Abstract
Glycolipid transfer proteins (GLTPs) are small (24 kD), soluble, ubiquitous proteins characterized by their ability to accelerate the intermembrane transfer of glycolipids in vitro. GLTP specificity encompasses both sphingoid- and glycerol-based glycolipids, but with a strict requirement that the initial sugar residue be beta-linked to the hydrophobic lipid backbone. The 3D protein structures of GLTP reveal liganded structures with unique lipid binding modes. The biochemical properties of GLTP action at the membrane surface have been studied rather comprehensively, but the biological role of GLTP remains enigmatic. What is clear is that GLTP differs distinctly from other known glycolipid-binding proteins, such as nonspecific lipid transfer proteins, lysosomal sphingolipid activator proteins, lectins, lung surfactant proteins as well as other lipid binding/transfer proteins. Based on the unique conformational architecture that targets GLTP to membranes and enables glycolipid binding, GLTP is now considered the prototypical and founding member of a new protein superfamily in eukaryotes.
Keywords: GLTP, lipid transfer protein structure, membrane interaction motif, glycosphingolipid, HET-C2, ACD11, FAPP2, cerebroside, ganglioside, sphingomyelin, cholesterol, phosphatidylcholine
1. Historic Overview --- Early Insights
The first lipid transfer protein found to selectively transfer glycosphingolipids (GSLs) between membranes was discovered in spleen and was called cerebroside transfer protein [1,2]. Although purification to homogeneity was hampered by protein instability that developed at the final steps of purification, characterization of the soluble protein revealed a molecular weight of ∼24 kD and a pl of ∼9.0. Soon after, protein–mediated glycolipid transfer activities were found in other tissues including pig and bovine brain [3,4]. The application of different liquid chromatographic approaches in various combinations eventually enabled GLTP to be purified to homogeneity in stable form from porcine and bovine brain [5,6]. Nonetheless, because GLTP was relatively sparsely expressed and the purifications were laborious and time consuming, insights into the structural features of GLTP were slow to emerge. Most early studies focused on defining the selectivity of GLTP for different glycolipids [2,7-10], the mechanism by which GLTP enabled glycolipids to be transferred between membranes [2,5,7,11] and the parameters that affected the kinetics of glycolipid transfer between membranes by GLTP [4,12-14].
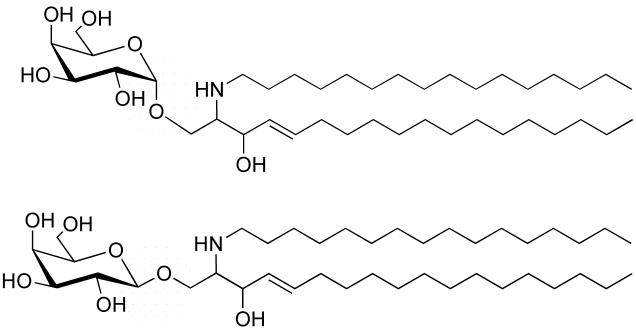
GLTP specificity was found to be broad encompassing both sphingoid-based and glycerol-based glycolipids in which the first sugar is β-linked to the nonpolar lipid moiety [8,9]. Figure 1 shows the structure galactosylceramide (GalCer) in both the alpha and beta configurations. While there is general agreement that high transfer rates are observed when glycolipids contain one or two sugars and that no transfer occurs with phosphoglycerides or neutral lipids, somewhat conflicting data have appeared with regards to how well GLTP can transfer anionic and complex neutral GSLs [2,7,8,10].
Figure 1.
N-Palmitoyl-galactosylceramide showing galactose linked in either alpha (upper) or beta (lower) configuration to the ceramide backbone. While the beta-linked form of GalCer (and GlcCer) are normal GSL constituents of mammals, the alpha-linked forms are not. The alpha-linked form of GalCer is found in Agelas maurifianus, a marine sponge. This glycolipid and its synthetic analog, KRN7000, have been used as specific activators of a subset of Natural Killer Cells known to be involved in the regulation of certain autoimmune diseases [118].
Even though early studies with porcine and bovine GLTP established the ability of the protein to selectively accelerate the intermembrane transfer of glycolipids in vitro, the mechanism of protein action remained poorly defined. Metz and Radin [2] reported that cerebroside transfer protein from bovine spleen bound small amounts of GalCer (∼4%), but also observed that the diffusion of the protein was reduced by mixing with liposomes or red cell ghosts that contained glycolipids. They speculated that the protein desorbs from the membrane surface as a protein/lipid complex that then rapidly dissociates in solution before reaching an acceptor membrane. Abe and Sasaki [5] proposed that porcine GLTP functions as a soluble intermediate that carries glycolipids between membranes, based on nondenaturing polyacryl-amide gel electrophoresis showing formation of a soluble GLTP/GalCer complex and Sephadex G-75 size exclusion chromatography showing no protein bound to vesicles containing glycolipid. However, total protein recovery from the size exclusion chromatography column was low and only 13 mol% of the porcine GLTP molecules acquired GalCer. Other attempts to confirm formation of soluble GLTP/glycolipid complexes resulted in conflicting data. Wong et al. [4] found that a substantial fraction (30−40%) of partially purified bovine brain GLTP coelutes with POPC:GalCer vesicles, but were unable to detect glycolipid binding by the protein. Brown et al. [6] reported weak association of bovine brain GLTP with pyrene-labeled glucosylceramide using fluorescence approaches, but found no evidence of protein binding to vesicles containing glycolipid. Sasaki and colleagues showed that porcine brain GLTP acquired pyrene-labeled GalCer from vesicles and formed a complex [12] but found no GLTP/glycolipid complex in the subphase beneath radiolabeled GalCer monolayers [11]. In all of these early studies, GLTP was isolated using different approaches from animal tissues known to contain various glycosphingolipids. Also, some difficulties were likely caused by the lengthy purification required for GLTP and instabilities that affected certain GLTP preparations [2,10].
The early challenges associated with GLTP, i.e. relatively low abundance in tissues and difficulties in achieving homogeneous stable GLTP preparations, limited the focus of GLTP research mostly to assessment of parameters that affected the kinetics of the GLTP-mediated transfer process. These analyses made use of two types of assays to assess protein activity. The glycolipid transfer process could be monitored using either radiolabeled glycolipid, which required separation of donor and acceptor membranes to quantify transfer [2,3] or using fluorescent glycolipid designed to produce a concentration-dependent change in emission signal response, enabling measurement of real-time transfer kinetics [4,12]. With the complementary assays, the effects of chemical agents on protein viability were investigated [4,12-14]. Studies of native and denatured porcine GLTP with sulfhydryl reagents under reducing and oxidizing conditions revealed three cysteines. One cysteine residue was relatively accessible suggesting a surface localization. The remaining two cysteine residues were postulated to form a disulfide bond required for maximal activity [13,14]. Competition analyses using various free sugar residues showed no interference with GLTP transfer activity [12]. In addition, the influence of the membrane environment containing the resident glycolipid was studied [4]. Rapid transfer of GlcCer by GLTP was observed only when the donor and acceptor phosphatidylcholine vesicles were both in the liquid-crystalline state [4]. Additional insights into these early studies of the 1980s are provided in several excellent and comprehensive reviews [15-18].
Despite the early challenges, the primary sequence was successfully determined by Edman degradation, revealing 208 amino acids in the porcine brain protein [19]. The sequence confirmed the presence of three cysteines, which had been postulated based on chemical reactivity to sulfhydryl reagents [13,14]. Cysteine 36 was found to react with iodoacetamide under nondenaturing conditions, while cysteines 112 and 176 presumably formed the intra-molecular disulfide reportedly needed for maximum transfer activity of glycolipid. Analyses of GLTP secondary structure by circular dichroism revealed high alpha-helical content (∼60%) along with ∼25% beta structure [14]. However, a comprehensive, detailed understanding of GLTP structure remained elusive.
2. Rebirth of GLTP Research
During the 1990s, interest in GLTP waned and nearly a decade passed without new advances. A major step forward occurred in 2000 with the successful molecular cloning of the full-length cDNA open reading frame from bovine and porcine gltp mRNA transcripts by bridge-overlapping extension PCR after successful production of partially coding cDNA clones by hot-start, semi-nested PCR and rapid amplification of cDNA ends PCR [20]. Heterologous expression in E. coli as a GST fusion protein and affinity purification resulted in fully active protein [20]. The encoded amino acid sequences (209 amino acids) were identical for the bovine and porcine GLTPs and matched closely with the porcine primary sequence determined by Edman degradation. Subsequent cloning and expression of human GLTP from skin fibroblasts revealed high homology (>98%) with the porcine and bovine proteins [21,22]. The ability to rapidly acquire relatively large amounts of pure, stable GLTP fueled a new wave of progress in the field. Chief among the recent advances have been an enhanced appreciation for how the membrane environment and lipid composition can affect GLTP accessibility to glycolipid, a more definitive and refined understanding of the mechanism of action used by GLTP to accomplish the intermembrane transfer of glycolipid, as well as detailed insights into the conformational architecture of GLTP in both the glycolipid-free and liganded forms.
3. Membrane Environment and Lipid Compositional Effects
By making use of resonance energy transfer (RET) between lipid fluorophores, real time insights have been gained into GLTP-mediated GSL transfer between bilayer vesicles [23]. In the RET assay involving anthrylvinyl (AV)-labeled GSLs (energy donor) and perylenoyl-labeled phosphatidylcholine or triglyceride (energy acceptor), both lipid probes are initially localized in close proximity in donor vesicles. Excitation using a wavelength selective for AV-galactosylceramide (AV-GalCer) results in energy transfer to 3-perylenoyl phosphatidylcholine, effectively quenching AV-GalCer emission. Upon addition of GLTP, the AV-GalCer emission intensity increases as a function of time as AV-GalCer is transferred to PC acceptor vesicles that are devoid of 3-perylenoyl phosphatidylcholine. Because of GLTP selectivity for glycolipids, almost no change in the initial emission signal is observed if AV-PC is substituted for AV-GalCer. Unlike the radiolabeled (3H-GSL) assay which requires separation of negatively-charged donors and neutral acceptors at discrete time intervals to assess GSL intervesicular transfer [6,24], the RET assay allows continuous real-time monitoring and still requires only small protein amounts (0.5 to 2.0 μg). Figure 2 illustrates a typical fluorescence signal response observed when GLTP transfer activity is monitored using a RET assay system in which DiO-C16 (3,3'-dihexadecyloxacarbocyanine perchlorate) serves as the nontransferable energy acceptor/quencher.
Figure 2.
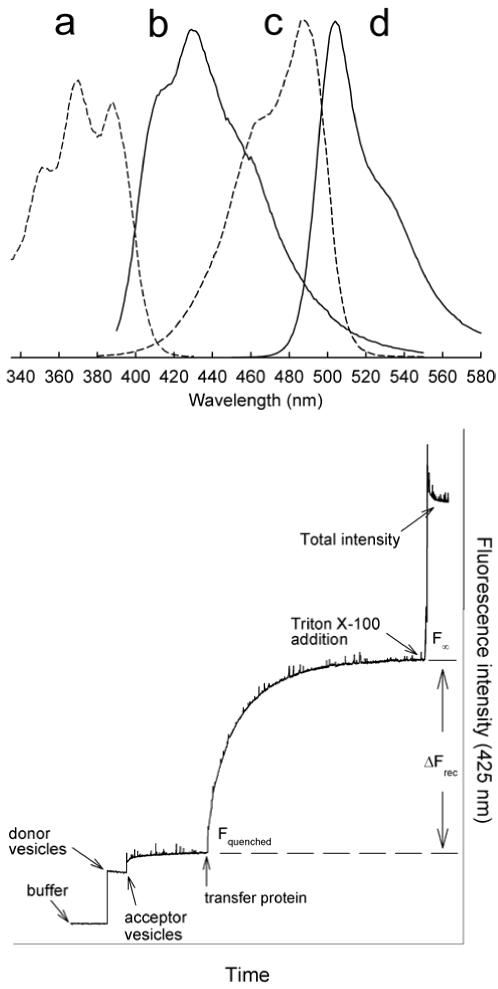
Typical resonance energy transfer assay for monitoring real-time kinetics of GLTP-mediated intervesicular transfer of glycolipid. (Upper panel). In this case, the fluorophore pairs undergoing energy transfer are anthrylvinyl-galactosylceramide (energy donor) and DiO-C16 (energy acceptor) in ethanol (Normalized spectras). (a) Excitation of anthrylvinyl-GalCer (λem=440 nm). (b) Emission of anthrylvinyl-GalCer at λex=370 nm. (c) Excitation of DiO-C16 (λem=580 nm). (d) Emission of DiO-C16 at λex=440 nm. (Lower Panel). Schematic shows the observed signal response during the movement of fluorescently labeled lipid between donor and acceptor bilayer vesicles. Donor vesicles contain a low molar ratio of a fluorescently labeled lipid substrate (i.e. anthrylvinyl-GalCer or BODIPY-glucosylceramide) and a non-transferable energy acceptor or quencher (i.e. perylenoyl-triglyceride or DiO-C16) in a suitable membrane matrix. Acceptor vesicles are usually added in 10-fold excess or more, and the system is allowed to equilibrate. Next, GLTP (1 μg) is added and the change in fluorescence intensity at the RET energy donor fluorescence maxima is recorded. The transfer reaction rate can be calculated from the obtained transfer curve by using the value for total fluorescence intensity obtained after Triton X-100 addition. The detergent causes a complete disruption of the vesicle system.
Using the preceding assay systems, investigations have been carried out to determine how changes in solution properties (e.g. ionic strength and pH) and in membrane properties (e.g. surface charge, phase state, curvature/bending stress, and glycolipid substrate concentration) affect the GLTP mediated transfer of glycolipids. The working hypothesis for the biochemical and biophysical studies has been that the physical environment generated by the lipids and their lateral distribution are key factors regulating the transfer activity of GLTP in vitro.
3.1 Glycolipid concentration in the membrane
The amount of glycolipid in the donor vesicles has little impact on the GLTP mediated transfer rates. Increasing the glycolipid concentration from 0.5 mol % up to 5.0 mol %, while keeping the concentration of the transfer protein constant (0.94 μg), increases the transfer rate of AV-GalCer only marginally, i.e. ∼15 % [24]. However, increasing the amount of transfer protein does indeed result in a faster transfer rate. The results are consistent with a transfer process that follows first-order kinetics.
3.2 Charge effects on GLTP
Previous studies showed that membrane surface charge has significant effects on lipid transfer protein catalyzed transfer of various lipids [25-27]. For instance, negatively charged donor and acceptor membranes reportedly inhibit the activity of the intracellular phosphatidylcholine transfer protein and non-specific lipid transfer protein. The phosphatidylcholine transfer activity of a yeast phosphatidylinositol / phosphatidylcholine transfer protein also is inhibited by negatively charged phospholipids and fatty acids [28]. Plasma-derived transfer proteins that facilitate the transfer of phospholipids and cholesterol esters between plasma lipoproteins are also sensitive to the lipoprotein surface charge [29,30]. In contrast to these inhibitory effects, negatively charged lipids stimulate the sphingolipid activator protein, SAP-C [31].
With bovine GLTP, there is a marked effect of different charged lipids on the rate of transfer of GalCer from a donor to acceptor vesicle population at neutral pH [24]. Compared to neutral donor vesicle membranes, introduction of negatively charged lipid at 5 or 10 mol % into the donor vesicles significantly decreases the transfer rate. Introduction of the same amount of negative charge into the acceptor vesicle membrane does not impede the transfer rate as effectively. Also, positive charge in the donor vesicle membrane is not as effective at slowing the transfer rate as is negative charge in the donor vesicle. Increasing the ionic strength of the buffer with NaCl significantly reverses the charge effects. At neutral pH, GLTP (pl = 9.0) is positively charged which promotes association with the negatively charged donor membrane.
3.3 Sphingomyelin, phosphatidylcholine and cholesterol effects on GLTP
Shipley and co-workers [32] showed that N-palmitoyl GalCer, below 23 mol %, is completely miscible in DPPC fluid or gel-phase bilayers, whereas Curatolo [33] found that cerebroside-POPC mixtures exhibit gel phase immiscibility over the compositional range 0-70 mol % cerebroside. Also, PCs and SMs are known to be highly miscible in each other when their chain lengths are similar in length [e.g.,34]. GalCer, at concentrations below 30 mole%, is miscible in both liquid-crystalline and gel-phase SM [35], whereas GalCer mixing in PC becomes less favorable with increasing unsaturation of the fatty acyl chains in PC. NMR studies of GalCer transbilayer distributions in PCs and SMs suggest preferential lateral interactions between GalCer and SM [36,37]. Interestingly, the GLTP-mediated transfer of AV-GalCer seems to be affected by its miscibility state in bilayer membranes composed of SM and PC. GLTP fails to transfer glycolipid from donor matrices with compositions that enhance miscibility of AV-GalCer in the matrix lipid, such as those high in SM and SM-like derivative content [38,39]. Interestingly, addition of cholesterol to such donor membranes does not significantly decrease the GLTP-mediated transfer of AV-GalCer any further. The preceding findings lead to the conclusion that interactions between glycolipid and SM are not affected by cholesterol. We postulate that SM interacts with GalCer in a way that restricts GLTP accessibility to the sugar headgroup, in contrast to cholesterol, which can order the hydrocarbon chains of SM and GalCer but does not prevent the glycolipid from being transferred by GLTP. Interestingly, GLTP can transfer glycolipids more easily to lipid raft-like membranes containing SM. It is therefore tempting to speculate that GLTP could be involved in raft assembly but not use lipid raft-like domains as a source for glycolipids.
Another notable feature of GLTP is its ability to acquire GSL from nonmembrane sources. In other words, the GSL ligand need not be dispersed among phosphoglycerides in a fluid-phase bilayer environment to be acquired by GLTP. Liganding of GalCer by GLTP has been shown after incubation with pure GalCer films adsorbed to glass and submerged in aqueous solution [41]. The GalCer in such pure films is expected to be tightly packed because the main thermotropic transition temperature of GalCer occurs at approximately 80°C [42]. Human GLTP also interacts efficiently with ganglioside GM1 when it is dispersed directly into aqueous buffer lacking phosphoglyceride, showing that GLTP can acquire glycolipid from nonbilayer sources [41]. Because the GM1 aqueous concentration was ∼5 μM, it appears likely that both GM1 micelles and monomers were available to interact with GLTP based on findings that purified GM1 has a cmc value in the 10−7 − 10−8 M range, when adequately corrected for nonspecific lipid adsorption to tube walls [43,44]. The data raise the possibility that GLTP can directly acquire monomeric GM1 from solution without interacting with GM1 micelles, as also has recently been suggested for human cytosolic sialidase interaction with gangliosides GM1, GD1a, and GM2 [45]. Nonetheless, the dramatically increased local concentration of GM1 within micelles could serve to enhance the efficiency of uptake upon interaction with GLTP. The finding that ganglioside GM1 needs not be dispersed in phosphoglyceride membranes to be acquired by GLTP raises the possibility that GLTP might be able to interact with other nonmembrane entities, i.e. cellular proteins, to acquire or deliver GSLs, analogous to the role played by endosomal sphingolipid activator proteins in delivering GSL antigens to CD1 protein receptors during antigen presentation processes [46-48].
The direct demonstration of interaction between ganglioside GM1 and GLTP resulting in formation of a soluble ganglioside GM1/GLTP complex [41], is also important because of earlier conflicting results. In some early studies of GLTP, complex glycolipids were reported not to be transferred efficiently by GLTP [2,3,8]; whereas, other reports indicated that GLTP can transfer GM1 between vesicles [7,10,49,50].
4. GLTP Mechanism of Action
The availability of homogenous, wild-type human GLTP by recombinant molecular biological approaches has enabled detailed analyses of GLTP action from kinetic and thermodynamic data obtained using the fluorescence transfer assay [22]. Assessment of GLTP-mediated transfer of glycolipid from both small (25 nm diameter) and large (100 nm diameter) unilamellar vesicles, i.e. SUVs and LUVs, over the temperature range of 30−44°C enabled determination of the initial GSL transfer rates (v0) and net transfer equilibrium constants (KEq). The initial GSL transfer rate constant (v0) exhibited a linear dependence with respect to changing GLTP concentration (0−143 nM range) for both SUVs and LUVs, consistent with a first order dependence on GLTP bulk concentration. Use of Arrhenius and van’t Hoff plots enabled evaluation of the thermodynamic parameters associated with the GLTP-GSL transition state complex and GSL net transfer, respectively. While the Gibbs’s free energy of the transition states (ΔG≠) were similar for both SUVs and LUVs, the activation energy barrier was higher for LUVs than SUVs and the initial transfer rates were lower for LUVs than for SUVs. Formation of a transition-state complex was predominantly enthalpy driven, whereas the net transfer of GSLs was mainly entropy driven. The rate-limiting step for GLTP action was not GLTP partitioning onto the donor vesicle surface but rather formation of GLTP-GSL complex and/or its release from the membrane, consistent with a shuttle/carrier mode of action depicted in Figure 3 [22]. Other mechanisms not supported by the data were a ‘Bound Transporter’ model, in which GLTP remains continuously bound to donor vesicles and mediates transfer by lowering the energy barrier for selective GSL desorption and a ‘Conduit’ model, in which GLTP forms a connecting conduit between donor and acceptor vesicles to mediate GSL transfer (see Figure 3 in [22]).
Figure 3.
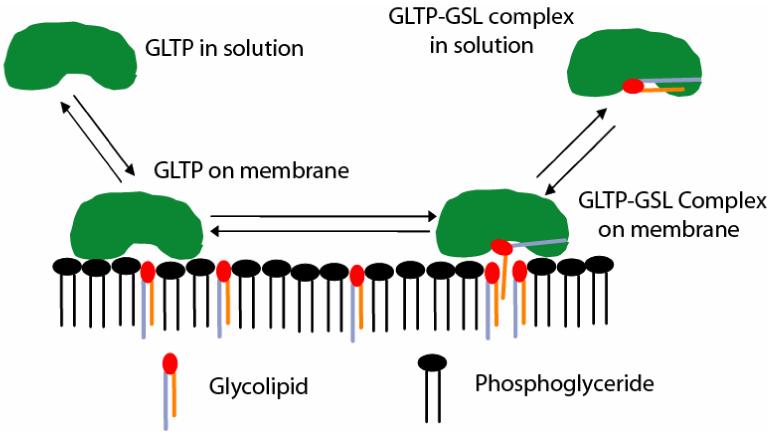
Mechanism of GLTP-mediated transfer of GSL between membranes in vitro. Glycolipid transfer kinetics, GLTP-membrane partitioning, and GLTP/GSL complex structural measurements support a mechanism in which GLTP acts as a GSL carrier that shuttles between membranes. Because GLTP in a GSL-free state displays a relatively weak protein/membrane-binding propensity [57], GLTP can be expected to readily partition on and off the membrane surface. The rapid lateral diffusions rates of lipids in fluid-phase membranes and the confinement of the glycolipid ligand to a membrane surface will likely enhance the capacity of GLTP to associate with a glycolipid molecule among other membrane lipids. The sugar moiety of the glycolipid acts as a primary specificity determinant, while the ceramide amide functional group orients the entry of the hydrocarbon chain(s) through the cleft-like gate. There is an increased interaction propensity centered about the cleft-like GLTP gate, that is surrounded by aromatic surface residues (e.g. W142, W96, Y153, Y157, Y207, Y81) and a half dozen Lys residues, which are known to interact favorably with membrane interfaces [91,92], thereby potentially facilitating the opening of the gate in the membrane-associated state and entry of the acyl chain into the hydrophobic channel of the GLTP. The accommodation limits of the hydrophobic tunnel, shown by the crystal structures of the GSL-GLTP complexes, strongly suggest that sphingosine is the last lipid part to enter GLTP and, most likely, the first to depart GLTP upon interaction with a membrane. GLTP is shown in green, phospholipids in black, and GSL with a red polar headgroup and the ceramide region made up of a lavender (longer) acyl chain and orange (shorter) sphingoid chain. Note that the depictions detailing the acquisition and liganding of glycolipid by GLTP (right-side of this figure) summarize structural data findings depicted in greater detail in Figures 4, 5, and 6.
Models for phospholipid transfer proteins developed previously [51-56] generally relied on ordinary two substrate enzyme-catalyzed reactions that could be described by ping-pong Bi-Bi mechanisms. While the models were well suited for transfer mechanisms involving single component lipid membranes, such as phosphatidylcholine transfer protein [51], with GLTP, a minor lipid component (e.g. GalCer) is usually transferred from a lipid membrane containing two or more lipids and in which the major lipid component (POPC) serves as a matrix and itself is not a ‘substrate’ for the lipid transfer protein. The situation is more similar, but not identical, to that of phosphatidylinositol transfer protein (PITP) which displays about a 15-fold preference for PI over PC. In modeling studies of PITP action [52,55] , it was assumed that the initial transfer velocity could be described solely in terms of donor and acceptor vesicle concentrations to model the functional unit of interaction with PITP. While this assumption may hold when PI concentrations are relatively high in the membrane vesicles, the situation becomes more complicated when the minor lipid component is present at low concentrations in the membrane (e.g. <15 mol%). This means that formation of a protein-lipid complex within the membrane interfacial environment may require lateral diffusion of either lipid or protein or both. This situation appears likely for GLTP and glycolipids which are found at low concentrations in most biomembranes but also have a tendency to locally concentrate in rafts and caveolae.
Direct confirmation of glycolipid liganding by GLTP by isolation and characterization of soluble GLTP-glycolipid complexes containing radioactive glycolipids has revealed a complexation stoichiometry of ∼1:1 between GLTP and glycolipid [41]. Isolation was achieved using size exclusion chromatography (SEC) to separate the soluble monomeric complex (∼25 kD) from vesicles containing glycolipid (>2,500 kD) or using an affinity-based separation involving binding of epitope-labeled GLTP (e.g. 6×HisGLTP) to Ni-NTA affinity resin to achieve rapid separation from glycolipid/PC vesicles. These approaches also have proven valuable for point mutational analyses of GLTP residues involved in GSL liganding [41].
During the kinetic analyses of GLTP action [22], the focus was on the glycolipid being transferred by GLTP, without the benefit of experimental evidence confirming direct interaction of GLTP with membrane vesicles. To address this shortcoming, Rao et al. [57] developed ways to directly measure interaction of GLTP with bilayer vesicles and further test the ‘carrier’ model of GLTP action. Using a fluorescence approach, protein-vesicle association was determined by RET from excited (285 nm) intrinsic tryptophan and tyrosine residues of GLTP to dansyl-labeled phosphatidylethanolamine (PE) in POPC vesicles. Because RET depends critically on the orientation and distance between the donor (tryptophan and tyrosine) and acceptor (dansyl-PE) energy transfer fluorophore pair and does not depend on strong interaction affinity, it is well suited for evaluating the partitioning of proteins to membranes, regardless of whether the interactions are strong or weak. By relying on the intrinsic fluorescence of the naturally-occurring tryptophan and tyrosine residues of GLTP, introduction of extraneous probes could be avoided at a site(s) that might perturb or alter the conformation of GLTP and/or its membrane interaction region. The GLTP partitioning data showed a saturation response, as GLTP increased, for vesicles containing dansyl PE. Moreover, GLTP partitioned to POPC vesicles lacking GSL. GLTP interaction with model membranes was nonpenetrating, as assessed by protein-induced changes in lipid monolayer surface pressure, and nonperturbing in that neither membrane fluidity nor order were affected, as monitored by anisotropy of 1,6-diphenyl-1,3,5-hexatriene (DPH) and 6-dodecanoyl-N,N-dimethyl-2-naphthylamine (Laurdan). Tryptophan anisotropy increased upon mixing with POPC vesicles lacking glycolipid suggesting the involvement of tryptophan residue(s) in the membrane interaction site of GLTP. Ionic strength, vesicle packing, and vesicle lipid composition were all found to affect GLTP partitioning to the membrane and led to the following conclusion: Conditions that increased the ratio of bound-to-unbound GLTP did not guarantee increased transfer activity, but conditions that decreased the ratio of bound-to-unbound GLTP always diminished transfer. Thus, Rao et al. [57] concluded that GLTP partitioning to the membrane is relatively weak, transient, and nonperturbing, can be strongly influenced by membrane lipid composition, and is likely to involve a GLTP membrane interaction site containing tryptophan. The data directly supported the transfer model illustrated in Figure 3. Upon translocation to the membrane interface, GLTP must find and recognize the carbohydrate moiety on GSL and then form a surface complex that is released into the bulk, thus acting as a GSL carrier that must desorb from the surface to accomplish GSL transfer [57].
Recently, the role of each of the three tryptophan residues of GLTP as components of a membrane interaction site of GLTP has been assessed by comprehensive fluorescence analyses of tryptophan mutants of human GLTP [58]. Changes in tryptophan emission intensity along with an accompanying blue shift in the emission wavelength maximum are commonly observed when peripheral proteins move from the aqueous milieu and become membrane associated [59]. West et al. [58] produced different tryptophan mutants of human GLTP. Single tryptophan mutants were constructed by site-directed mutagenesis where two of the three tryptophans (W) of wild-type human GLTP were substituted with phenylalanine (F) and accordingly named W85 GLTP (W96F and W142F), W96 GLTP (W85F and W142F) and W142 GLTP (W85F and W96F). Wild-type GLTP and W96 GLTP were both able to transfer AV-GalCer, but the W85 GLTP and W142 GLTP variants did not show any transfer activity, indicating that the tryptophan in position 96 is crucial for transfer activity. Interaction of wild-type GLTP and W96 GLTP with vesicles containing glycolipid resulted in a blue-shifted, tryptophan emission wavelength maximum, while no blue shift was observed for the W85 GLTP and W142 GLTP variants. The quantum yield of tryptophan emission was highest for the W96 GLTP protein whereas W85 GLTP, W142 GLTP and wild-type GLTP showed a lower and almost similar quantum yield. The lifetime and anisotropy decay of the different tryptophan mutants also changed upon binding to vesicles containing GalCer. Again wild-type GLTP and W96 GLTP showed similar behavior in the presence of vesicles containing glycolipids. However, the value of W142 GLTP was low (KSV = 3.63) indicating a relatively hydrophobic surrounding and suggesting that the W142 residue, to some extent, comes into contact with the hydrophobic part of the membrane during the binding and transfer process of glycolipids. What is clear is that the W96 residue is part of the GLTP membrane interaction site that helps GLTP to function as it scoots along the membrane surface in search of a glycolipid ligand.
5. GLTP Conformational Structure
Prior to the point mutational assessment of the role of the three tryptophan residues in the membrane interaction of bovine GLTP [58], the intrinsic fluorescence response of the tryptophan residues in GTLP had been characterized in a study describing the first cloning of human GLTP [21]. Excitation at 295 nm yielded an emission maximum of 347 nm indicating a relatively polar average environment for the three tryptophans of GLTP. Quenching with acrylamide at physiological ionic strength or with potassium iodide resulted in linear Stern-Volmer plots, consistent with accessibility of emitting tryptophans to soluble quenchers. Reversible conformational changes induced by urea and accompanying changes in GLTP activity were investigated by monitoring changes in the fluorescence of intrinsic tryptophan or added 1-anilinonaphthalene-8-sulfonic acid (1,8 ANS) after rapid dilution of urea. Incubation of GLTP with glycolipid liposomes resulted in diminished tryptophan emission intensity (∼20−40%) and a blue-shifted (∼12 nm) emission maximum, λmax. The persistence of the blue-shifted emission maximum in the recovered soluble GLTP, after separation from glycolipid liposomes, provided the first indication of tryptophan involvement in the glycolipid liganding site of GLTP [21].
A quantum leap forward in GLTP research occurred in late 2004 with the publication of the first high resolution structure of GLTP [60]. The crystal structures of human GLTP, in GSL-free (apo) (1.65 Å) and lactosylceramide (LacCer)-bound (1.95 Å) form, revealed a novel conformational architecture among proteins that transfer or bind lipids. GLTP utilizes an all α-helix conformation, achieved without intramolecular disulfide bridges, to form a two-layer ‘sandwich motif’ that accommodates glycolipid. The GLTP architecture markedly contrasts that of other lipid binding and transfer proteins which generally use motifs dominated by β-sheet, i.e. β-grooves/concave cups and β-barrels, or helical bundles stabilized by multiple disulfide-bridges, i.e. saposin-folds. Such proteins include sphingolipid activator proteins, CD1 proteins, ceramide transfer protein, phosphoglyceride transfer proteins, other START-related proteins, nonspecific lipid transfer proteins, fatty acid binding proteins, lipocalins, and plant lipid transfer proteins [48,61-75]. The novel architecture now defines GLTP as the prototypical and founding member of a new protein superfamily (see http://supfam.org/SUPERFAMILY/cgibin/scop.cgi?sunid=110004 and http://scop.mrc-lmb.cam.ac.uk/scop-1.69/data/scop.b.b.bja.b.b.A.html) characterized by a novel protein fold for lipid binding/transfer and for membrane interaction [76,77].
In the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do), eleven structures are currently deposited for GLTP, four in glycolipid-free form and seven complexed with various GSLs [60,78,79]. All structures support the original observations of Malinina et al. [60] showing that the two-layer, α-helical topology of GLTP enables complexation of GSL in sandwich-like fashion within a single GSL liganding site, and occurs via adaptive recognition, involving a surface recognition center for the sugar headgroup and a molded-to-fit, hydrophobic tunnel for the hydrocarbon chains of the ceramide moiety (Figure 4). Comparative structural analyses of apo-GLTP and the LacCer-GLTP complex, including crystallographic B-factor distributions, suggest that a cleft-like gating mechanism, in which conformational changes to two interhelical loops and one α-helix facilitate entry and exit of the lipid chains in the membrane-associated state when the GSL headgroup is anchored to the sugar headgroup recognition center [60].
Figure 4.
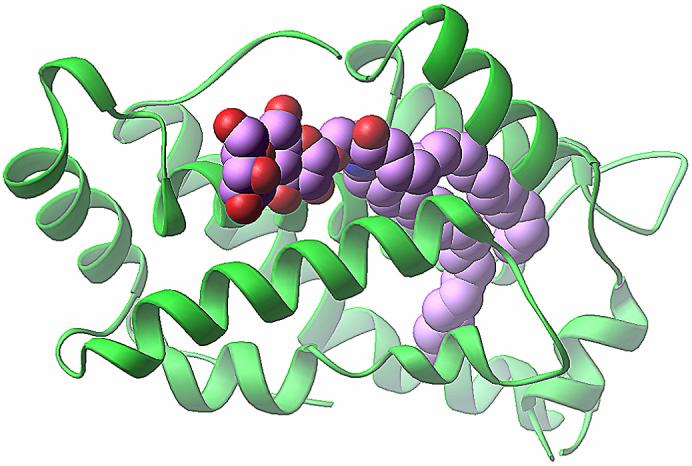
Crystal structure of the 18:1 LacCer-GLTP complex. The GLTP is shown in a green ribbon representation. The carbon atoms of the LacCer are shown in a lavender-colored space-filling representation. The red- and blue-colored atoms in LacCer represent oxygen and nitrogen, respectively. Adapted from Figure S1 (panel C) of Malinina et al. [79].
Mapping of the headgroup recognition center and hydrophobic tunnel of GLTP enabled identification of amino acid side chains that hydrogen bond and make hydrophobic contacts with the bound glycolipid (Figure 5). The initial sugar linked to ceramide undergoes the majority of anchoring within the GLTP recognition center, located in a depression on the protein surface, via a network of hydrogen bonding and hydrophobic tethering interactions. The hydrogen bonding involves aspartic acid 48 (D48), asparagines 52 (N52), and lysine 55 (K55) of α-helix 2 and tyrosine 207 (Y207) near the C-terminus. Tryptophan 96 (α-helix 4) serves as a platform for stacking of the sugar ring, thereby facilitating orientation for optimal hydrogen bonding interactions involving perimeter OH-2 and OH-3 groups of the sugar and the D48 and N52 residues of GLTP, respectively. The same headgroup interaction pattern has been observed in complexes of bovine GLTP and ganglioside GM3 in which the initial glucose residue is anchored identically to the sugar recognition center of the protein surface, but the other two sugars are unobservable because of disorder [78]. In recently reported complexes of human GLTP with different GalCers [79], the same hydrogen bonding pattern involving D48, N52, and K55 of α-helix 2 and Y207 near the C-terminus was observed again. The stacking of W96 (α-helix 4) against the b face of the galactose ring also was similar to that of the glucose ring in the 18:1 LacCer complex. The only difference in the liganding between the single sugar headgroup of GalCer and the two-sugar headgroup of 18:1 LacCer, involved K55, which hydrogen bonds with the OH3 and OH4 hydroxyls of the initial sugar ring, attached to the ceramide in GalCer, rather than with the OH3 hydroxyl of the second sugar residue in the lactose of the 18:1 LacCer.
Figure 5.
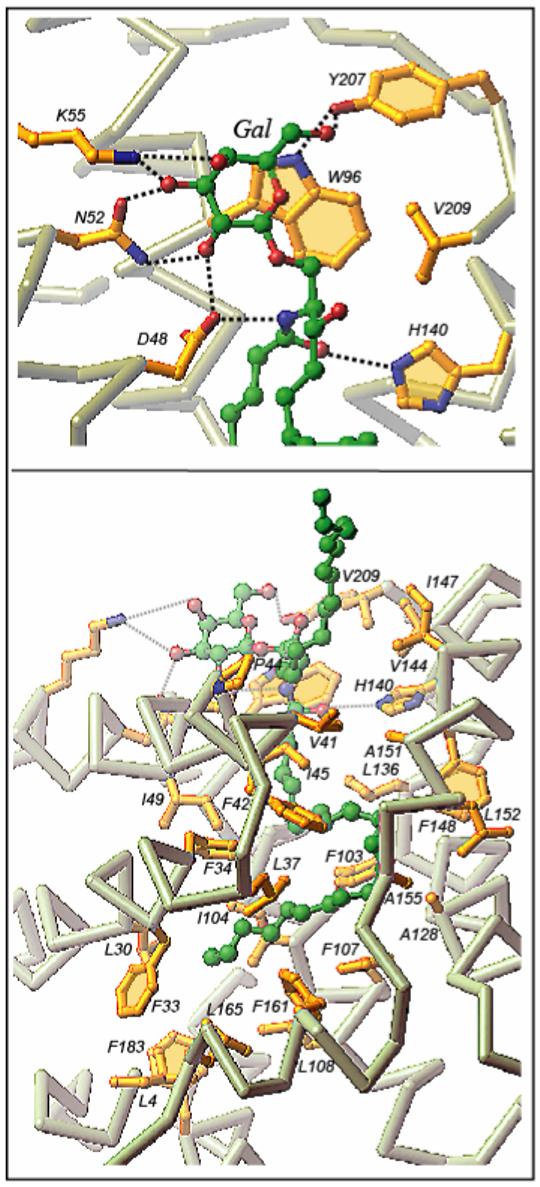
GSL-GLTP interactions in the 24:1 GalCer-GLTP complex. (Upper panel) 24:1 GalCer headgroup (sugar and amide) interactions with GLTP recognition center residues. Hydrogen bonds are shown by black dashed lines. The bound GSL atoms colored green, red and blue represent carbon, oxygen and nitrogen atoms, respectively. The GLTP Cα-backbone is colored light grey, the side chains are shown in gold, and oxygen and nitrogen are in red and blue, respectively. (Lower panel) 24:1 GalCer ceramide chain interactions with the GLTP hydrophobic tunnel residues. The longer acyl chain occupies the tunnel while the shorter sphingosine chain is directed outwards. Reproduced from Figure 2 of Malinina et al. [79].
The tripartite interaction cluster consisting of aspartate, asparagine and tryptophan has been observed previously in the sugar-binding site of other proteins like in the Escherichia coli galactose chemoreceptor protein [80]. This same triad of residues has been proposed to be part of the sphingolipid binding domains associated with helix-turn-helix structural motifs that are observed in the V3 loop of the HIV-1 gp120 protein, the prion protein, the Alzheimer beta-amyloid, and the pancreatic bile salt dependent lipase which are known to bind GSLs [81,82].
All hydrogen bond classes observed in previous protein-carbohydrate interactions [83] are evident when GLTP interacts with the GSL sugar-amide. Among the classes are bidendate hydrogen bonding involving N52, bifurcated hydrogen bonds involving D48 (and K55 in case of galactose), as well as the propensity of the OH2 and OH3 sugar hydroxyl groups and amino acid side chains (e.g. Y207) to form ‘cooperative’ hydrogen bonds. The liganding interaction pattern, involving sugar stacking against an aromatic residue to enable formation of hydrogen bond tethers with peripheral sugar hydroxyls, observed in the all-α-helical conformation of GLTP is a conserved feature of other carbohydrate binding proteins, including all known galactose-specific binding proteins, all of which have α + β or all β conformational architectures [83-86].
The orientation of the ceramide amide group of GSL during liganding is controlled by a pair of hydrogen bonds involving D48 and histidine 140 (H140), with alignment of the initial segment of the sphingoid base being facilitated by van der Waals contacts with valine 209 (V209) (Figure 5). This situation occurs in all GSL/GLTP complexes examined thus far [60,78,79]. The interactions are significant because they result in a conserved orientation of the GSL ceramide lipid tails through the cleft-like gate when accessing the hydrophobic tunnel of GLTP with the GSL acyl chain always entering prior to the sphingosine chain (Figures 3 & 5).
The hydrophobic tunnel that encapsulates the GSL hydrocarbon chains is characterized by two noteworthy features. First, the tunnel is highly nonpolar. No water molecules reside in the channel, which is lined by the side chains of nonpolar phenylalanine, leucine, isoleucine, and alanine residues, together with a few valine and proline residues (Figure 5). Second, a large portion of the tunnel is conformationally adaptable and expands during GSL acquisition to accommodate the hydrocarbon chains. Moreover, localized in the lower part of the hydrophobic tunnel of apo-GLTP, i.e. glycolipid-free GLTP, is an extraneous hydrocarbon molecule, containing at least 6 carbon atoms and clearly observable in electron density maps. Airenne et al. [78] reported finding decanoic acid (10-carbon chain) within the hydrophobic tunnels of two glycolipid-free bovine GLTPs expressed in E. coli. It appears likely that the extraneous hydrocarbon is acquired as a result of heterologous expression in E. coli, which does not express GSLs. Similar observations have been reported for other GSL binding proteins [63] and other lipid binding/transfer proteins [73], consistent with a possible chaperone-like function for the extraneous lipids.
The ease with which the ceramide region of GSL can be accommodated within the hydrophobic channel of GLTP appears to be influenced by structural features of the lipid chains themselves. Complexation of 18:1 LacCer by GLTP resulted in encapsulation of both ceramide chains in roughly parallel fashion within the hydrophobic tunnel [60] (Figure 4). However, in the GLTP/GM3 complex, Airenne et al. [78] found a significant portion of the sphingosine chain to be disordered and proposed that it might remain outside the hydrophobic tunnel. Definitive evidence of the ‘sphingosine-out’ binding mode was recently provided for certain GLTP/GSL complexes. Strong chain ordering that resulted from cross-bridging between sphingoid chains of partner monomers in the crystalline complexes enabled complete, uninterrupted electron density mapping of the chains. Moreover, the acyl chain structural features were systematically altered to perturb the optimal fitting of the ceramide hydrocarbon chains within the hydrophobic channel [79]. Since entry of ceramide chains of the hydrophobic tunnel occurs by a highly conserved and oriented mechanism with the acyl chain always entering first, it is the trailing sphingosine chain that is always more prone to being obstructed from entry into the tunnel. Space limitations within the confines of the hydrophobic tunnel does place restrictions on the acyl chain conformation that can interfere with the entry of the sphingoid chain. A particularly striking illustration is seen when GLTP is complexed with GalCer containing nervonoyl acyl chains. Accommodation of the long, 24-carbon acyl chain of GalCer within the tunnel dictates a serpentine acyl conformation which interferes with entry of the sphingosine chain of 24:1 GalCer into the tunnel (Figures 5 & 6). Interestingly, the degree of disorder associated with the N-terminus also appears to affect acyl conformation within the tunnel. When more disordered, as seems to be case when N-terminally tagged with 6×-His, the N-terminus fails to block a portal near the bottom of the tunnel, enabling the ganglioside GM3 acyl chain to pass through it [78].
Figure 6.
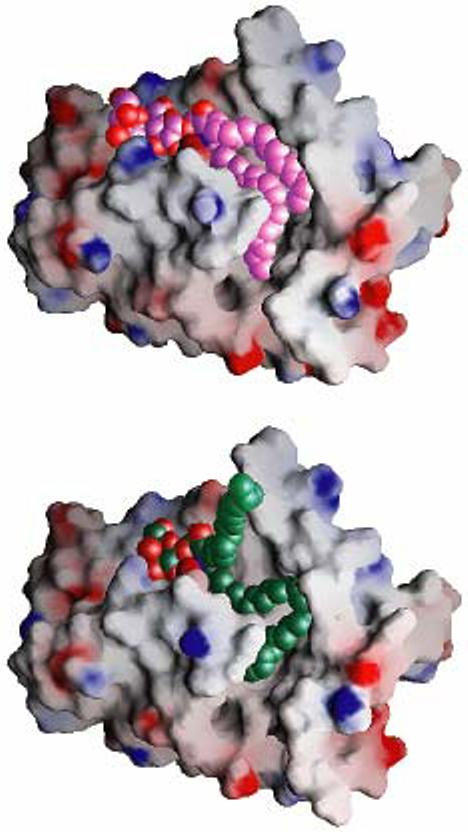
‘Gate-removed’ electrostatic surface views of the GLTP hydrophobic tunnel accommodating GSLs. The GLTP is shown in an electrostatic surface representation (blue, positive; red, negative; grey, neutral), with gate residues 33 and 35 to 45 removed to make the tunnel and its contents visible. (Upper panel) Structure of the 18:1 LacCer-GLTP complex exhibiting the sphingosine-in mode. The carbon atoms of the 18:1 LacCer are shown in lavender color and space filling representation. Both lipid chains optimally fit into the available space of the hydrophobic tunnel. (Lower panel) Structure of the 24:1 GalCer-GLTP complex exhibiting the sphingosine-out mode. The carbon atoms of the 24:1 GalCer are shown in a green space filling representation. The long acyl chain, bent in a serpentine fashion, occupies the available space of the hydrophobic tunnel, resulting in an outward positioning of the sphingosine chain. Reproduced from Figure 3 of Malinina et al. [79].
Because the sphingosine-out conformation has been directly observed in four out of five of the GLTP-GSL complexes solved by Malinina et al. [79] and has been implicated in GM3/bovine GLTP complex, in which the last 10 of 18 carbons of the GM3 sphingosine chain are unobservable because of disorder [78], the sphingosine-out conformation may be preferred by GLTP. The observations establish a structural basis for the accommodation of various GSL species with different acyl chemical structures by GLTP and support a concerted sequence of events during GSL acquisition and/or release in which the sphingosine chain is the last to enter and first to leave the GLTP liganding site (e.g. Figure 3).
The sphingosine-out conformer in GSL-GLTP complexes can be compared to monoacylation of a soluble protein, except that the exposed hydrocarbon segment of sphingosine (10−12 methylenes) is even shorter than protein acylation achieved by myristoylation (14:0) or by palmitoylation (16:0) events. It is well established that attachment of a single myristoyl group to a soluble protein only marginally affects its partitioning affinity for nonpolar (membrane) surfaces [87,88]. Generally, a second acylation event involving palmitate (16 carbons) is needed for soluble proteins to form strong, stable associations with membranes. Also, there are examples of proteins containing surface grooves that accommodate significant stretches of nonpolar hydrocarbons on their surfaces while remaining monomeric [89,90]. Thus, it is not surprising that GLTP and its GSL-bound complexes are monomeric in solution, as monitored by sedimentation analysis or size exclusion chromatography, even when a substantial portion of the GSL sphingosine chain (10−12 methylenes) is outside the hydrophobic tunnel [79].
6. GLTP Membrane Interaction Motif
The uniqueness of the GLTP conformational architecture resides not only in its structural motif compared to other lipid-binding and carbohydrate-binding proteins but also in its membrane interaction domain. While the surface region surrounding the glycolipid liganding site has many nonpolar amino acids as well as multiple tyrosines, tryptophans, and lysines [57,58,60,78], typical residues of membrane interaction motifs [91,92], the GLTP conformational fold that spatially organizes the residues clearly differs from other known membrane targeting motifs, such as the protein kinase C homology −1 and −2 (C1, C2) domains associated with many phospholipases and protein kinases, the FYVE, pleckstrin homology (PH), and phox (PX) domains that bind to membranes containing PI derivatives [93-95]. Recent studies have begun to provide direct experimental evidence implicating select residues (e.g. Trp) as components of the GLTP membrane interaction motif [57,58,79]. While there is general agreement that the membrane interaction region is located around the glycolipid binding site of GLTP, some details remain unsettled. Whereas West et al. [58] proposed roles for all three Trp residues in GLTP based on point mutational analyses, modeling studies using the Optimal Docking Area algorithm to identify regions on the GLTP surface that display a favorable energy change upon replacement of the water environment for a lower dielectric (but still polar) medium, such as the surface of a membrane [79] as well as predictions of the Orientations of Proteins in Membranes (OPM) Database, (http://opm.phar.umich.edu/families.php?family=117), which uses a computational approach to optimize the spatial arrangement of protein structures in lipid bilayers [96,97] include W96 and W142 (Figure 7). What is clear is that additional study will be needed to fully characterize the GLTP membrane interaction site. Understanding the structural details of the GLTP membrane interaction site is expected to provide better insights into how GLTP acquires and/or releases GSLs while interacting with the membrane matrix in a transient, nonperturbing way during the transfer process [57].
Figure 7.
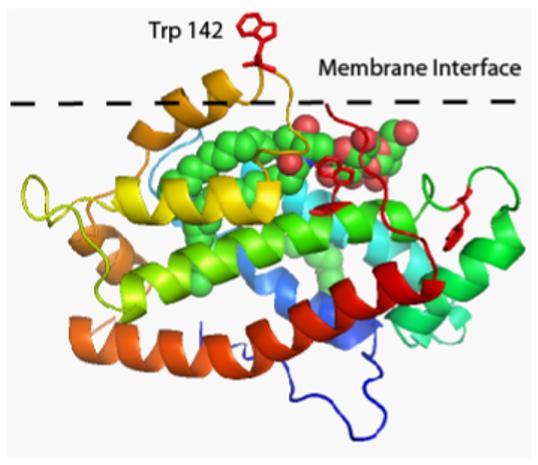
Putative membrane interaction region of GLTP, showing the location of the three tryptophan residues (red color). LacCer is shown in space-filling mode with carbon, oxygen, and nitrogen atoms colored green, red, and blue, respectively. The membrane interface is represented by the black dashed line with Trp142 penetrating the membrane surface. The figure was adapted from the Orientations of Proteins in Membranes (OPM) Database website, (http://opm.phar.umich.edu/families.php?family=117), which uses a computational approach to optimize the spatial arrangement of protein structures in lipid bilayers [96,97].
7. GLTP Homologues/Orthologues
Orthologs of GLTP do occur in nonmammalian organisms. The vast majority of GLTP orthologs has been indirectly deduced from different genomes and expressed cDNA sequences encoding uncharacterized proteins (e.g., [20,78,98]). Recently, the structure of an algae protein (Galdieria sulphuraria) with GLTP-like conformational structure has been resolved to 1.38Å by x-ray diffraction (PDB 2I3F;http://www.uwstructuralgenomics.org/gallery/2i3f.pdf). In other cases, molecular modeling and hydropathy analyses provide evidence for orthologs having structural similarity with GLTP [78,99]. Such proteins include HET-C2 of the filamentous fungi, Podospora anserina, and ACD11 in the model plant, Arabidopsis, which both are of similar size and have limited sequence homology to GLTP. Shown in Figure 8 is a stereo view of the GLTP structure superimposed on representative GLTP-like proteins. Functional analyses of these GLTP orthologs suggest important roles in processes linked to programmed cell death [98-100].
Figure 8.
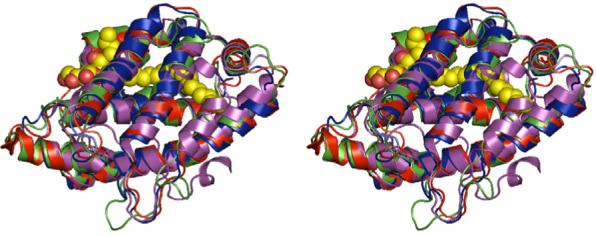
Stereo view of the X-ray structure (1.36 Å) of bovine GLTP (shown in red) with bound ganglioside GM3 (in yellow), superimposed are the structurally modeled HET-C2 homologue (in blue) and the Arabidopsis thaliana GLTP-like protein ACD11 (in green). The crystal structure (PDB 2I3F) of the GLTP-like protein from the algae Galdieria sulphuraria is shown in lavender.
HET-C2 plays a decisive role in regulating thecompatibility and stability of heterokaryons that form by hyphal fusion between different individual organisms during the universally key process of self-nonself recognition among metazoan somatic cells [99-102]. The resulting heterokaryons contain genetically distinct nuclei within a common cytoplasm. If their nuclear components possess different specificities at any of several het loci, incompatibility interactions are triggered resulting in growth cessation and cell destruction by a lytic, apoptotic-like process. The genetic basis of this vegetative incompatibility has been studied in various ascomycetes including Neurospora, Aspergillus, and Podospora and been found to share evolutionary features with other eukaryotes, such as the MHC complex in humans and the S locus in plants. The cloned het-c2 allele encodes a 208 amino acid protein with significant homology to GLTP (29% identity and an additional 30% similarity) [20,100]. Mattjus et al. [99] showed that purified rHET-C2 does mediate intervesicular transfer of monohexosylceramides, albeit at a slower rate than GLTP. Whether other glycolipids or sphingolipids are transferred by HET-C2 is currently not known. What is clear is that the transfer activity of HET-C2 is mitigated much less than bovine GLTP by negatively-charged membranes, but the HET-C2-mediated glycolipid transfer was more strongly affected than GLTP by the type of anionic phosphoglyceride (e.g., POPG vs. DPPA) [99]. The response appears to be partly, but not entirely, related to the differing isoelectric points of native HET-C2 and GLTP. At neutral pH, HET-C2 (pI = 6.0) is expected to have only a modest attraction for negatively charged vesicles, compared to GLTP (pI = 9.0), which has a net positive charge.
Because no x-ray or NMR structural data exist for HET-C2, the degree of structural homology between HET-C2 and GLTP remains to be clearly established. However, recent homology models, constructed with the MODELLER35 program based on the pair-wise alignment derived from the multiple alignment and the crystal structures of human LacCer-GLTP and bovine apo-GLTP, respectively, indicate that key residues in the sugar-binding pocket and the hydrophobic channel of GLTP are well conserved in HET-C2 [78]. However, the modeled structure of HET-C2 lacks the positively charged patch (surrounding the glycolipid binding site) on its surface that is present in the crystal structure of GLTP, perhaps accounting for the experimental differences in sensitivity to membrane negative charge [78].
ACD11 was identified by transposon knockout of an Arabidopsis gene encoding a protein with similarity to mammalian GLTP (25% identity; 42% similarity) [98]. The transposon knockout exhibits accelerated cell death (ACD) with a pattern of cell death very similar to that induced by Fumonisin B1 [103]. Fumonisin B1 is a sphinganine analog that disrupts sphingolipid metabolism by inhibiting ceramide synthase [104]. ACD in plants is often associated with overexpression of pathogenesis-related genes. Microarray analyses (10k cDNAs) of the ACD-knockout mutant indicate that loss of the GLTP-like protein leads to expression of numerous disease-related genes [105] and suggest that ACD11 is needed for repression of the death response. While ACD11 does not stimulate the intermembrane transfer of monohexosylceramides or ceramides, this protein does accelerate the intermembrane transfer of sphingosine, an important intermediate metabolite involved in intracellular signaling processes [98]. As with HET-C2, no definitive structural information exists for ACD11. However, homology models constructed with the MODELLER35 program indicate neither the sugar-binding nor the gate residues are well conserved in ACD11 compared to GLTP [78]. In addition, the surface potential map of the ACD11 model differs markedly from those of GLTP and HETC2. The analyses provide a structurally-based explanation for why ACD11 is unable to transfer glycolipids despite its limited sequence homology to GLTP.
Airenne et al. [78] have reported that A. thaliana contains three other putative GLTP-like sequences (sequence accession numbers: Q6NLQ3, O22797 and Q9LU33) that not only have similar sequence identity as ACD11 to mammalian GLTPs but also contain key, conserved sugar-binding and gate residues lining the hydrophobic channel as in GLTP and unlike ACD11. One of the putative A. thaliana GLTPs (Q6NLQ3) has been expressed in E. coli and shows glycolipid transfer activity in vitro.
The GLTP domain also appears to serve as a functionally conserved building block used in other human proteins. Studies of carrier vesicle trafficking from the Golgi complex to the cell surface have identified an ubiquitously expressed protein effector of phosphatidylinositol 4-phosphate (PtdIns(4)P), the four-phosphate-adaptor protein 2 (FAPP2). This protein localizes to the trans-Golgi network (TGN) on nascent carrier vesicles, and interacts with PtdIns(4)P and the small GTPase ADP-ribosylation factor (ARF) through its pleckstrin homology (PH) domain. Human FAPP2 is a 507 amino acid protein, encoded by a gene on chromosome 7, containing a putative glycolipid transfer protein (GLTP) domain [106,107]. BLAST alignments show the 194 amino acid, carboxy-terminus of FAPP2 to be 33% identical and 46% similar to GLTP (209 amino acids). What remains unclear is the level of structural homology that exists between GLTP and the FAPP2 carboxy terminal region as well as the capacity of FAPP2 to bind/transfer glycolipids. Investigations of the in vitro glycolipid transfer ability of FAPP2 are underway (Mattjus & De Matteis, unpublished observations).
8. Potential Roles of GLTP in Disease Processes
BLAST searches of the NCBI Human Genome Database against cDNA sequences derived from GLTP mRNA transcripts of human skin fibroblasts (GenBank AF209074) and glioma cells (AY372530, AY372531, AY372532) localize the GLTP gene to loci 12q24.11 on chromosome 12 (Chung and Brown, unpublished observation). The GLTP gene has not yet been linked to any known human pathological conditions. However, in mouse mammary epithelial cells transformed by an activated β-catenin, expression of the gltp gene is known to be affected [108]. Using microarray analysis, the gltp gene was found to be one of a dozen genes, out of 6000 screened mammary tissue cDNA clones, that become transcriptionally up-regulated in transformed mouse mammary epithelium that have undergone either partial or complete transdifferentiation to squamous metaplasias.
9. Epilogue.
The recent realization that GLTP is the protypical member of a new protein superfamily places renewed importance on elucidating the in vivo functionality of the protein. It is tempting to speculate that GLTP could perform a function in cells similar to what is measured in vitro, i.e. transfer of GSLs. While this remains a possibility, there are important topological issues that must be kept in mind. GLTP has long been suspected of residing in the cell cytosol [16,20], a localization that has recently been confirmed experimentally in transfected HeLa cells overexpressing GLTP (Tuuf & Mattjus, submitted). A cytosolic localization for GLTP has important consequences regarding which GSLs are likely to be accessible to the protein because the synthesis of most GSLs occurs at the luminal side of the Golgi apparatus [109,110]. The exceptions are monohexosylceramides, i.e. glucosylceramide (GlcCer) and galactosylceramide (GalCer), which are synthesized on the cytosolic side of the Golgi and ER, respectively [111]. This topology would render GlcCer and GalCer potentially accessible to GLTP. Indeed, a substantial fraction of GlcCer is known to be transported to the plasma membrane by non-Golgi mediated pathways [112,113]. However, further complicating the situation, but providing other potential GSL ‘substrate pools’ for GLTP within the cell, is the emerging realization that GSLs can localize to intracellular sites, i.e. nuclear and mitochondrial membranes, other than the lumen of the Golgi and the external face of the plasma membrane [110,114,115]. Exactly how GLTP might be targeted within the cell is not clear but might involve a vesicle-independent mechanism that shares some similarities with that used by ceramide transfer protein, i.e. CERT [66,67] and oxysterol binding proteins [116]. These proteins appear to operate at or near membrane contact sites formed between the endoplasmic reticulum and the trans-Golgi network. Membrane contact sites, which are intermembrane gaps of ∼20 nm, are also known to occur between endoplasmic reticulum and many other membranes, including the plasma membrane, mitochondria, endosomes, and peroxisomes [116,117]. One function for the contact sites is facilitation of the trafficking of small molecules, especially lipids [116]. It seems likely that lipid transfer proteins could play a role in the transfer of lipids across the short-range distances at such contact sites and that GLTP could be one of these lipid transfer proteins.
Another potential in vivo role for GLTP, that has been proposed previously [20, 41], is that of an intracellular sensor of glucosylceramide. Interestingly, experimental evidence directly supporting a role for GLTP as a regulatory sensor in GSL homeostasis in vivo comes from recent work by Tuuf and Mattjus (submitted) who have investigated de novo sphingolipid synthesis using sphingosine metabolic labeling in HeLa cells overexpressing GLTP. Compared to mock-transfected HeLa cells, GLTP overexpressers show an increase in glucosylceramide synthesis and a decrease in sphingomyelin synthesis. No changes were detected in the ganglioside GM1 and GM3 or in the lacto- and galactosylceramide levels.
In any case, proteins with the capability to alter the intracellular distributions of glycolipid by directly transferring GSL to specific sites within cells or to act as regulatory sensors that affect GSL metabolism can be expected to be of utmost importance in cells because of the life-and-death processes in which GSLs (and their metabolites) are known to participate, i.e. cell proliferation, differentiation, development, and apoptosis. Although GLTP appears to be a prime candidate for playing a key role in such in vivo processes, ’the jury is still out’ on these timely issues that are currently at the forefront of GLTP research efforts. Definitive insights into such matters will likely be gained by application of siRNA approaches and whole organism transgenic technologies.
Acknowlegements
We wish to express our gratitude to former and present members of the REB and PM labs. Special thanks to Dinshaw J. Patel (Memorial Sloan Kettering Cancer Center) for essential collaborative contributions enabling elucidation of GLTP structure as Co-PI of NIH/NCI CA121493, to Lucy Malinina for leading contributions in solving the structure of GLTP, and to Julian G. Molotkovsky (Shemyakin-Ovhininnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences) for synthesizing many of the sphingolipid fluorophores used to investigate GLTPs over the past decade.
Financial support: REB, United States Public Health Service (NIH-GM45928 & NIH-CA121493) and the Hormel Foundation; PM, Academy of Finland, Sigrid Jusélius Foundation, Magnus Ehrnrooth Foundation, Svenska Kulturfonden, Medicinska Understödsföreningen Liv och Hälsa r.f., and Åbo Akademi University.
Glossary
Abbreviations:
- GLTP
glycolipid transfer protein
- GSLs
glycosphingolipids
- GalCer
galactosylceramide
- LacCer
lactosylceramide
- POPC
palmitoyl-oleoyl-phosphatidylcholine
- GlcCer
glucosylceramide
- SM
sphingomyelin
- SUVs
small unilamellar vesicles
- LUVs
large unilamellar vesicles
- RET
resonance energy transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metz RJ, Radin NS. Glucosylceramide uptake from spleen cytosol. J. Biol. Chem. 1980;255:4463–4467. [PubMed] [Google Scholar]
- 2.Metz RJ, Radin NS. Purification and properties of a cerebroside transfer protein. J. Biol. Chem. 1982;257:12901–12907. [PubMed] [Google Scholar]
- 3.Abe A, Yamada K, Sasaki T. A protein purified from pig brain accelerates the intermembranous translocation of mono- and dihexosylceramides, but not the translocation of phospholipids. Biochem. Biophys. Res. Commun. 1982;104:1386–1393. doi: 10.1016/0006-291x(82)91403-6. [DOI] [PubMed] [Google Scholar]
- 4.Wong M, Brown RE, Barenholz Y, Thompson TE. Glycolipid transfer protein from bovine brain. Biochemistry. 1984;23:6498–6505. doi: 10.1021/bi00321a035. [DOI] [PubMed] [Google Scholar]
- 5.Abe A, Sasaki T. Purification and some properties of the glycolipid transfer protein from pig brain. J. Biol. Chem. 1985;260:11231–11239. [PubMed] [Google Scholar]
- 6.Brown RE, Jarvis KL, Hyland KJ. Purification and characterization of glycolipid transfer protein from bovine brain. Biochim. Biophys. Acta. 1990;1044:77–83. doi: 10.1016/0005-2760(90)90221-i. [DOI] [PubMed] [Google Scholar]
- 7.Brown RE, Stephenson FA, Markello T, Barenholz Y, Thompson TE. Properties of a specific glycolipid transfer protein from bovine brain. Chem. Phys. Lipids. 1985;38:79–93. doi: 10.1016/0009-3084(85)90059-3. [DOI] [PubMed] [Google Scholar]
- 8.Yamada K, Abe A, Sasaki T. Specificity of the glycolipid transfer protein from pig brain. J. Biol. Chem. 1985;260:4615–4621. [PubMed] [Google Scholar]
- 9.Yamada K, Abe A, Sasaki T. Glycolipid transfer protein from pig brain transfers glycolipids with beta-linked sugars but not with alpha-linked sugars at the sugar-lipid linkage. Biochim. Biophys. Acta. 1986;879:345–349. [PubMed] [Google Scholar]
- 10.Gammon CM, Vaswani KK, Ledeen RW. Isolation of two glycolipid transfer proteins from bovine brain: reactivity towards gangliosides and neutral glycosphingolipids. Biochemistry. 1987;26:6239–6243. doi: 10.1021/bi00393a043. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T, Demel RA. Net mass transfer of galactosylceramide facilitated by glycolipid transfer protein from pig brain: a monolayer study. Biochemistry. 1985;24:1079–1083. doi: 10.1021/bi00326a002. [DOI] [PubMed] [Google Scholar]
- 12.Abe A, Yamada K, Sakagami T, Sasaki T. A fluorimetric determination of the activity of glycolipid transfer protein and some properties of the protein purified from pig brain. Biochim. Biophys. Acta. 1984;778:239–44. doi: 10.1016/0005-2736(84)90364-x. [DOI] [PubMed] [Google Scholar]
- 13.Abe A, Sasaki T. Sulfhydryl groups in glycolipid transfer protein: formation of an intramolecular disulfide bond and oligomers and Cu2+-catalyzed oxidation. Biochim. Biophys. Acta. 1989;985:38–44. doi: 10.1016/0005-2736(89)90100-4. [DOI] [PubMed] [Google Scholar]
- 14.Abe A, Sasaki T. Formation of an intramolecular disulfide bond of glycolipid transfer protein. Biochim. Biophys. Acta. 1989;985:45–50. doi: 10.1016/0005-2736(89)90101-6. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki T. Glycolipid-binding proteins. Chem. Phys. Lipids. 1990;38:63–77. doi: 10.1016/0009-3084(85)90058-1. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki T, Abe A, Roerink F. Glycolipid transfer protein in animal cells. In: Hilderson HJ, editor. Subcellular Biochemistry. Vol. 16. Plenum Press; New York: 1990. pp. 113–127. (Intracellular Transfer of Lipid Molecules). [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T. Glycolipid transfer protein and intracellular traffic of glucosylceramide. Experientia. 1990;46:611–616. doi: 10.1007/BF01939700. [DOI] [PubMed] [Google Scholar]
- 18.Bankaitis VA, Cartee RT, Fry MR, Kagiwada S. Phospholipid Transfer Proteins: Emerging Roles in Vesicle Trafficking, Signal Transduction, and Metabolic Regulation. Springer; New York: 1996. pp. 51–72. [Google Scholar]
- 19.Abe A. Primary structure of glycolipid transfer protein from pig brain. J. Biol. Chem. 1990;265:9634–9637. [PubMed] [Google Scholar]
- 20.Lin X, Mattjus P, Pike HM, Windebank AJ, Brown RE. Cloning and expression of glycolipid transfer protein from bovine and porcine brain. J. Biol. Chem. 2000;275:5104–5110. doi: 10.1074/jbc.275.7.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X-M, Malakhova ML, Lin X, Pike HM, Chung T, Molotkovsky JG, Brown RE. Human glycolipid transfer protein: Probing conformation using fluorescence spectroscopy. Biochemistry. 2004;43:10285–10294. doi: 10.1021/bi0495432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao CS, Lin X, Pike HM, Molotkovsky JG, Brown RE. Glycolipid transfer protein mediated transfer of glycosphingolipids between membranes: A model for action based on kinetic and thermodynamic analyses. Biochemistry. 2004;43:13805–13815. doi: 10.1021/bi0492197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattjus P, Molotkovsky JG, Smaby JM, Brown RE. A fluorescence resonance energy transfer approach for monitoring protein-mediated glycolipid transfer between vesicle membranes. Anal. Biochem. 1999;268:297–304. doi: 10.1006/abio.1998.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattjus P, Pike HM, Molotkovsky JG, Brown RE. Charged membrane surfaces impede the protein-mediated transfer of glycosphingolipids between phospholipid bilayers. Biochemistry. 2000;39:1067–1075. doi: 10.1021/bi991810u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dicorleto PE, Fakharzadeh FF, Searles LL, Zilversmit DB. Stimulation by acidic phospholipids of protein-catalyzed phosphatidylcholine transfer. Biochim. Biophys. Acta. 1977;468:296–304. doi: 10.1016/0005-2736(77)90122-5. [DOI] [PubMed] [Google Scholar]
- 26.Machida K, Ohnishi SI. A spin-label study of phosphatidylcholine exchange protein. Regulation of the activity by phosphatidylserine and calcium ion. Biochim. Biophys. Acta. 1978;507:156–164. doi: 10.1016/0005-2736(78)90382-6. [DOI] [PubMed] [Google Scholar]
- 27.Somerharju PJ, Brockerhoff H, Wirtz KWA. A new fluorimetric method to measure protein-catalyzed phospholipid transfer using 1-acyl-2parinaroylphosphatidylcholine. Biochim. Biophys. Acta. 1981;649:521–528. doi: 10.1016/0005-2736(81)90155-3. [DOI] [PubMed] [Google Scholar]
- 28.Szolderits G, Hermetter A, Paltauf F, Daum G. Membrane properties modulate the activity of a phosphatidylinositol transfer protein from the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1989;986:301–309. doi: 10.1016/0005-2736(89)90481-1. [DOI] [PubMed] [Google Scholar]
- 29.Desrumaux C, Athias A, Masson D, Gambert P, Lallemant C, Lagrost L. Influence of the electrostatic charge of lipoprotein particles on the activity of the human plasma phospholipid transfer protein. J. Lipid Res. 1998;39:131–142. [PubMed] [Google Scholar]
- 30.Huuskonen J, Olkkonen VM, Jauhiainen M, Sareneva T, Somerharju PJ, Ehnholm C. Oxidative modification of HDL3 in vitro and its effect on PLTP-mediated phospholipid transfer. Biochim. Biophys. Acta. 1998;1391:181–192. doi: 10.1016/s0005-2760(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 31.Wilkening G, Linke T, Sandhoff K. Lysosomal degradation on vesicular membrane surfaces. Enhanced glucosylceramide degradation by lysosomal anionic lipids and activators. J. Biol. Chem. 1998;273:30271–30278. doi: 10.1074/jbc.273.46.30271. [DOI] [PubMed] [Google Scholar]
- 32.Ruocco MJ, Shipley GG, Oldfield E. Galactocerebroside-phospholipid interactions in bilayer membranes. Biophys. J. 1983;43:91–101. doi: 10.1016/S0006-3495(83)84327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curatolo W. The interactions of 1-palmitoyl-2-oleylphosphatidylcholine and bovine brain cerebroside. Biochim. Biophys. Acta. 1986;861:373–376. doi: 10.1016/0005-2736(86)90441-4. [DOI] [PubMed] [Google Scholar]
- 34.Maulik PR, Shipley GG. N-palmitoyl sphingomyelin bilayers: Structure and interactions with cholesterol and dipalmitoylphosphatidylcholine. Biochemistry. 1996;35:8025–8034. doi: 10.1021/bi9528356. [DOI] [PubMed] [Google Scholar]
- 35.Johnston DS, Chapman D. A calorimetric study of the thermotropic behaviour of mixtures of brain cerebrosides with other brain lipids. Biochim. Biophys. Acta. 1988;939:603–614. doi: 10.1016/0005-2736(88)90108-3. [DOI] [PubMed] [Google Scholar]
- 36.Mattjus P, Malewicz B, Valiyaveettil JT, Baumann WJ, Bittman R, Brown RE. Sphingomyelin modulates the transbilayer distribution of galactosylceramide in phospholipid membranes. J. Biol. Chem. 2002;277:19476–19481. doi: 10.1074/jbc.M201305200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malewicz B, Valiyaveettil JT, Jacob K, Byun H-S, Mattjus P, Baumann WJ, Bittman R, Brown RE. The 3-hydroxy group and 4,5-trans double bond of sphingomyelin are essential for modulation of galactosylceramide transmembrane asymmetry. Biophys. J. 2005;88:2670–2680. doi: 10.1529/biophysj.104.057059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattjus P, Klein A, Pike HM, Molotkovsky JG, Brown RE. Probing for preferential interactions among sphingolipids in bilayer vesicles using the glycolipid transfer protein. Biochemistry. 2002;41:266–273. doi: 10.1021/bi015718l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylund M, Mattjus P. Protein mediated glycolipid transfer is inhibited from sphingomyelin membranes but enhanced to sphingomyelin containing raft like membranes. Biochim. Biophys. Acta. 2005;1669:87–94. doi: 10.1016/j.bbamem.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Nylund M, Kjellberg MA, Molotkovsky JG, Byun HS, Bittman R, Mattjus P. Molecular features of phospholipids that affect glycolipid transfer protein-mediated galactosylceramide transfer between vesicles. Biochim. Biophys. Acta. 2006;1758:807–812. doi: 10.1016/j.bbamem.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 41.Malakhova ML, Malinina L, Pike HM, Kanack AT, Patel DJ, Brown RE. Point mutational analysis of the liganding site in human glycolipid transfer protein. Functionality of the complex. J. Biol. Chem. 2005;280:26312–26320. doi: 10.1074/jbc.M500481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koynova R, Caffrey M. Phases and phase transitions of the sphingolipids. Biochim. Biophys. Acta. 1995;1255:213–236. doi: 10.1016/0005-2760(94)00202-a. [DOI] [PubMed] [Google Scholar]
- 43.Sonnino S, Cantù L, Corti M, Acquotti D, Venerando B. Aggregative properties of gangliosides in solution. Chem. Phys. Lipids. 1994;71:21–45. doi: 10.1016/0009-3084(94)02304-2. [DOI] [PubMed] [Google Scholar]
- 44.Cantù L, Corti M, Del Favero E, Raudino A. Physical aspects of non-ideal mixing of amphiphilic molecules in solution: the interesting case of gangliosides. J. Phys. Condens. Matter. 1997;9:5033–5055. [Google Scholar]
- 45.Tringali C, Papini N, Fusi P, Croci G, Borsani G, Preti A, Tortora P, Tettamanti G, Venerando B, Monti E. Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ganglioside molecules. J. Biol. Chem. 2004;279:3169–31379. doi: 10.1074/jbc.M308381200. [DOI] [PubMed] [Google Scholar]
- 46.Zhou D, Cantu C, Sagiv Y, Schrantz N, Kulkarni AS, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L. Editing of CD1-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou D, Mattner J, Cantu C, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu Y, Yamashita T, Teneberg S, Wang D, Proia R, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 48.Kolter T, Winau F, Schaible UE, Leippe M, Sandhoff K. Lipid-binding proteins in membrane digestion, antigen presentation, and antimicrobial defense. J. Biol. Chem. 2005;280:41125–41128. doi: 10.1074/jbc.R500015200. [DOI] [PubMed] [Google Scholar]
- 49.Brown RE, Sugár IP, Thompson TE. Spontaneous transfer of gangliotetraosylceramide between phospholipid vesicles. Biochemistry. 1985;24:4082–4091. doi: 10.1021/bi00336a042. [DOI] [PubMed] [Google Scholar]
- 50.Brown RE, Thompson TE. Spontaneous transfer of ganglioside GM1 between phospholipid vesicles. Biochemistry. 1987;26:5454–5460. doi: 10.1021/bi00391a036. [DOI] [PubMed] [Google Scholar]
- 51.van den Besselaar AM, Helmkamp GM, Jr., Wirtz KWA. Kinetic model of the protein-mediated phosphatidylcholine exchange between single bilayer liposomes. Biochemistry. 1975;14:1852–1858. doi: 10.1021/bi00680a008. [DOI] [PubMed] [Google Scholar]
- 52.Helmkamp GM, Jr., Wirtz KWA, van Deenen LLM. Phosphatidylinositol exchange protein. Effects of membrane structure on activity and evidence for a ping-pong mechanism. Arch. Biochem. Biophys. 1976;174:590–600. doi: 10.1016/0003-9861(76)90388-x. [DOI] [PubMed] [Google Scholar]
- 53.Machida K, Ohnishi S. Effect of bilayer membrane curvature on activity of phosphatidylcholine exchange protein. Biochim. Biophys. Acta. 1980;596:201–209. doi: 10.1016/0005-2736(80)90355-7. [DOI] [PubMed] [Google Scholar]
- 54.Bozzato RP, Woolley D, Tinker DO. Catalytic properties of the yeast phospholipid transfer protein. Biochem. Cell Biol. 1987;65:203–210. doi: 10.1139/o87-026. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimura T, Welti R, Helmkamp GM., Jr General kinetic model for protein-mediated phospholipid transfer between membranes. Arch. Biochem. Biophys. 1988;266:299–312. doi: 10.1016/0003-9861(88)90262-7. [DOI] [PubMed] [Google Scholar]
- 56.Lalanne F, Ponsin G. Mechanism of the phospholipid transfer protein-mediated transfer of phospholipids from model lipid vesicles to high density lipoproteins. Biochim. Biophys. Acta. 2000;1487:82–91. doi: 10.1016/s1388-1981(00)00087-1. [DOI] [PubMed] [Google Scholar]
- 57.Rao CS, Chung T, Pike HM, Brown RE. Glycolipid transfer protein interaction with bilayer vesicles: Modulation by changing lipid composition. Biophys. J. 2005;89:4017–4028. doi: 10.1529/biophysj.105.070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West G, Nylund M, Slotte JP, Mattjus P. Membrane interaction and activity of the glycolipid transfer protein. Biochim. Biophys. Acta. 2006;1758:1732–1742. doi: 10.1016/j.bbamem.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 59.Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 60.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruhn H. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 2005;389:249–257. doi: 10.1042/BJ20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snook CF, Jones JA, Hannun YA. Sphingolipid-binding proteins. Biochim. Biophys. Acta. 2006;1761:927–946. doi: 10.1016/j.bbalip.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Schubert Wright C, Zhao Q, Rastinejad F. Structural analysis of lipid complexes of GM2-activator protein. J. Mol. Biol. 2003;331:951–964. doi: 10.1016/s0022-2836(03)00794-0. [DOI] [PubMed] [Google Scholar]
- 64.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat. Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 65.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat. Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 66.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 67.Hanada K. Discovery of the molecular machinery CERT for endoplasmic reticulum-to-Golgi trafficking of ceramide. Mol. Cell. Biochem. 2006;286:22–31. doi: 10.1007/s11010-005-9044-z. [DOI] [PubMed] [Google Scholar]
- 68.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 69.Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure (Camb.) 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Wirtz KWA. Phospholipid transfer proteins revisited. Biochem. J. 1997;324:353–360. doi: 10.1042/bj3240353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soccio RE, Breslow JL. StAR-related lipid transfer (START) proteins: Mediators of intracellular lipid metabolism. J. Biol. Chem. 2003;278:22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- 72.Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 73.Hamilton JA. Fatty acid interactions with proteins: what X-ray crystal and NMR solution structures tell us. Prog. Lipid Res. 2004;43:177–199. doi: 10.1016/j.plipres.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Flower DR. Experimentally determined lipocalin structures. Biochim. Biophys. Acta. 2000;1482:46–56. doi: 10.1016/s0167-4838(00)00147-3. [DOI] [PubMed] [Google Scholar]
- 75.Kader J-C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 76.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. http://supfam.org/SUPERFAMILY/cgibin/scop.cgi?sunid=110004 [DOI] [PubMed] [Google Scholar]
- 77.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. http://scop.mrc-lmb.cam.ac.uk/scop-1.69/data/scop.b.b.bja.b.b.A.html [DOI] [PubMed] [Google Scholar]
- 78.Airenne TT, Kidron H, Nymalm Y, Nylund M, West GP, Mattjus P, Salminen TA. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J. Mol. Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 79.Malinina L, Malakhova ML, Kanak AT, Lu M, Abagyan R, Brown RE, Patel DJ. The liganding mode of glycolipid transfer protein is controlled by glycosphingolipid structure. PLoS Biol. 2006;4:e362. doi: 10.1371/journal.pbio.0040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vyas NK, Vyas MN, Quiocho FA. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- 81.Mahfoud R, Garmy N, Maresca M, Yahi N, Puigserver A, Fantini J. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and Hiv-1 proteins. J. Biol. Chem. 2002;277:11292–11296. doi: 10.1074/jbc.M111679200. [DOI] [PubMed] [Google Scholar]
- 82.Aubert-Jousset E, Garmy N, Sbarra V, Fantini J, Sadoulet M-O, Lombardo D. The combinatorial extension method reveals a sphingolipid binding domain on pancreatic bile salt-dependent lipase: role in secretion. Structure (Camb.) 2004;12:1437–1447. doi: 10.1016/j.str.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 83.Quiocho FA, Vyas NK. Atomic interactions between protein/enzymes and carbohydrates. In: Hecht SM, editor. Bioorganic Chemistry: Carbohydrates. Oxford Univ. Press; New York: 1999. pp. 441–457. [Google Scholar]
- 84.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate interactions. Annu. Rev. Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 85.Sujatha MS, Balaji PV. Identification of common structural features of binding sites in galactose-specific proteins. Proteins: Struct. Func. Bioinform. 2004;55:44–65. doi: 10.1002/prot.10612. [DOI] [PubMed] [Google Scholar]
- 86.Sujatha MS, Sasidhar YU, Balaji PV. Energetics of galactose- and glucose-aromatic amino acid interactions: Implications for binding in galactose-specific proteins. Protein Sci. 2004;13:2502–2514. doi: 10.1110/ps.04812804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murray D, Ben-Tal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- 88.Pool CT, Thompson TE. Chain length and temperature dependence of the reversible association of model acylated proteins with lipid bilayers. Biochemistry. 1998;37:10246–10255. doi: 10.1021/bi980385m. [DOI] [PubMed] [Google Scholar]
- 89.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van den Berg B, Black PN, Clemons WM, Jr., Rapoport TA. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- 91.White SH, Wimley WC. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 92.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 93.Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 94.Hurley JH. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho W, editor. Lipid-Binding Domains. Biochim. Biophys. Acta. 2006;1761:803–968. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 96.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. http://opm.phar.umich.edu/families.php?family=117 [DOI] [PubMed] [Google Scholar]
- 97.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: A computational approach. Protein Science. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jorgensen LB, Brown RE, Mundy J. Knockout of Arabidopsis accelerated-cell-death 11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mattjus P, Turcq B, Pike HM, Molotkovsky JG, Brown RE. Glycolipid intermembrane transfer is accelerated by HET-C2, a filamentous fungus gene product involved in the cell-cell incompatibility response. Biochemistry. 2003;42:535–542. doi: 10.1021/bi026896x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saupe SJ, Descamps C, Turcq B, Begueret J. Inactivation of the Podospora anserina vegetative incompatibility locus het-c2, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5927–5931. doi: 10.1073/pnas.91.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Begueret J, Turcq B, Clave C. Vegetative incompatibility in filamentous fungi: het genes begin to talk. Tends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 102.Loubradou G, Turcq B. Vegetative incompatibility in filamentous fungi: a roundabout way of understanding the phenomenon. Res. Microbiol. 2000;151:239–245. doi: 10.1016/s0923-2508(00)00145-5. [DOI] [PubMed] [Google Scholar]
- 103.Stone JM, Heard JE, Asai T, Ausubel FM. Simulation of fungal-mediated cell death by Fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell. 2000;12:1811–1822. doi: 10.1105/tpc.12.10.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H, Jones C, Ciacci-Zanella J, Holt T, Gilchrist DG, Dickman MB. Fumonisins and Alternaria alternata lycopersici toxins: sphingangine analog mycotoxins induce apoptosis in monkey kidney cells. Proc. Natl. Acad. Sci. USA. 1996;93:3461–3465. doi: 10.1073/pnas.93.8.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielson HB, Lacy M, Austin MA, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy JA. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 106.Godi A, DiCampli AD, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell membrane traffic by binding ARF and PtdIns(4)P. Nature Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 107.De Matteis MA, Godi A. Protein-lipid interactions in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta. 2004;1666:264–274. doi: 10.1016/j.bbamem.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 108.Renou J-P, Bierie B, Miyoshi K, Cui Y, Djiane J, Reichenstein M, Shani M, Hennighausen L. Identification of genes differentially expressed in mouse mammary epithelium transformed by an activated β-catenin. Oncogene. 2003;22:4594–4610. doi: 10.1038/sj.onc.1206596. [DOI] [PubMed] [Google Scholar]
- 109.van Meer G, Lisman Q. Sphingolipid transport: rafts and translocators. J. Biol. Chem. 2002;277:25855–25858. doi: 10.1074/jbc.R200010200. [DOI] [PubMed] [Google Scholar]
- 110.Futerman AH. Intracellular trafficking of sphingolipids: relationship to biosynthesis. Biochim. Biophys. Acta. 2006;1758:1885–1892. doi: 10.1016/j.bbamem.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 111.Burger KNJ, van der Bijl P, van Meer G. Topology of sphingolipid galactosyltransferases in ER and Golgi: Transbilayer movement of monohexosyl sphingolipids is required for higher glycosphingolipid synthesis. J. Cell Biol. 1996;133:15–28. doi: 10.1083/jcb.133.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warnock DE, Lutz MS, Blackburn WA, Young WW, Jr., Baenziger JU. Transport of newly synthesized glucosylceramide to the plasma membrane by a non-Golgi pathway. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2708–2712. doi: 10.1073/pnas.91.7.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zegers MMP, Kok JW, Hoekstra D. Use of photoactivatable sphingolipid analogues to monitor lipid transport in mammalian cells. Biochem. J. 1997;328:489–498. doi: 10.1042/bj3280489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ledeen RW, Wu G. Sphingolipids of the nucleus and their role in nuclear signaling. Biochim. Biophys. Acta. 2006;1761:588–598. doi: 10.1016/j.bbalip.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 115.Morales A, Colell A, Mari M, Garcia-Ruiz C, Fernandez-Checa JC. Glycosphingolipids and mitochondria: role in apoptosis and disease. Glycoconj. J. 2004;20:579–588. doi: 10.1023/B:GLYC.0000043294.62504.2c. [DOI] [PubMed] [Google Scholar]
- 116.Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell. Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 117.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Reports. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Van Kaer L. α-Galactosylceramide therapy for autoimmune diseases: prospects and obstacles. Nat. Rev. Immunol. 2005;5:31–42. doi: 10.1038/nri1531. [DOI] [PubMed] [Google Scholar]