Abstract
Purpose
The molecular mechanism underlying retinal ganglion cell (RGC) differentiation is not fully understood. In this study, the role of the basic helix-loop-helix (bHLH) genes ath5 and NSCL1 in RGC differentiation was examined, by testing whether their coexpression would promote RGC differentiation to a greater extent than either gene alone.
Methods
The replication-competent avian RCAS retrovirus was used to coexpress ath5 and NSCL1 through an internal ribosomal entry site. The effect of the coexpression on RGC differentiation was assayed in vivo in the developing chick retina and in vitro in RPE cell cultures derived from day 6 chick embryos.
Results
Coexpression of ath5 and NSCL1 in RPE cells cultured in the presence of bFGF promoted RPE transdifferentiation toward RGCs, and the degree of transdifferentiation was much higher than with either gene alone. Cells expressing RGC markers, including RA4, calretinin, and two neurofilament-associated proteins, displayed processes that were remarkably long and thin and often had numerous branches, characteristics of long-projecting RGCs. In the developing chick retina, retroviral expression of NSCL1 resulted in a moderate increase in the number of RGCs, results similar to retroviral expression of ath5. Coexpression of ath5 and NSCL1 yielded increases in RGCs greater than the sum of their increases when expressed separately.
Conclusions
Both in vitro and in vivo data indicate that the combination of ath5 and NSCL1 promotes RGC differentiation to a greater degree than either gene alone, suggesting a synergism between ath5 and NSCL1 in advancing RGC development.
Many aspects of the molecular regulation of retinal ganglion cell (RGC) differentiation remain unclear. Published studies have shown that genes encoding transcription factors of the homeodomain family1–4 and the basic helix-loop-helix (bHLH) family, including Ath5 and NSCL1, play a role in regulating gene expression during RGC development in the vertebrate retina. Ath5 is expressed in immature RGCs, and their expression becomes undetectable in mature RGCs5,6 (Ma et al., manuscript submitted). Chicken NSCL1, like its mammalian counterpart,7,8 is specifically expressed in the nervous system.9 In the developing retina, NSCL1 is first expressed in differentiating ganglion cells during early development and then in Müller glia during late development.10 Retroviral expression of NSCL1 in the developing chick retina causes premature cessation of mitotic activity, massive cell death, and microphthalmia.10
The function of ath5 in RGC development has been the subject of extensive investigation. Xenopus ath5 mRNA injected into Xenopus embryos at the two-cell stage promotes retinal progenitor cell differentiation into RGCs.11 In the embryonic chick retina, retroviral expression of chick and mouse ath5 increases the number of RGCs.6 Targeted deletion of ath5 in mouse results in a lack, or a dramatic reduction, of RGCs, indicating that ath5 is indispensable for RGC development.12,13 Kay et al.14 have reported a lack of RGCs in the zebrafish ath5 mutant, lackritz. Nonetheless, the role that ath5 plays during vertebrate retinal development is likely to be multifaceted. Brown et al.5 reported that lipofection of mouse ath5 into the frog retina resulted in an increase in bipolar cells. Through lineage tracing, Yang et al.15 found that photoreceptors and amacrine cells, in addition to RGCs, may express ath5 at certain times in their lives. Our recent RPE transdifferentiation assays indicate that chick ath5 (also referred to as cath5) can lead to the photoreceptor pathway (Ma et al., manuscript submitted).
We have previously investigated the roles of ath5 and NSCL1 in RGC genesis using the RPE transdifferentiation assay we developed16–20 (Ma et al., manuscript submitted). The assay takes advantage of the simplicity and plasticity of RPE cells to allow us to examine the proneural activities of various genes or factors. The neural retina not only naturally expresses several proneural genes, but also contains different types of cells, creating molecularly and biochemically heterogeneous cellular contexts and intercellular environments. The non-neural RPE, in contrast, is a single-layered structure that lacks the expression of many proneural genes. Furthermore, the RPE is developmentally related to the retina. RPE tissue at early developmental stages may be triggered to transdifferentiate into a neural retina,21–24 and cultured RPE cells can be induced to undergo transdifferentiation into what closely resembles retinal neurons.16–19 An RPE cell transdifferentiation assay showed that ath5 and NSCL1 promote bFGF-initiated RPE transdifferentiation toward RGCs, but alone they are insufficient to induce this change; nonetheless, the extent of transdifferentiation is rather limited.20
In this study, we examined whether ath5 and NSCL1 act synergistically in advancing RGC differentiation. Ath5 and NSCL1 were coexpressed through an internal ribosomal entry site (IRES) and a replication-competent retrovirus. We found that retroviral coexpression of ath5 and NSCL1 in the developing chick retina increased the number of cells that expressed RGC markers more than the sum of the increases caused when the two genes were express separately. In RPE cell cultures, cells morphologically resembling RGCs were readily observed in dishes infected by retrovirus coexpressing ath5 and NSCL1. However, in the absence of bFGF, ath5 and NSCL1 coexpression did not alleviate the insufficiency in inducing RPE transdifferentiation toward RGCs.
Methods
Generation of Recombinant Retrovirus Coexpressing ath5 and NSCL1
The IRES (nucleotide 2175–2760 in pFB-Neo; Stratagene, La Jolla, CA) was subcloned into a modified version of the adeno-associated virus (AAV) vector (Stratagene), onto which a cassette of ath5-IRES-NSCL1 was built. The translation initiation of ath5 was under the regulation of the Src oncogene sequence originating from the shuttle vector Cla12Nco,25 and the translation initiation of NSCL1 was regulated by the IRES. The ath5-IRES-NSCL1 cassette was then inserted into an replication-competent avian RCAS proviral vector25 for coexpression of ath5 and NSCL1 (Fig. 1). Previously described procedures were followed for transfecting chick embryonic fibroblast cells with recombinant proviral DNA and for generating high-titer viral stocks.16 RCAS retroviruses expressing other genes were generated as described (RCAS-ath5,20 RCAS-NSCL1,9 RCAS-GFP16). Titers of the viral stocks ranged from 5 × 107 to 2 × 108 pfu/mL.
Figure 1.
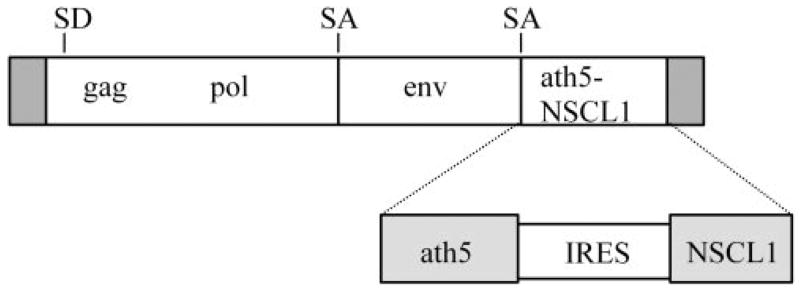
Schematic illustration of RCAS coexpressing ath5 and NSCL1. SD, splicing donor; SA, splicing acceptor.
Microinjection of Retrovirus into the Subretinal Space
To access the developing chick embryos, an opening of approximately 1 × 1 cm was made in the eggshell. Through the opening, approximately 1 μL of concentrated viral suspension was placed into the neural tube and the subretinal space of day 2 embryos, using a micropipette attached to a pneumatic pump with a positive-pressure delivery system. The injected viral suspension contained fast green, allowing us to monitor visually the spreading into the subretinal space. Injection was repeated two more times during the day; this procedure routinely produces a thorough infection of the retina.10,16,26 Viral infection was later detected with a specific antibody against viral protein p27.
Dissociation of Retinal Cells
Retinas were harvested at embryonic day (E)8. At this time, essentially all the RGCs have been generated and the developmental ganglion cell death that takes place between E10 and E12 has not yet begun. The right-eye (injected) retinas from three or more embryos were pooled, and their cells were dissociated by incubation with trypsin/EDTA followed by mechanical trituration. Dissociated retinal cells were seeded onto polyornithine-treated substratum for 4 hours in a humidified 37°C incubator with 5% CO2. Cells were then fixed with ice-cold 4% paraformaldehyde for 30 minutes at room temperature and subjected to double immunostaining to identify retinal ganglion cells (Brn3A+27 or Islet-1+28) that had been infected with the virus (p27+). To achieve a cell density suitable for counting, cells equivalent to on thirtieth of an E8 retina were seeded into each 35-mm dish.
Cultures of Dissociated RPE Cells
RPE was isolated from day-6 chick embryos and cultured as dissociated cells, as previously described.16,20 When used, bFGF was added to the culture medium at the inception of culture at a final concentration of 10 ng/mL. When the cells reached approximately 50% confluence, which usually took approximately 3 to 4 days, the culture was inoculated with RCAS virus expressing the various genes: ath5, NSCL1, ath5-NSCL1, or GFP as a negative control. Cells in the infected cultures were fixed with 4% paraformaldehyde for 30 minutes and then subjected to immunocytochemical analysis.
Immunocytochemistry
Monoclonal antibody RA4 (1:1000 dilution) was a gift from Steven McLoon (University of Minnesota, Minneapolis, MN). Monoclonal antibody (clone HM-2; 1:200 dilution) against microtubule-associated protein (MAP)-2 was purchased from Sigma-Aldrich (St. Louis, MO). Monoclonal antibody against Brn3A (1:200 dilution) was purchased from Chemicon (Temecula, CA). Three monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank (Iowa University, Iowa City, IA): anti-Islet-1 (clone 39.4D5, 1:100; developed by Thomas Jessell, Columbia University, New York, NY), 3A10 (1:100; developed by Thomas Jessell), and 4H6 (1:100; developed by Willi Halfter, University of Pittsburgh, PA). Antibody against viral protein p27 was purchased from Spafas (Preston, CT). Standard immunocytochemistry was performed with alkaline phosphatase-conjugated secondary antibodies (Vector Laboratories, Burlingame, CA) or fluorophore-conjugated secondary antibodies (Molecular Probes, Eugene, OR), as described by the manufacturers.
Cell Counting and Analysis
The number of cells with typical neural morphology in each well of a 24-well plate was scored from 10 viewing areas under a 20× objective. The counts from three wells were used to calculate the mean and SD with a computer program (Origin 7.0; Microcal, Northampton, MA). Each experiment included samples infected individually with RCAS-ath5-NSCL1, RCAS-ath5, RCAS-NSCL1, and RCAS-GFP as a negative control. Experiments were repeated at least three times in their entirety, from RPE dissection to cell counting.
For dissociated retinal cells, the number of single-labeled and double-labeled cells was counted from 20 viewing areas in one 35-mm dish with a 20× objective. The number in the 20 view areas was summed, and the means and SDs of three dishes of each experiment were calculated (Origin 7.0; Microcal). Experiments were repeated at least three times in their entirety, from microinjection to cell counting. Data from RCAS-ath5-NSCL1 were compared with those from RCAS-ath5, RCAS-NSCL1, and RCAS-GFP.
To analyze the number of RGCs on E7 retinal sections, sections were viewed under a 40× objective; 10 viewing areas of infected regions and 10 viewing areas of adjacent, uninfected regions from eight sections per retina were used. The total number of Islet-1+ cells and Brn3A+ cells in the selected view areas were scored and used to calculate the percentage of change in the virally infected regions versus that in the adjacent, uninfected regions. The mean and SD of three retinas were calculated (Origin 7.0; Microcal).
Results
Enhanced Transdifferentiation into RGCs by ath5 and NSCL1 Coexpression in RPE Cell Cultures
To assay whether coexpression of ath5 and NSCL1 would be sufficient to trigger RPE transdifferentiation into RGCs, we examined RPE cell cultures infected with RCAS-ath5-NSCL1 for the de novo generation of neuronlike cells bearing similarities to RGCs. Previously, we showed that a single proneural bHLH gene, neurogenin2, can induce such transdifferentiation when ectopically expressed in cultured RPE cells through RCAS retroviral transduction.19 Using the same transdifferentiation system, however, we found that coexpression of ath5 and NSCL1 did not induce RPE transdifferentiation toward RGCs. Cultures infected with RCAS-ath5-NSCL1 contained no more cells positive for monoclonal antibody RA4 than did control cultures infected with RCAS-GFP (data not shown). RA4 is an early marker for ganglion cells in the chick retina.29 Thus, simple coexpression of ath5 and NSCL1 did not initiate an RGC differentiation program in cultured RPE cells.
We then examined whether coexpression of ath5 and NSCL1 would promote RGC differentiation when the RPE cells were cultured in the presence of bFGF. Previously, we showed that bFGF can initiate RPE transdifferentiation toward RGCs, but the extent of transdifferentiation is very limited, because essentially all RA4+ cells maintain the fibroblast-like morphology typically of RPE cells in low-density cultures. When the cells are manipulated to express ath5 or NSCL1 through retroviral transduction, not only are RA4+ cells more numerous, but they show altered morphology, such as distinguishable processes20 (Figs. 2B, 2C; dashed arrows). Nonetheless, the processes on the RA4+ cells in cultures treated with bFGF and with RCAS-ath5 or RCAS-NSCL1 often are devoid of branches and are shorter and wider than typical neuronal processes (Figs. 2B, 2C; dashed arrows). Only on rare occasions do RA4+ cells exhibit typical RGC morphology with long, thin, and often branching processes.
Figure 2.
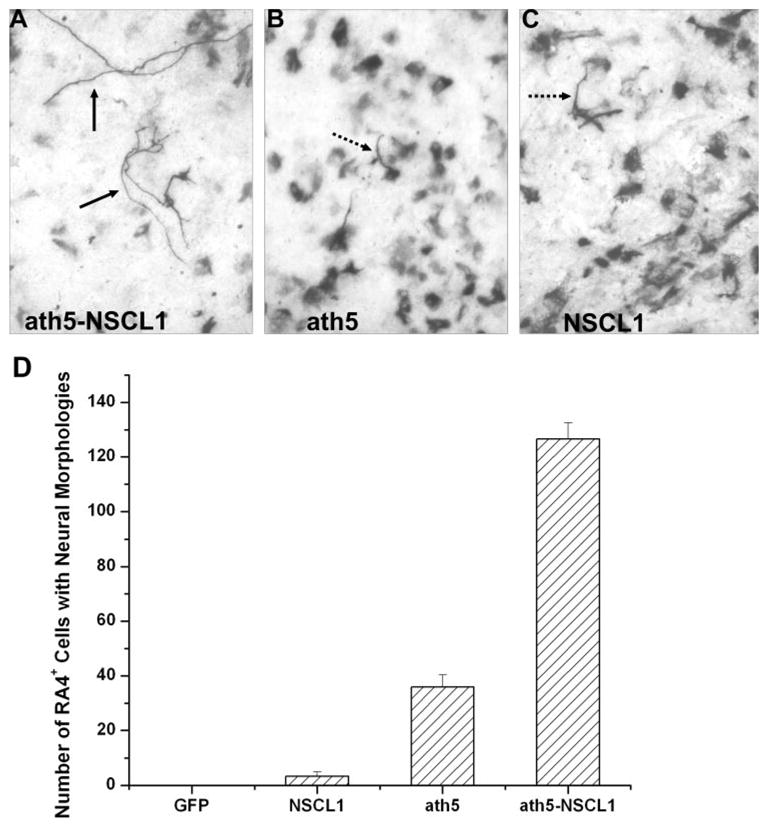
RA4+ cells in bFGF-primed RPE cell cultures infected with retroviruses expressing various genes. (A–C) Morphology of RA4+ cells in cultures infected with RCAS-ath5-NSCL1, RCAS-ath5, or RCAS-NSCL1. Dashed and solid arrows: distinguishable processes. (D) The number of RA4+ cells with typical neural morphology. Shown are the mean and SD in 10 viewing areas under a 20× objective.
When bFGF-primed RPE cells were infected with RCAS-ath5-NSCL1, some of the RA4+ cells exhibited morphology remarkably resembling typical RGC neurons (Fig. 2A). These cells displayed elaborate cellular processes (Fig. 3). Some cells showed multiple processes emanating from the cell bodies; some had two opposing processes, with one of them forming articulate branches at the end, indicative of fine axons reaching different targets (Fig. 3). The various morphologies of the RA4+ cells suggested that different subtypes of RGCs might be present in the bFGF-primed RPE cells ectopically coexpressing ath5 and NSCL1. No cells with typical neural morphology were observed in the GFP control. A small number of such cells were detected when RPE cells were infected with either RCAS-ath5 or RCAS-NSLC1 (3.3 ± 1.5 and 36.0 ± 4.3, respectively, in 10 viewing areas under a 20× objective; Fig. 2D). With coexpression of ath5 and NSCL1, the number increased to 126.7 ± 5.9, which is more than three times greater than the sum of either gene alone.
Figure 3.
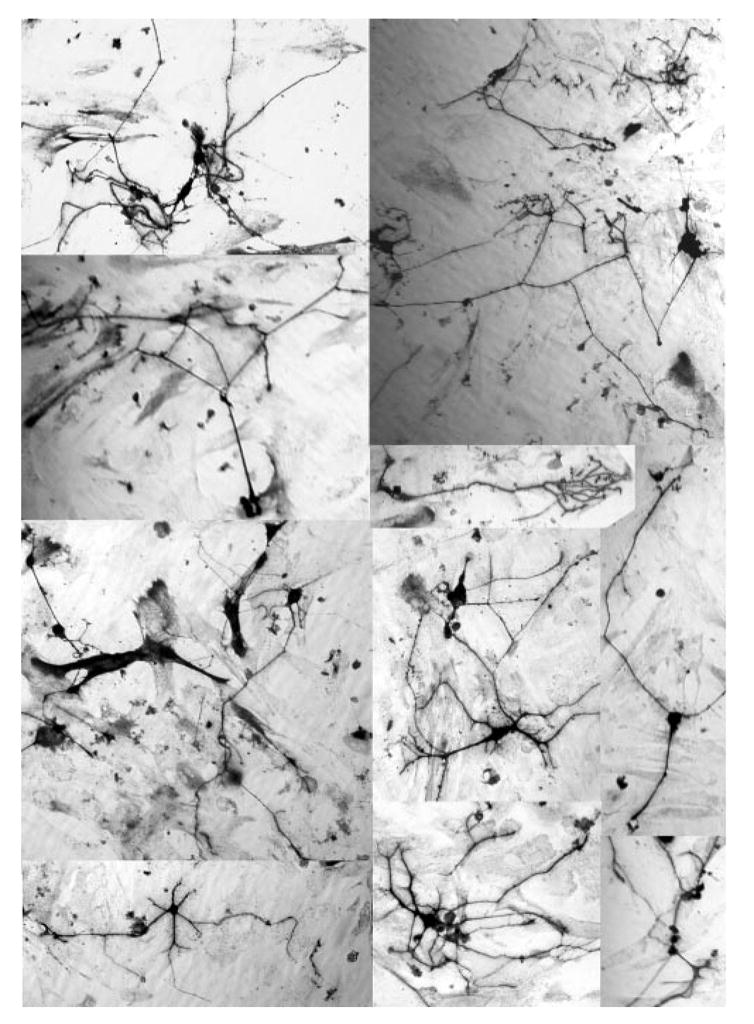
The neuronlike morphology of RA4+ cells from bFGF-primed RPE cell cultures ectopically coexpressing ath5 and NSCL1.
In addition to the RA4 antigen, cells undergoing transdifferentiation expressed other RGC markers, including calretinin, MAP2, the 160-kDa neurofilament protein recognized by monoclonal antibody 4H6, and the neurofilament-associated antigen recognized by monoclonal antibody 3A10 (Fig. 4). Some of the immunopositive cells exhibited typical neuronal morphology. We did not find detectable levels of expression of Islet-1 or Brn3A (data not shown).
Figure 4.
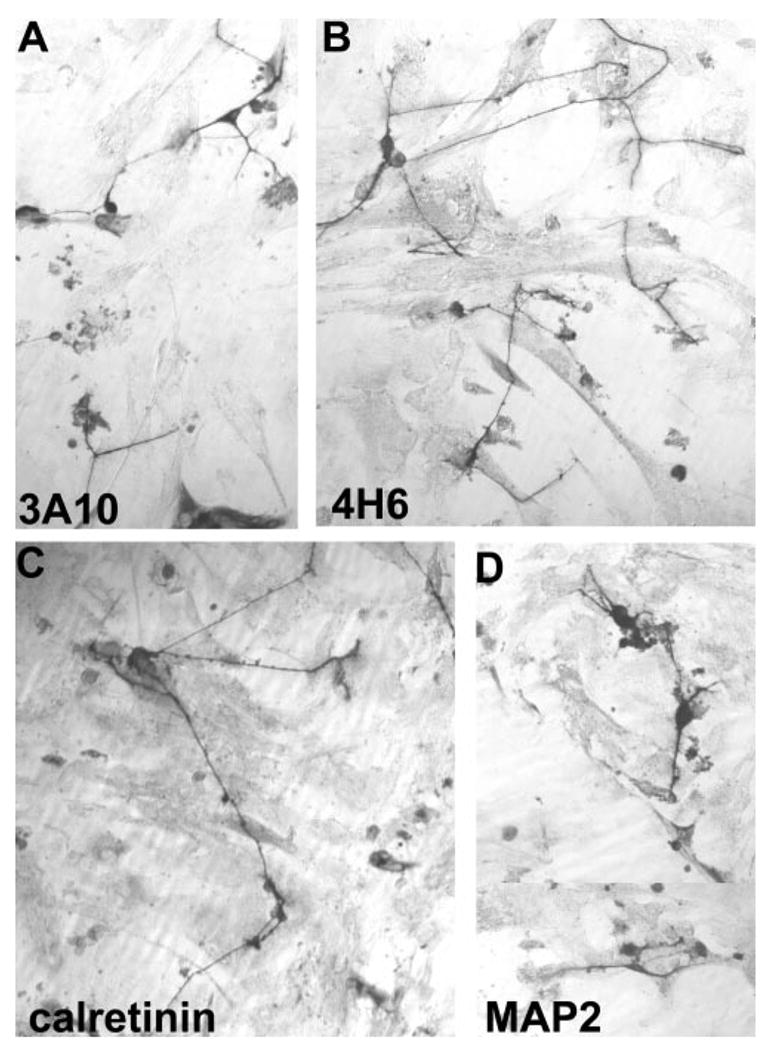
The neuronlike morphology of cells immunopositive for (A) 3A10, (B) 4H6, (C) calretinin, and (D) MAP2. RPE cultures were primed with bFGF and infected with RCAS-ath5-NSCL1.
Increase in RGCs in Retinas Infected with RCAS-ath5-NSCL1
To address whether ath5 and NSCL1 act synergistically in vivo to promote RGCs differentiation, we examined whether ath5 and NSCL1 coexpression would result in an increase in cells that expressed RGC markers in the developing retina. RCAS-ath5-NSCL1 was used to achieve coexpression of ath5 and NSCL1 in the developing retina by microinjecting the virus into the subretinal space at E2, the optic cup stage. Retinas were harvested for analysis of the number of RGCs at E7 to E8, when ganglion cell genesis is essentially finished. Another reason for using E7-to-E8 retina was to avoid complications caused by cell death that we have previously observed to take place beginning at E9 in retinas misexpressing NSCL1.10 To facilitate cell identification and data analysis, we used monoclonal antibodies that recognize nuclear proteins Islet-128 and Brn3A.27
In initial experiments, we performed repeated microinjection, each time with ~1 μL of virus (titer ~ 5 × 107 pfu/mL) placed into the subretinal space and the neural tube, aiming at a thorough infection of the retina.10,16,26 Because RCAS is replication-competent and because a small amount of the injected viral suspension may overflow from the injection site onto the embryo, a widespread viral infection in the embryos, including systemic regions, often occurs several days after the injection.30
Chick embryos injected with RCAS coexpressing ath5 and NSCL1 developed normally. They appeared indistinguishable from those not subjected to experimental manipulation and those injected with RCAS-ath5 or RCAS-GFP. This was unexpected, as we had observed that a similar microinjection of RCAS-NSCL1 results in severe growth retardation and embryonic lethality with no embryos surviving longer than E13. In addition, retroviral NSCL1 expression in the developing retina causes microphthalmia and gross distortion of retinal lamination.10 None of these effects was observed with embryos coexpressing ath5 and NSCL1. The size and appearance of the eye and the lamination of the retina were indistinguishable from the controls (data not shown). The phenotypic disparities between expression of NSCL1 and coexpression of ath5 and NSCL1 could be due to a lower NSCL1 expression level in the coexpression construct, since the level of protein synthesis regulated through the IRES is believed to be low,31 or, alternatively, to a possible attenuation of the teratogenic effect of NSCL1 by ath5.
Despite their normal appearance, retinas infected with RCAS-ath5-NSCL1 contained more RGCs. To reduce ambiguity associated with comparing retinal sections of different eyes, we generated partially infected retinas using a single, instead of a repetitive, microinjection. A partially infected retina allows for a direct comparison between virally infected and uninfected regions at adjacent locations in the same retinal section. We found that E7 retinas that had been infected with RCAS-ath5-NSCL1 contained more Islet-1+ cells in the infected regions than the adjacent, uninfected regions (Figs. 5G, 5H). An increase in Islet-1+ cells was also observed in regions infected with RCAS-ath5 (Figs. 5C, 5D) and RCAS-NSCL1 (Figs. 5E, 5F), whereas the number of Islet-1+ cells appeared similar between infected and uninfected regions in embryos microinjected with RCAS-GFP (Figs. 5A, 5B). Cell counting revealed that the increase due to ath5 and NSCL1 coexpression was greater than that of expressing ath5 or NSCL1 alone. On average, coexpression of ath5 and NSCL1 resulted in a more than 45% increase in the number of Islet-1+ cells, whereas expression of either alone resulted in increases of ~20% and ~10%, respectively (Fig. 5I).
Figure 5.
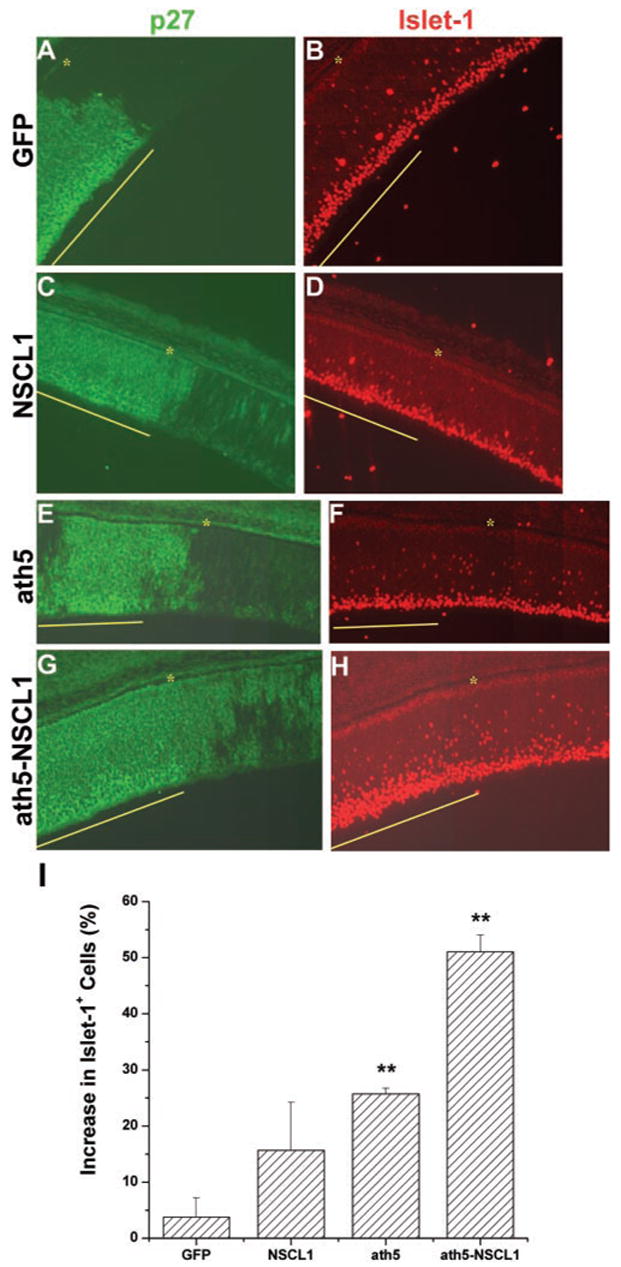
The increase in Islet-1+ cells in E7 retinas partially infected with retrovirus expressing ath5-NSCL1, ath5, or NSCL1. Each section was double stained to show the regions infected by the virus (p27 immunostaining; A, C, E, G) and expression of Islet-1 (B, D, F, H). RCAS expressing GFP (A, B) was used as the control. Yellow line: virally infected regions; (*) RPE. (I) The percentage increase in the Islet-1+ cells in the infected regions compared with that in adjacent, uninfected regions. **Statistically significant difference at P ≤ 0.01 compared with the GFP sample.
The number of Brn3A+ cells also increased with expression of either gene alone and with coexpression of ath5 and NSCL1. Coexpression of ath5 and NSCL1 in the developing retina resulted in a 30% increase in cells expressing Brn3A, whereas expression of either ath5 or NSCL1 alone resulted in an increase of approximately 5% (Fig. 6). The increase in Brn3A+cells with coexpression was more than the sum of the increases by the expression of either gene alone.
Figure 6.
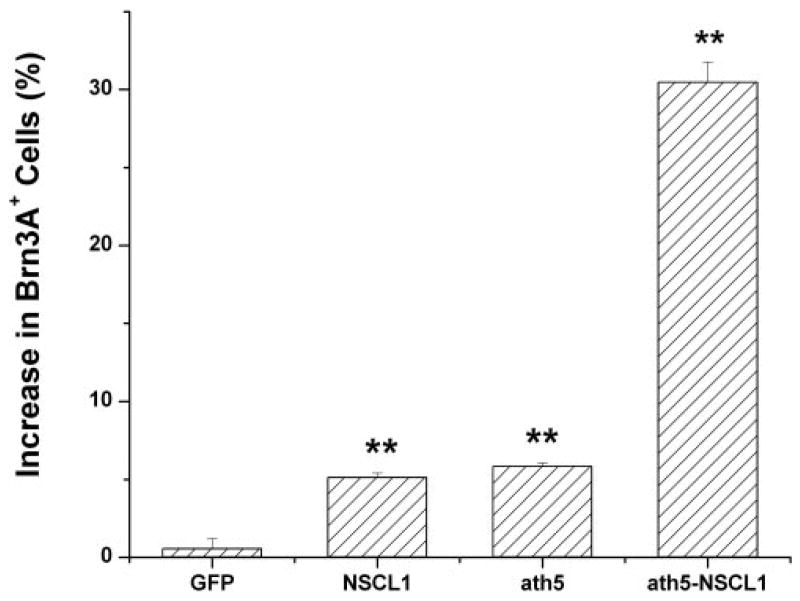
The percentage of increase in Brn3A+ cells in regions infected with the virus compared with that in uninfected regions in E7 retinas. **Statistically significant difference at P ≤ 0.01 compared with the GFP sample.
The number of RGCs was also scored, using dissociated E8 retinal cells to avoid histologic variations. In the control retina, 6.92% ± 0.87% of the cells infected with RCAS-GFP developed into RGCs and expressed Islet-1 (Fig. 7A). In the experimental retinas, 9.58% ± 0.28% and 10.48% ± 0.61%, respectively, of retina cells infected with RCAS-NSCL1 or RCAS-ath5 were Islet-1+, increases of 38% and 51%, respectively, over the control. When retina was infected with RCAS-ath5-NSCL1, 15.73% ± 0.87% of the infected cells were Islet-1+, an increase of 127% over the control (Fig. 7A). Similarly, the percentage of infected retinal cells that expressed Brn3A increased from 4.4% ± 0.51% in the GFP retinas to 7.39% ± 0.51% in the NSCL1 (68% increase), 7.35% ± 0.41 (67% increase) in ath5 retinas, and 10.03% ± 0.64% (128% increase) in the ath5 + NSCL1 retinas (Fig. 7B). All the increases were statistically significant at P ≤ 0.01 compared with that of GFP, and the increase in coexpression over either gene alone was also significant at P ≤ 0.01. The increases in Islet-1+ cells in the coexpressing retinas were higher than the sum of single expression of the two genes.
Figure 7.
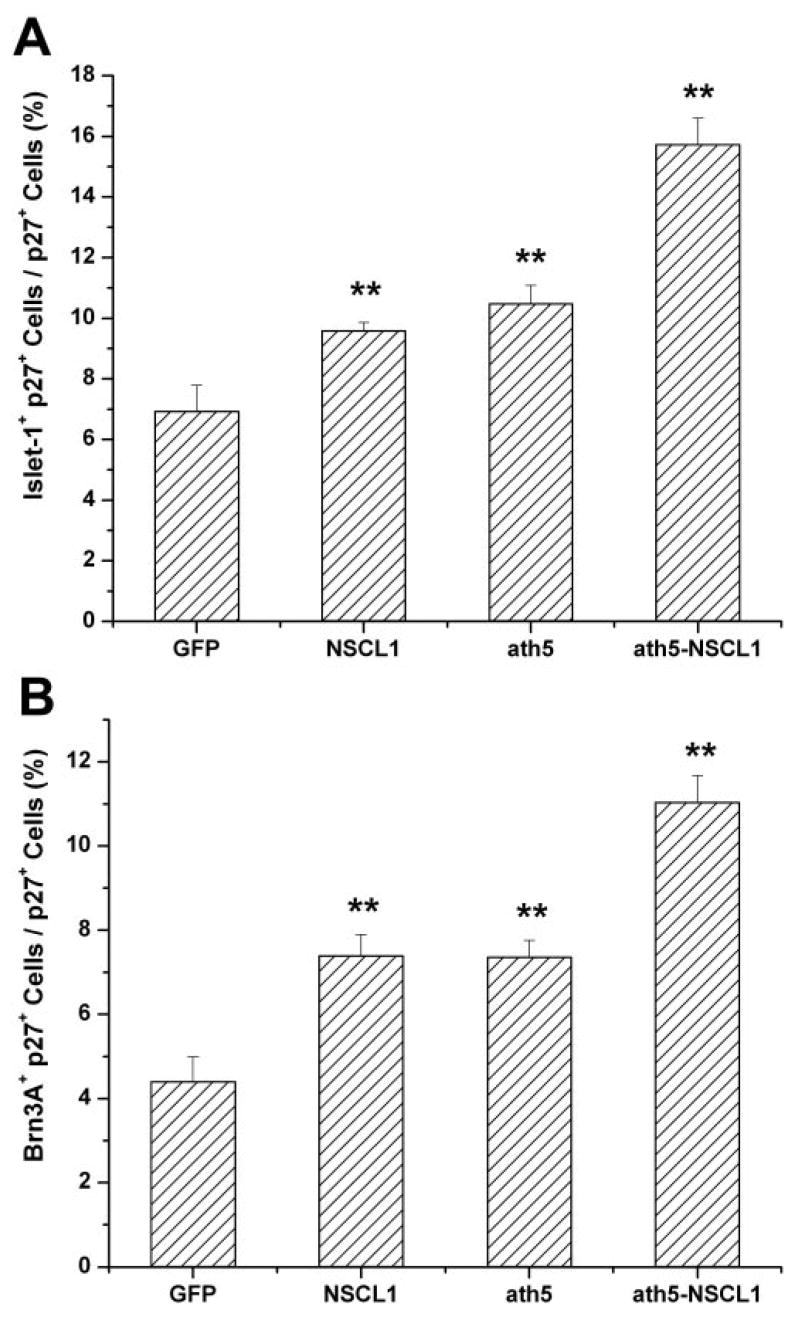
The percentage of p27+ cells that expressed a ganglion cell marker: Islet-1 (A) or Brn3A (B). **Statistically significant difference at P ≤ 0.01 compared with the GFP sample.
Discussion
In the developing retina, coexpression of ath5 and NSCL1 resulted in an increase in the number of RGCs, whereas smaller increases were observed with expression of ath5 alone or NSCL1 alone. Intuitively, these increases suggest that ath5 and NSCL1, particularly when coexpressed, recruit more multipotent progenitors to differentiate into RGCs and thus may have played a role in determination of RGC fate. However, it is also possible that the increase was due to enhanced RGC differentiation that had been potentiated by other factor(s). In this scenario, cells fated to be RGCs remained uncommitted in the absence of ath5 and NSCL1, and their predetermined differentiation along the RGC pathway was promoted by ath5 and NSCL1 expression. This possibility is favored by the observation that ath5 and NSCL1 caused an increase in the number of RA4+ cells in RPE cell culture in the presence of bFGF, which by itself induces de novo expression of RA4.18,20 In the absence of bFGF, neither gene nor a combination of the two had an effect on inducing RPE transdifferentiation. Thus, coexpression of ath5 and NSCL1 in the developing retina may have resulted in enhanced commitment to RGCs through advancing an RGC cell differentiation program initiated by another factor, such as bFGF, in cells that otherwise could be lured into other pathways by other genes or environmental cues.
The morphologic transdifferentiation observed with ath5 and NSCL1 coexpression was robust, and the resemblance to RGCs is indicative of the cooperation among ath5, NSCL1, and bFGF in bringing about neural differentiation that may involve a spectrum of genes. Note, however, that not every gene that is expressed by RGCs in the developing retina was expressed at a detectable level in the transdifferentiating cells. The cells did not, for instance, stain positively with antibodies against Islet-1 or Brn3A. This could be because of several factors, singly or in combination. First, it is possible that ath5 and NSCL1 induce a portion of genes needed for RGC development, and thus their coexpression inherently could elicit only a limited number of genes. Second, the RPE cells provided a different cellular context than retinal cells, and thus coexpression of ath5 and NSCL1 in RPE cells was unable to induce the entire repertoire of genes that are otherwise downstream of ath5 and NSCL1 in retinal cells. It should be pointed out, nonetheless, that previous studies have indicated that RPE cells have the capacity to transdifferentiate into cells with characteristics of RGCs. For instance, ectopic expression of ngn2 in cultured RPE cells induces de novo production of cells morphologically and molecularly resembling RGCs,19 and bFGF induces RPE tissues in young chick embryos to transdifferentiate into a neural retina encompassing retinal neurons including RGCs.21–24 Third, dissociated RPE cells used in this study were more fastidious than intact RPE tissue, and thus certain neural traits would only be detected with factors of high inducing power. Notably, studies have shown that dissociated RPE cells failed to manifest the neural transdifferentiation phenomenon that was observed with RPE tissue.23 Fourth, the ATH5 and NSCL1 protein levels in the RPE cells were insufficient to induce certain genes. This is supported by the lack of embryonic lethality of the coexpression construct. Fifth, the microenvironment under our experimental conditions was not permissive for RGC development. The observed increases in Islet-1+ and Brna3A+ cells in the developing retina coexpressing ath5 and NSCL1 argues that the developing retina provides additional factors that are lacking in culture conditions and that are required for expression of these genes and for RGC differentiation.23,29–35 Together with those factors, ath5 and NSCL1 may regulate a spectrum of targets associated with RGC development and differentiation. In light of this, in vitro- generated cells may manifest a greater degree of differentiation on exposure to the native microenvironment than when grown in culture.
Ectopic coexpression of ath5 and NSCL1 in cultured RPE cells in the absence of bFGF did not induce de novo expression of RA4, an outcome similar to ectopic expression of ath5 or NSCL1 alone. Thus, combining ath5 and NSCL1 did not promote an RGC fate under the experimental conditions. Coexpression of ath5 and NSCL1 promoted bFGF-initiated RGC differentiation and to a higher degree than either gene alone. These findings, together with the significant increase in RGCs in the developing retina with coexpression over single expression, suggest that ath5 and NSCL1 probably function synergistically in advancing the RGC differentiation program.
The absence of embryonic lethality after ath5 and NSCL1 coexpression was unexpected because, of more than 100 embryos misexpressing NSCL1 thus far generated in our laboratory, none have survived longer than embryonic day (E)13.30 The absence of embryonic lethality could be due to a lower level of NSCL1 protein synthesized under IRES regulation.31 This seems consistent with the observation that Islet-1+ cells were detected in RPE culture infected with RCAS-NSCL1,20 in which NSCL1 translation was under the Src oncogene control, and were not detected with the coexpression construct, in which NSCL1 translation was under IRES control. Apparently, under IRES regulation, NSCL1 protein was synthesized at a sufficient level to exert a biological effect as manifested by the increased RGC differentiation in vivo and in vitro. Alternatively, the absence of embryonic lethality after ath5 and NSCL1 coexpression could be due to ath5’s having a certain rescue effect and thus attenuating the teratogenic effect of NSCL1 on embryonic development. More studies are needed to verify this.
Generating RGCs in vitro is a tantalizing prospect for both basic and clinical research. The present studies show de novo generation of cells that closely resembled RGCs morphologically from bFGF-primed cultured RPE cells coexpressing ath5 and NSCL1. It remains to be further investigated whether these cells have the potential to mature into functional RGCs, the development of which is likely to require intricate interplay between intrinsic and extrinsic factors present in the developing retina.
Acknowledgments
The authors thank Stephen Hughes for the retroviral vector RCAS (B/P) and the shuttle vector Cla12Nco.
Supported by National Eye Institute Grant EY11640, EyeSight Foundation of Alabama Grant 01-7, Research to Prevent Blindness Dolly Green Scholar Award, unrestricted grants to UAB Department of Ophthalmology from Research to Prevent Blindness.
Footnotes
Disclosure: W. Xie, None; R.-T. Yan, None; W. Ma, None; S.-Z. Wang, None
References
- 1.Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- 2.Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang SW, Gan L, Martin SE, Klein WH. Abnormal polarization and axon outgrowth in retinal ganglion cells lacking the POU-domain transcription factor Brn-3b. Mol Cell Neurosci. 2000;16:141–156. doi: 10.1006/mcne.2000.0860. [DOI] [PubMed] [Google Scholar]
- 4.Wang SW, Mu X, Bowers WJ, et al. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002;129:467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- 5.Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci USA. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begley CG, Lipkowitz S, Gobel V, et al. Molecular characterization of NSCL, a gene encoding a helix-loop-helix protein expressed in the developing nervous system. Proc Natl Acad Sci USA. 1992;89:38–42. doi: 10.1073/pnas.89.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown L, Espinosa R, Le Beau MM, Siciliano MJ, Baer R. HEN1 and HEN2: a subgroup of basic helix-loop-helix genes that are coexpressed in a human neuroblastoma. Proc Natl Acad Sci USA. 1992;89:8492–8496. doi: 10.1073/pnas.89.18.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C-M, Yan R-T, Wang S-Z. Misexpression of a bHLH gene, cNSCL1, results in abnormal brain development. Dev Dyn. 1999;215:238–247. doi: 10.1002/(SICI)1097-0177(199907)215:3<238::AID-AJA6>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C-M, Yan R-T, Wang S-Z. Misexpression of cNSCL1 disrupts retinal development. Mol Cell Neurosic. 1999;14:17–27. doi: 10.1006/mcne.1999.0765. [DOI] [PubMed] [Google Scholar]
- 11.Kanekar S, Perron M, Dorsky R, et al. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- 12.Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SW, Kim BS, Ding K, et al. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Yan R-T, Wang S-Z. neuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J Neurobiol. 1998;36:485–496. [PMC free article] [PubMed] [Google Scholar]
- 17.Yan R-T, Wang S-Z. Expression of an array of photoreceptor genes in chick embryonic RPE cell cultures under the induction of neuroD. Neurosci Lett. 2000;280:83–86. doi: 10.1016/s0304-3940(99)01003-4. [DOI] [PubMed] [Google Scholar]
- 18.Yan R-T, Wang S-Z. Differential induction of gene expression by basic fibroblast growth factor and neuroD in cultured retinal pigment epithelial cells. Vis Neurosci. 2000;17:157–164. doi: 10.1017/s0952523800171172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R-T, Ma W-X, Wang S-Z. neurogenin2 elicits the genesis of retinal neurons from cultures of non-neural cells. Proc Natl Acad Sci USA. 2001;98:15014–15019. doi: 10.1073/pnas.261455698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma W, Yan R-T, Xie W, Wang S-Z. bHLH genes cath5 and cNSCL1 promote bFGF-stimulated RPE cells to transdifferentiate towards retinal ganglion cells. Dev Biol. 2004;265:320–328. doi: 10.1016/j.ydbio.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Coulombre JL, Coulombre AJ. Regeneration of neural retina from the pigmented epithelium in the chick embryo. Dev Biol. 1965;12:79–92. doi: 10.1016/0012-1606(65)90022-9. [DOI] [PubMed] [Google Scholar]
- 22.Park CM, Hollenberg MJ. Basic fibroblast growth factor induces retinal regeneration in vivo. Dev Biol. 1989;134:201–205. doi: 10.1016/0012-1606(89)90089-4. [DOI] [PubMed] [Google Scholar]
- 23.Pittack C, Jones M, Reh TA. Basic fibroblast growth factor induces retinal pigment epithelium to generate neural retina in vitro. Development. 1991;113:577–588. doi: 10.1242/dev.113.2.577. [DOI] [PubMed] [Google Scholar]
- 24.Guillemot F, Cepko CL. Retinal fate and ganglion cell differentiation are potentiated by acidic FGF in an in vitro assay of early retinal development. Development. 1992;114:743–754. doi: 10.1242/dev.114.3.743. [DOI] [PubMed] [Google Scholar]
- 25.Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C-M, Yan R-T, Wang S-Z. A novel homeobox gene and its role in the development of retinal bipolar cells. Mech Dev. 2002;116:85–94. doi: 10.1016/s0925-4773(02)00148-x. [DOI] [PubMed] [Google Scholar]
- 27.Xiang M, Zhou L, Macke JP, et al. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- 29.Waid DK, McLoon SC. Immediate differentiation of ganglion cells following mitosis in the developing retina. Neuron. 1995;14:117–124. doi: 10.1016/0896-6273(95)90245-7. [DOI] [PubMed] [Google Scholar]
- 30.Yan R-T, Wang S-Z. Embryonic abnormalities from misexpression of cNSCL1. Biochem Biophys Res Commun. 2001;287:949–955. doi: 10.1006/bbrc.2001.5690. [DOI] [PubMed] [Google Scholar]
- 31.Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci USA. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Barnstable CJ. Differential effects of bFGF on development of the rat retina. Brain Res. 1996;723:169–176. doi: 10.1016/0006-8993(96)00237-5. [DOI] [PubMed] [Google Scholar]
- 33.Fischer AJ, Dierks BD, Reh TA. Exogenous growth factors induce the production of ganglion cells at the retinal margin. Development. 2002;129:2283–2291. doi: 10.1242/dev.129.9.2283. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XM, Yang XJ. Regulation of retinal ganglion cell production by Sonic hedgehog. Development. 2001;128:943–957. doi: 10.1242/dev.128.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenkamp DL, Frey RA. Extraretinal and retinal hedgehog signaling sequentially regulate retinal differentiation in zebrafish. Dev Biol. 2003;258:349–363. doi: 10.1016/s0012-1606(03)00121-0. [DOI] [PubMed] [Google Scholar]


