Abstract
A significant amount of literature was dedicated to hypotheses concerning the origin of ancient introns and exons, but accumulating evidence indicates that new exons are also constantly being added to evolving genomes. Several mechanisms contribute to the creation of novel exons in metazoan genomes, including whole gene and single exon duplications, but perhaps the most intriguing are events of exonization, where intronic sequences become exons de novo. Exonizations of intronic sequences, particularly those originating from repetitive elements, are now widely documented in many genomes including human, mouse, dog, and fish. Such de novo appearance of exons is very frequently associated with alternative splicing, with the new exon-containing variant typically being the rare one. This allows the new variant to be evolutionarily tested without compromising the original one, and provides an evolutionary strategy for generation of novel functions with minimum damage to the existing functional repertoire. This review discusses the molecular mechanisms leading to exonization, its extent in vertebrate genomes, and its evolutionary implications.
Keywords: alternative splicing, exonization
INTRODUCTION
The human genome contains about 200,000 exons (Lander et al. 2001), a number similar to that found in the mouse and other mammalian genomes (Waterston et al. 2002). This number is not static, but rather being constantly changed through a dynamic evolutionary process in which exons are newly created and lost. New exons were reported to arise from gene duplication, exon duplication (Kondrashov and Koonin 2001), and other mechanisms. This review, however, focuses on the unique process of exonization by exaptation (Brosius and Gould 1992), where genomic sequences that did not originally function as exons are adopted into exonic, coding sequences.
In 1994, Makalowski and coworkers described 17 events in which intronic transposed elements were inserted within the coding regions of human genes (Makalowski et al. 1994). A further analysis revealed that these elements were incorporated via splicing, i.e., their sequences contained splice sites leading to their recognition as exons (Makalowski et al. 1994; Makalowski 2000). Following the sequencing of the human genome as well as millions of expressed sequence tags (ESTs), hundreds of additional such events were found (Nekrutenko and Li 2001; Sorek et al. 2002). With the accumulation of many recently sequenced vertebrate genomes, reports on exonizations become more abundant and now include thousands of new exons in rodents (Wang et al. 2005), dogs (Wang and Kirkness 2005), and multiple other vertebrate genomes such as cow and fish (Alekseyenko et al. 2007; Krull et al. 2007).
MOLECULAR MECHANISMS LEADING TO EXONIZATION
The molecular mechanisms leading to exonization of intronic sequences were studied in detail for Alu elements. These elements are primate-specific retrotransposons that appear in the human genome over one million times and constitute >10% of it (Lander et al. 2001). They are abundant in intergenic regions and in introns, and are frequently exonized creating new, primate-specific exons (Sorek et al. 2002).
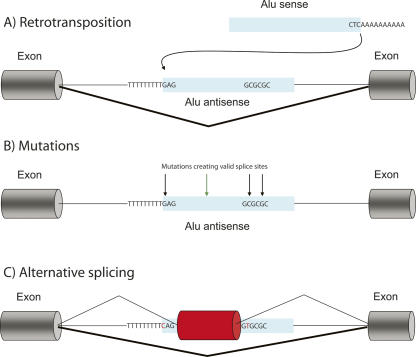
The consensus Alu sequence carries multiple sites that are similar, but not identical, to real splice sites (denoted pseudosplice sites) (Makalowski et al. 1994; Sorek et al. 2002) (Fig. 1). Specifically, the antisense orientation of the Alu sequence contains a preceding poly-T stretch that mostly serves as the polypyrimidine tract required upstream of the 3′ splice site. This poly-T originates from the terminal poly-A existing in many retrotransposons, which might explain why so many detected exonization events are associated with mobile retroposed elements (see below). Following insertion of the Alu to an intron in the antisense orientation, mutations that change the pseudosplice sites to real splice sites occur, so that the splicing machinery now recognizes part of the Alu as a bona fide exon (Fig. 1).
FIGURE 1.
Schematic model for exonization of an Alu element. (A) Alu is inserted into introns of primate genes by retrotransposition. (B) During the course of evolution, mutations within pseudo -splice sites in the intronic Alu activate these sites (black arrows). Mutations changing splicing regulatory elements are also possible (green arrow). (C) Following these mutations, part of the Alu sequence is recognized as a new exon (“exonized”), and spliced into the transcript. Typically, the Alu-containing transcript is the minor splice form, as in most cases the created splice sites are weak. Most exonizations involve the antisense orientation of the Alu sequence, presumably because of the preceding long poly-T that serves as a strong poly-pyrimidine tract necessary for the 3′SS recognition.
Several studies have characterized the series of mutations needed to occur within an Alu element in order to have it give birth to a new exon (Lev-Maor et al. 2003; Sorek et al. 2004a; Krull et al. 2005). By comparing between exonized and nonexonized Alus, it was shown that the number of mutations leading to exonization is surprisingly small, and that these mutations are generally concentrated in the 3′ and the 5′ splice site regions (Lev-Maor et al. 2003; Sorek et al. 2004a). Other studies showed that mutations changing exonic splicing enhancers or silencers can also influence exonization (Lei et al. 2005; Lei and Vorechovsky 2005). Such understandings have enabled researchers to synthetically exonize an intronic Alu element by introducing the right mutations in its sequence (Lev-Maor et al. 2003).
The precise steps needed for exonization and function-gaining in primate-specific, Alu-derived exonized sequences were further explored by Schmitz and colleagues (Singer et al. 2004; Krull et al. 2005). Their studies show that the transposon insertion in the genome and its actual exonization can be events separated millions of years apart, and depend both on the formation of active splice sites and, in some cases, the formation of an open reading frame (ORF). For example, by sequencing DNA from 13 different primates, Singer et al. (2004) reconstructed the key events leading to the generation of an alternative 5′ exon in the human tumor necrosis factor receptor gene (p75TNFR). Following the genomic integration of Alu ∼50 million years ago, an A→G substitution led to the creation of an alternative ATG start codon in this Alu. However, the actual exonization was facilitated only 25 million years later, when a 7-base-pair (bp) deletion resulted in an uninterrupted ORF, and a C→T substitution created a functional 5′ splice site (Singer et al. 2004); thus, multiple, stepwise small changes enabled this functional exonization.
ASSOCIATION WITH ALTERNATIVE SPLICING
In principle, insertion of an exon (typically sized over 100 bp) in the middle of the coding region of a gene is expected to be deleterious, since, in most cases, the exon would interrupt the frame or introduce a premature stop codon (Sorek et al. 2004b). Nevertheless, thousands of exonization events were recorded in human, mouse, and other vertebrates. How, then, could such exonizations be tolerated? The answer lies in alternative splicing.
Newly born exons are alternatively spliced at a much higher frequency than old exons (Alekseyenko et al. 2007). Specifically, nearly all exonized Alu elements in the human genome are alternatively spliced (Sorek et al. 2002), as well as 87% of newly formed exons in rodents (Wang et al. 2005). In addition, newly created exons are spliced into the transcript at a low frequency, i.e., only a small fraction of the transcripts contain the new exon (Sorek et al. 2002; Modrek and Lee 2003; Alekseyenko et al. 2007). Thus, the original transcript remains intact, making the effect of the exon insertion neutral or only minorly deleterious (Xing and Lee 2006). This allows exonized sequences to increase the coding and regulatory versatility of the transcriptome but at the same time maintain the intactness of the original proteomic repertoire. As evolution proceeds, some of the tested exons might be fixed, and their expression then becomes more pronounced. Indeed, it was shown that inclusion levels of new exons become higher for older exonizations (Zhang and Chasin 2006), and that this increase of inclusion levels is correlated with mutations creating stronger splice sites (Sela et al. 2007).
What leads to the observed excess of alternatively spliced new exons over constitutively spliced ones? It is probable that pseudosplice sites that mutate into strong constitutive splice sites are selected against because of their interference with normal protein production. In contrast, mutational changes that create weak splice sites leading to alternatively spliced new exons are tolerated. Indeed, several genetic disorders were documented where a constitutively exonized sequence was inserted in the coding region of a human gene. For example, a G-to-C mutation in an Alu sequence within intron 3 of the ornithine delta-aminotransferase (OAT) gene led to the creation of a strong 5′ splice site, consequently causing a constitutive insertion of a novel exon between exons 3 and 4. The insertion caused a premature stop codon, which led to OAT deficiency (Mitchell et al. 1991). In a similar manner, mutations in the genes COL4A3 and GUSB resulted in constitutive exonizations of Alu sequences leading to Alport (Knebelmann et al. 1995) and Sly (Vervoort et al. 1998) syndromes, respectively. These examples indicate that constitutive exonizations are only rarely seen because they are strongly selected against.
Rarely, new exons could be constitutively spliced without having a deleterious effect. This could happen, for example, following gene duplication, where one of the duplicated copies maintains the original function while the other is free to obtain new exons. Such an event was documented for the bovine BCNT gene, where shortly after gene duplication in the ruminant lineage, one of the copies acquired an endonuclease domain from a ruminant-specific retrotransposon (Iwashita et al. 2003). Conversly, exonizations leading to alternatively spliced new exons could also lead to genetic disorders. This might happen in genes where the exact dosage of the transcript has to be strictly maintained. Such an event was documented when a C→T mutation at position 156 of an Alu sequence in intron six of the CTDP1 gene created a weak 5′ splice site, leading to a new alternative exon and resulting in CCFDN syndrome (Varon et al. 2003).
FUNCTIONAL CONSEQUENCES OF EXONIZATIONS
Although over 3400 new exons were predicted in humans (Sela et al. 2007), analysis of newly created coding cassette exons showed that most of them (over 79%) create frameshifts or introduce premature stop codons (Sorek et al. 2004b; Sela et al. 2007). This contrasts the behavior of human–mouse conserved cassette exons, only 27% of which destroy the reading frame (Sorek et al. 2004b). Moreover, although exonized transposable elements were detected in 4% of transcript data derived from cDNAs (Nekrutenko and Li 2001), they are found only in 0.1% of functional proteins in the Protein Data Bank (PDB) (Gotea and Makalowski 2006). This indicates that most exonized sequences are nonfunctional, and are tolerated in transcripts simply because they are alternatively spliced and the frequency of their inclusion is low.
Nevertheless, growing evidence indicates that a minority of exonized sequences can acquire function after a period of natural evolution. For example, events of exaptation of old transposable elements into functional proteins in humans were documented for the CAPN1, GZMA, and PTPN1 genes (Gotea and Makalowski 2006). Another example was described in the human RNA editing enzyme ADAR2, where exon 8 is a new Alu-derived primate-specific exon that is alternatively spliced in high inclusion levels (Lev-Maor et al. 2003) in a tissue-regulated manner (Gerber et al. 1997). The new exon is inserted in the catalytic domain of ADAR2, and, while the exon-containing variant has the same substrate specificity as the original one, it has an altered catalytic activity (Gerber et al. 1997). Interestingly, a mouse-specific exonization occurred at the same position in the mouse ADAR2 gene, suggesting an event of parallel evolution favoring the creation of such novel splice variant in two different lineages (Rueter et al. 1999).
Perhaps the most convincing example of a bona fide function acquired through exonization was recently described by Bejerano et al. (2006). This study examined one of the most evolutionarily conserved (“ultraconserved”) elements in mammalian genomes, which encompasses a coding cassette exon in the human PCBP2 gene and its flanking intronic regions. This sequence spans 223 bp that are 100% conserved between human, mouse, and rat. Bejerano and colleagues (2006) managed to trace back the origin of this exon to a short interspersed repetitive element (SINE) that was active in the ancestor of humans and fish and is still highly active in the Indonesian coelacanth “living-fossil” fish. Moreover, this same SINE (called LF-SINE) is the source for 16 additional coding exons that are highly conserved in tetrapods, 15 of which are alternatively spliced. The extreme conservation of these exons across vertebrate species is a strong indicator that they acquired a highly important function. This also implies that a large number of exons that we currently view as fixed, highly conserved exons across vertebrates were, in fact, exapted from an intronic sequence in the ancestral organism.
THE ROLE OF REPETITIVE SEQUENCES
As is well evident from the examples described in this review, repetitive elements, and specifically SINEs, are a major source for exonized sequences. Over 90% of new exons in humans are from repetitive sequences (totaling ∼3400 novel exons) (Sela et al. 2007), with the primate-specific Alu SINE responsible for 62% of these events (Zhang and Chasin 2006). Similarly, 65% of new exons in rodents are derived from repetitive elements (Zhang and Chasin 2006). In the canine genome, the SINEC_Cf repeats were shown to contribute multiple exons through exonization (Wang and Kirkness 2005). New exons originating from repeat sequences were also identified in cow and zebrafish genomes (Alekseyenko et al. 2007).
Although repetitive elements compose over 40% of the human genome (Lander et al. 2001), this number still does not explain their excessive overrepresentation (90%) in sequences creating new exons. A possible explanation for the propensity of SINEs in producing new exons is the existence of a long poly-A tail at the 3′ end of many SINE repeats. Intronic SINEs inserted in the antisense orientation to a transcribed gene will therefore carry a 5′ poly-T, which then serves as the poly pyrimidine tract (PPT) portion of the 3′ splice site during exonizations (Fig. 1). In fact, the PPT is the largest constraint on the de novo creation of functional splice sites, which, apart from the obligate GT/AG nucleotides, are poorly defined. Accordingly, 85% of Alu exonizations are formed from Alus found in the antisense orientation (Sorek et al. 2002), as well as all ancient LF-SINE exonizations (Bejerano et al. 2006). In mouse, the MIR, B1, B2, and B4 repeats are biased toward exonization of their antisense strand (Krull et al. 2007; Sela et al. 2007). SINEC_Cf exonizations in the dog genome also predominantly occur in the antisense orientation and utilize the reverse poly-A as the PPT of their 3′ splice sites (Wang and Kirkness 2005). Therefore, it appears that the inherent poly-A tail of SINE repeats is one of the most important features facilitating their frequent exonizations.
Despite the dominance of repetitive elements in the production of new exons, exonization of nonrepetitive intronic sequences occurs in nonnegligible rates (Alekseyenko et al. 2007). It was recently shown that such purely intronic sequences can be recruited into new exons by mutations creating functional splice sites, similar to the process shown to occur in repeats (Alekseyenko et al. 2007). Similarly, cases of exonization were documented where one of the splice sites originated from a SINE, and the other was recruited from a nearby, nonrepeat intronic sequence (Sorek et al. 2002). Thus, exonization by mutational shift into splice sites seems to be a general mechanism, which does not necessarily depend on the origin of the exonized sequence.
RNA EDITING-MEDIATED EXONIZATION
There is now evidence that exonization events can rely on mechanisms other than mutational changes in splice sites (Lev-Maor et al. 2007) (Fig. 2). One such mechanism is A-to-I RNA editing, which alters the nucleotide sequence of RNA transcripts from that encoded by genomic DNA. This single-nucleotide editing process, catalyzed by enzymes of the ADAR family, changes selected adenosine residues to inosine, a nucleotide recognized by most biological machineries as a guanosine (G) nucleotide. Editing targets are typically located within double-stranded RNA (dsRNA), to which the ADAR enzymes bind (Bass 2002).
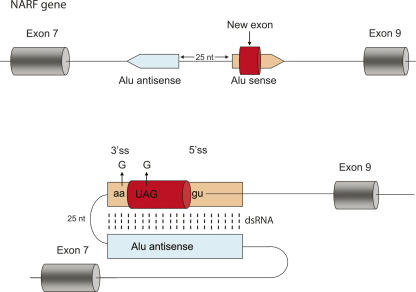
FIGURE 2.
The birth of an exon through RNA editing (Lev-Maor et al. 2007). Shown is a schematic illustration of the genomic region spanning exons 7–9 of the human NARF gene (not to scale). Exons are depicted as cylinders. The Alu element that is the source of the new exon is orange; an intronic, antisense orientation Alu sequence (light blue) is 25 bp upstream of the exonized Alu. Sense and antisense Alus fold to form a double-stranded RNA (dsRNA) secondary structure, thus allowing RNA editing to take place (lower panel). RNA editing changes an AA dinucleotide into a functional AG 3′ splice site and also changes a UAG stop codon into a UGG Trp codon. Thus, RNA editing leads to the creation of a new functional exon.
A recent study (Lev-Maor et al. 2007) describes how RNA editing can lead to exonization in the nuclear prelamin A recognition factor (NARF), a protein that interacts with the carboxyterminal tail of prelamin A and localizes to the nuclear lamina (Barton and Worman 1999). Exon 8 in the NARF gene is derived from a recently exonized Alu sequence, and is alternatively spliced in a tissue-dependent manner, being eightfold more abundant in brain tissue than in skeletal muscle (Lev-Maor et al. 2007). The intron flanking this exon on the upstream side ends by AA, a 3′ splice site that usually cannot be recognized by the spliceosome. However, the second A is changed by RNA editing to AI acting as a valid AG splice site, therefore enabling exonization. In addition, the sequence of this new exon contains an in-frame TAG stop codon that is efficiently edited to TGG (tryptophane) to keep the reading frame intact. A key to these editing events is the existence of another Alu element found 25 bp upstream to the exonized Alu in the reverse orientation (Fig. 2). This Alu provides the template for creation of the Alu–Alu dsRNA that is required for RNA editing (Lev-Maor et al. 2007). Thus, RNA editing leads to the exonization of an otherwise silent intronic sequence. This demonstrates how exonizations can be facilitated by molecular biological mechanisms that were originally designed to function in other processes.
EVOLUTIONARY SIGNIFICANCE OF EXONIZATION
In contrast to bacterial genomes that are very compact (and intronless), many eukaryotic genomes display considerable size plasticity. The human genome is 10% larger than the mouse genome, and eight times larger than the Fugu (pufferfish) genome, although these three genomes have a similar number and repertoire of genes (Aparicio et al. 2002; Waterston et al. 2002). Much of the size differences can be attributed to insertions of lineage-specific transposable elements in introns and intergenic regions (Lander et al. 2001; Waterston et al. 2002). The vast majority of this additional DNA is nonfunctional; in fact, comparisons between the human and other mammalian genomes led to the conclusion that only ∼5% of the human genome is under purifying selection (Waterston et al. 2002). However, it seems that despite the heavy burden that the additional DNA imposes on the organism (both in terms of excessive DNA replication and the need to transcribe excess intronic RNA), this constant addition of DNA to the genome is tolerated.
Why would an organism “allow” such expansion of its genome? The answer to this question is probably complex, but may reside, in part, in the benefits offered by the possibilities of exaptation. Exaptation is an evolutionary event where a genomic element is adapted to a function different than its original (Brosius and Gould 1992). Exonization, being a private case of exaptation, holds a huge evolutionary advantage: the new exon is almost always alternatively spliced (Sorek et al. 2002; Wang et al. 2005), leaving the original form intact and hence usually not harmful (Xing and Lee 2006). Therefore, the new exon could be evolutionarily “tested” without deleterious consequences to the organism. Moreover, each coding exonization event leads to an insertion of over 100 novel nucleotides into the protein coding region, a change several-fold more radical than a single nucleotide substitution. Using the exonization process, evolution can therefore seek new radical solutions with minimal jeopardy to the existing ones (Gilbert 1978; Xing and Lee 2006), and although the vast majority of these evolutionary “experiments” are not expected to be advantageous, a minority would eventually be fixed and acquire function (Bejerano et al. 2006; Gotea and Makalowski 2006). This may partially explain the observed genome expansion tolerance in many intron-containing organisms: the more DNA residing in introns, the more chance there is for the occurrence of splice-like sites and subsequent advantageous exonizations.
In this light, there is also an evolutionary advantage in keeping the basic splice signals relatively simple, so that a random sequence could easily be mutated into the right combination of 5′ and 3′ splice sites leading to exonization. Indeed, in the compact genome of the yeast Saccharomyces cerevisiae, where exonization is not thought to play an evolutionary role, the splice signals are much more defined than those of mammals (Ast 2004).
Exonizations, therefore, seem to be a highly efficient way to try and ultimately fix new functional modules with minimal risk for deleterious effects. Much of the observed exonizations are lineage specific. For example, 62% of new exons in human are associated with primate-specific Alu retroposons, and 28% of new exons in rodents are derived from rodent-specific SINEs (Zhang and Chasin 2006). Canine-specific elements were also documented as a source for dog-specific exonizations (Wang and Kirkness 2005). Therefore, it seems that in vertebrates, and especially mammals, the exonization mechanism is being used as a major source for accelerated, lineage-specific evolution, and is perhaps a key driving force to eventual speciations.
CONCLUSIONS
Despite the initial regard of exonization as a negligible phenomenon, evidence has been accumulating that the ability of vertebrate genomes to adopt foreign sequences as new exons is remarkable. We now know that exonizations are constantly reshaping the functional repertoire of genes in many genomes. Specific future attention is needed in order to uncover exonizations that lead to genetic disorders. Such events have been so far discovered serendipitously but are hypothesized to be much more frequently causing diseases than currently appreciated (Lev-Maor et al. 2003). The extent by which such harmful exonizations occur, as well as whether and how did species-specific exonizations directly contribute to speciation throughout the evolutionary history of vertebrates, are yet to be explored.
ACKNOWLEDGMENTS
The author thanks Zohar Biron, Galit Lev-Maor, and Gil Ast for stimulating discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.682507.
[Rotem Sorek was the winner of the 2006 RNA Society/Scaringe Young Science Award.]
REFERENCES
- Alekseyenko, A.V., Kim, N., Lee, C.J. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Ast, G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Barton, R.M., Worman, H.J. Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 1999;274:30008–30018. doi: 10.1074/jbc.274.42.30008. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano, G., Lowe, C.B., Ahituv, N., King, B., Siepel, A., Salama, S.R., Rubin, E.M., Kent, W.J., Haussler, D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Brosius, J., Gould, S.J. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.”. Proc. Natl. Acad. Sci. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A., O'Connell, M.A., Keller, W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Gotea, V., Makalowski, W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Iwashita, S., Osuda, N., Itoh, T., Sezaki, M., Oshima, K., Hashimoto, E., Kitagawa-Arita, Y., Takahashi, I., Musui, T., Hashimoto, K., et al. A transposable element-mediated gene divergence that directly produces a novel type bovine Bcnt protein including the endonuclease domain of RTE-1. Mol. Biol. Evol. 2003;20:1556–1563. doi: 10.1093/molbev/msg168. [DOI] [PubMed] [Google Scholar]
- Knebelmann, B., Forestier, L., Drouot, L., Quinones, S., Chuet, C., Benessy, F., Saus, J., Antignac, C. Splice-mediated insertion of an Alu sequence in the COL4A3 mRNA causing autosomal recessive Alport syndrome. Hum. Mol. Genet. 1995;4:675–679. doi: 10.1093/hmg/4.4.675. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Koonin, E.V. Origin of alternative splicing by tandem exon duplication. Hum. Mol. Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- Krull, M., Brosius, J., Schmitz, J. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- Krull, M., Petrusma, M., Makalowski, W., Brosius, J., Schmitz, J. Functional persistence of exonized mammalian-wide interspersed repeat elements. Genome Res. 2007 doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lei, H., Vorechovsky, I. Identification of splicing silencers and enhancers in sense Alus: A role for pseudoacceptors in splice site repression. Mol. Cell. Biol. 2005;25:6912–6920. doi: 10.1128/MCB.25.16.6912-6920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, H., Day, I.N., Vorechovsky, I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Levanon, E.Y., Paz, N., Eisenberg, E., Ast, G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W. Genomic scrap yard: How genomes utilize all that junk. Gene. 2000;259:61–67. doi: 10.1016/s0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- Makalowski, W., Mitchell, G.A., Labuda, D. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Mitchell, G.A., Labuda, D., Fontaine, G., Saudubray, J.M., Bonnefont, J.P., Lyonnet, S., Brody, L.C., Steel, G., Obie, C., Valle, D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: A role for Alu elements in human mutation. Proc. Natl. Acad. Sci. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Lee, C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Nekrutenko, A., Li, W.H. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Sela, N., Mersch, B., Gal-Mark, N., Lev-Maor, G., Hotz-Wagenblatt, A., Ast, G. Comparative analysis of transposed elements' insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, S.S., Mannel, D.N., Hehlgans, T., Brosius, J., Schmitz, J. From “junk” to gene: Curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., Graur, D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Lev-Maor, G., Reznik, M., Dagan, T., Belinky, F., Graur, D., Ast, G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004a;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Shamir, R., Ast, G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004b;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Varon, R., Gooding, R., Steglich, C., Marns, L., Tang, H., Angelicheva, D., Yong, K.K., Ambrugger, P., Reinhold, A., Morar, B., et al. Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat. Genet. 2003;35:185–189. doi: 10.1038/ng1243. [DOI] [PubMed] [Google Scholar]
- Vervoort, R., Gitzelmann, R., Lissens, W., Liebaers, I. A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- Wang, W., Kirkness, E.F. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Zheng, H., Yang, S., Yu, H., Li, J., Jiang, H., Su, J., Yang, L., Zhang, J., McDermott, J., et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Lee, C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Zhang, X.H., Chasin, L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl. Acad. Sci. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]