Dubrovsky, E.B., Berger, E.M., Ambros, V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Tarasov, V., Jung, P., Verdoodt, B., Lodygin, D., Epanchintsev, A., Menssen, A., Meister, G., Hermeking, H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G(1)-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ach, R.A., Curry, B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, Y., Formentini, A., Chien, M., Weir, D., Russo, J., Ju, J., Kornmann, M., Ju, J. Prognostic values of microRNAs in colorectal cancer. Biomarker Insights. 2006a;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- Xi, Y., Shalgi, R., Fodstad, O., Pilpel, Y., Ju, J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 2006b;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]