Abstract
rRNA plays an important role in function of peptidyl transferase, the catalytic center of the ribosome responsible for the peptide bond formation. Proper placement of the peptidyl transferase substrates, peptidyl-tRNA and aminoacyl-tRNA, is essential for catalysis of the transpeptidation reaction and protein synthesis. In this report, we define a small set of rRNA nucleotides that are most likely directly involved in binding of tRNA in the functional sites of the large ribosomal subunit. By binding biotinylated tRNA substrates to randomly modified large ribosomal subunits from Escherichia coli and capturing resulting complexes on the avidin resin, we identified four nucleotides in the large ribosomal subunit rRNA (positions G2252, A2451, U2506, and U2585) whose modifications prevent binding of a peptidyl-tRNA analog in the P site and one residue (U2555) whose modification interferes with transfer of peptidyl moiety to puromycin. These nucleotides represent a subset of positions protected by tRNA analogs from chemical modification and significantly narrow the number of 23S rRNA nucleotides that may be directly involved in tRNA binding in the ribosomal functional sites.
A crucial step in protein biosynthesis is the peptidyl transferase (PT) reaction in which a peptidyl moiety from peptidyl-tRNA, positioned in the ribosomal P site, is transferred to the amino group of aminoacyl-tRNA, bound in the A site, resulting in formation of a new peptide bond. For catalysis to take place, both donor and acceptor tRNAs must be properly positioned in the P and A sites of the PT center of the large ribosomal subunit. rRNA is likely to play an important, and maybe primary, role in the binding and correct placing of the tRNA in the ribosome catalytic center (see ref. 1 for review). Crosslinking of tRNA derivatives and chemical footprinting identified several residues in domains IV and V of 23S rRNA as being located close to the acceptor stem of tRNA derivatives bound in the A, P, and E sites of the ribosome (2–6). A Watson–Crick interaction between one of the protected positions, G2252, and C74 at the 3′ end of tRNA was demonstrated by site-directed mutagenesis (7). However, mutational and biochemical studies failed to define the importance of the other rRNA nucleotides that might interact with tRNA.
Although they provide important information about the ribosomal environment of bound tRNA, RNA footprinting and crosslinking techniques are unable to establish which residues of rRNA are essential for tRNA binding as opposed to merely being in the tRNA vicinity. In contrast, the modification interference approach makes it possible to identify rRNA nucleotides that are likely to form functional contacts with ribosomal ligands. In this case, tRNA is complexed with ribosomes containing randomly modified rRNA so that nucleotide modifications that interfere with complex formation can be identified. Surprisingly, when applied to interaction of tRNA with the small ribosomal subunit, this approach showed that much fewer nucleotides were essential for tRNA binding than was revealed by RNA footprinting (8). In this study, we used the modification interference strategy to identify sites in 23S rRNA that are involved in tRNA binding to the large ribosomal subunit. The two key elements of our experiments were forming a complex of peptidyl-tRNA analog with the large ribosomal subunit containing randomly modified 23S rRNA and separating the resulting complex from the reaction mixture by capturing it on the insoluble carrier.
Peptidyl transfer is an intrinsic feature of the large ribosomal subunit. In the presence of 33% methanol, the PT reaction can be catalyzed by the isolated large ribosomal subunit alone (9). In this assay, known as “fragment reaction,” peptidyl-tRNA can be replaced by N-acetyl-aminoacyl tRNA or its 3′ terminal oligonucleotide fragment, whereas the acceptor substrate can be substituted by puromycin, an analog of aminoacyl-tRNA 3′ terminus. The authentic nature of the PT reaction, catalyzed by the large subunit in the fragment reaction conditions, is demonstrated by its sensitivity to antibiotics (9, 10). In a complex, formed under the fragment reaction conditions between tRNA and the 50S subunit, only the acceptor end of tRNA is involved in interaction with the PT center. Therefore, the tRNA anticodon can be chemically altered without affecting interaction with the 50S subunit. Introduction of a biotin derivative in the tRNA anticodon made possible the isolation of a complex between tRNA and randomly modified 50S ribosomal subunits on the avidin resin and allowed us to identify 23S rRNA nucleotides that are essential for binding of tRNA in ribosomal functional sites.
MATERIALS AND METHODS
Aminoacylation and Biotinylation of tRNATyr.
Escherichia coli tRNATyr (type I) (Subriden RNA, Rollingbay, WA) was aminoacylated with [3H]tyrosine (final specific activity, 6 Ci/mmol; 1 Ci = 37 GBq) (American Radiolabeled Chemicals, St. Louis) and N-acetylated by incubation with acetic acid-N-hydroxysuccinimide ester as previously described (2, 11). N-Ac-Tyr-tRNATyr was HPLC-purified, ethanol-precipitated, resuspended in 10 mM sodium acetate, pH 5.5, and stored at −80°C.
For biotinylation, 1 nmol of N-Ac-Tyr-tRNATyr was incubated in the dark in 100 μl of 100 mM sodium acetate solution (pH 5.5) containing 10 mM KIO4 for 30 min at 20°C. The reaction was quenched by addition of Na2SO3 to the final concentration of 20 mM and incubating 5 min at 0°C. tRNA was ethanol-precipitated, dried, and resuspended in 100 μl of 100 mM sodium acetate (pH 5.5) containing 10 mM biotin-amidocaproyl hydrazide (Sigma) and 1 mM MnCl2 (12). After incubation for 1 hr in the dark at 20°C, biotinylated tRNA was purified by gel-filtration on a Sephadex G25 column, ethanol-precipitated, and stored as a dry pellet at −80°C. Before use, the pellet was resuspended in 10 mM sodium acetate (pH 5.5), the absorbance at A260 was determined, and the extent of aminoacylation was assayed by precipitation with trichloracetic acid. No significant deacylation of N-Ac-Tyr-tRNATyr occurred under biotinylation conditions used.
Chemical Modification of Ribosomal Subunits and tRNA Binding.
Large ribosomal subunits were prepared from E. coli strain MRE600 as previously described (13).
Chemical modification of 50S subunits with kethoxal (Research Organics), dimethyl sulfate (Aldrich), or 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide (Aldrich) was carried out essentially as described in ref. 14. Heat-activated 50S subunits (500 pmol), modified in 200 μl of the appropriate buffer for 10 min at 37°C, were purified by gel-filtration on a 5 ml Sephadex G-50 column equilibrated in the fragment reaction (FR) buffer (50 mM Tris⋅HCl, pH 7.5/400 mM NH4Cl/20 mM MgCl2). Modified ribosomal subunits were directly used in tRNA-binding experiments. An aliquot of modified subunits was kept on ice during the modification-interference experiment and served as “modified control” for the primer extension. In a standard modification-interference assay, 120 pmol of freshly modified 50S subunits was combined on ice with 120 pmol biotinylated N-Ac-Tyr-tRNATyr (N-Ac-Tyr-tRNABiotTyr) in a total volume of 200 μl FR buffer containing 0.1 mM sparsomycin. After addition of 100 μl methanol, the reaction mixture was incubated on ice for 10 min.
The extent of complex formation between the large ribosomal subunit and N-Ac-Tyr-tRNABiotTyr was assessed by filter binding. In a standard binding assay, 10 pmol of heat-activated 50S subunits was combined on ice with various amounts of N-Ac-Tyr-tRNABiotTyr in 40 μl of FR buffer with or without addition of sparsomycin (final sparsomycin concentration was 0.1 mM). After addition of 20 μl methanol, the reaction mixtures were incubated for 15 min on ice. At the end of incubation, 240 μl of ice-cold FR buffer/methanol (2:1 vol/vol) mixture (FR/M buffer) was added and solutions were filtered through MultiScreen-HA Millipore filters. The filters were rinsed three times with 300 μl of an ice-cold FR/M buffer and dried, and the amount of bound radioactivity was measured in a liquid scintillation counter.
Binding of N-Ac-Tyr-tRNABiotTyr to 50S Subunits and Trapping of the Complex on the Avidin Resin.
To block nonspecific binding of the ribosomal subunits to the resin, 30 μl of the Tetralink avidin resin (binding capacity, 30 pmol of biotin per μl of resin) (Promega) was incubated 15 min on ice in 180 μl of the FR/M buffer, containing 0.3 mg/ml BSA (New England Biolabs). The blocked resin was washed with 1 ml FR/M buffer and resuspended in 80 μl of the same solution.
Eighty microliters of Tetralink avidin resin suspension (Promega) was then added to the reaction mixture containing a complex of large ribosomal subunit with N-Ac-Tyr-tRNABiotTyr or tRNABiotTyr, and incubation was continued on ice for 15 min with occasional gentle stirring. The resin suspension was diluted to 1 ml with cold FR/M buffer containing 0.1 mM sparsomycin, loaded on a Quick-Snap disposable chromatography column (Isolab), and washed twice by 1 ml of the same cold buffer. Bound subunits were eluted by 400 μl of elution buffer (300 mM sodium acetate, pH 5.5/5 mM EDTA/0.5% SDS), and rRNA was extracted according to ref. 14.
In the experiments in which 50S subunits were released from the column by puromycin treatment, binding of tRNA, capturing of the complex on the resin, and subsequent washes were performed as described above with the exception that sparsomycin was omitted from all the buffers. The subunits were eluted from the resin by a slow passage of 600 μl of the cold FR/M buffer containing 200 μM puromycin. Eluted 50S subunits were ethanol-precipitated and resuspended in 400 μl of elution buffer (300 mM sodium acetate, pH 5.5/5 mM EDTA/0.5% SDS), and rRNA was isolated by phenol extraction.
Analysis of Distribution of Modified Residues in 23S rRNA.
The distribution of modified residues in 23S rRNA and the extent of modification of individual bases were analyzed by primer extension (14). The concentration of rRNA extracted from untreated, modified, and selected subunits was adjusted after a test primer extension. Subsequent primer extension analysis was performed by using a set of DNA primers that allowed scanning of the entire 23S rRNA molecule.
Footprinting experiments on 50S subunits or 70S ribosomes in the presence of 33% methanol were performed as described previously (3). The extent of modification of individual nucleotides was determined by quantification of band intensities on gels using a phosphorimager (AMBIS).
RESULTS
To investigate which bases in 23S rRNA are essential for positioning tRNA in the functional sites of the large ribosomal subunit, we developed an experimental system that allowed us to trap a complex of tRNA with the large ribosomal subunit on the insoluble carrier. We then used this system to identify base modifications that interfere with tRNA binding.
23S rRNA Residues Essential for Binding of Peptidyl tRNA in the Ribosomal P Site.
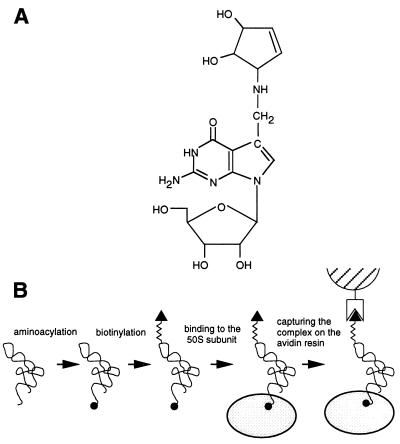
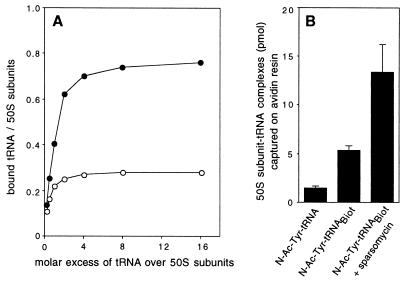
tRNATyr contains a chemically reactive Q base at position 34 of the anticodon loop (Fig. 1A) (15). The cis-glycol group of N-Ac-Tyr-tRNATyr Q base was oxidized by periodate and conjugated to biotin via a flexible linker (Fig. 1B). N-acetyl-aminoacyl-tRNA is an analog of peptidyl tRNA and binds preferentially to the P site of the ribosome. Several tests indicate that biotinylation of the anticodon did not affect specificity of tRNA interaction with the 50S ribosomal subunit. Similar to unmodified N-Ac-Tyr-tRNATyr, the biotinylated derivative of N-Ac-Tyr-tRNA (N-Ac-Tyr-tRNABiotTyr) binds to the large ribosomal subunit in the fragment reaction conditions and the binding is significantly stimulated by sparsomycin, an antibiotic that enhances binding of peptidyl tRNA to the P site (16) (Fig. 2A). N-Ac-Tyr-tRNABiotTyr bound to the large ribosomal subunit was fully reactive with puromycin, as determined by filter binding, demonstrating that N-Ac-Tyr-tRNABiotTyr was bound specifically in the P site.
Figure 1.
(A) The chemical structure of the Q-nucleoside present in the anticodon loop of tRNATyr. (B) An outline of an experimental scheme leading to formation of a complex between N-acetyl-aminoacyl-tRNA and the large ribosomal subunit and capturing the complex on the resin. N-Acetyl-tyrosyl residue is shown as a solid circle, biotin as a triangle, the large ribosomal subunit as a gray oval, and the avidin resin as a hatched semicircle.
Figure 2.
(A) Binding of N-Ac-Tyr-tRNABiotTyr to the large ribosomal subunit in the fragment reaction conditions. The amount of bound tRNA was estimated by nitrocellulose filter binding. Open circles represent binding in the absence of sparsomycin; solid circles show binding in the presence of 0.1 mM sparsomycin. (B) Trapping of the complex of large ribosomal subunits with N-acetyl-aminoacyl-tRNA on the avidin resin. 32P-labeled 50S subunits were incubated with nonbiotinylated N-Ac-Tyr-tRNA, N-Ac-Tyr-tRNABiot, or N-Ac-Tyr-tRNABiot in the presence of sparsomycin and passed through the avidin resin, and, after washing, the captured material was eluted and counted.
Both N-acetyl-aminoacyl-tRNA, bound in the P site of 70S ribosomes, and its 3′ terminal oligonucleotide fragment, bound in the fragment reaction conditions to the P site of large ribosomal subunits, protect a distinct set of 23S rRNA residues from chemical modification (2, 3) (Table 1). In the presence of 33% methanol, N-Ac-Tyr-tRNABiotTyr strongly protected U2506, U2584, U2585, A2439, A2451, and, at a lower level, G2251, G2252, and G2253 in 23S rRNA (Table 1). These protections overlap with those induced in 23S rRNA by peptidyl-tRNA bound to the 70S ribosome (2) and are identical to protections caused by the 3′ terminal tRNA fragment bound in the P site of the 50S subunit (3). Thus, interaction of N-Ac-Tyr-tRNABiotTyr with the P site of the large subunit is functionally and structurally indistinguishable from interactions of unmodified tRNA.
Table 1.
Protection of bases in 23S rRNA by tRNA or tRNA fragments bound to the ribosomal P site
| tRNA substrate | Ribosomes or subunits* | Protected bases† | Ref. |
|---|---|---|---|
| N-Ac-Phe-tRNAPhe | 70S | A1916, A1918, U1926, G2252, G2253, A2439, A2451, G2505, U2506, U2584, U2585, A2602↑‡ | 2 |
| CACCA-(Ac-Phe)§ | 70S¶ | G2251‖, G2252, | 3 |
| UACCA-(Ac-Leu) | G2253, A2439, | ||
| CAACCA-(Ac-Met) | A2451, U2506, U2584, U2585, A2602↑ | ||
| N-Ac-Tyr-tRNABiotTyr | 50S** | G2251, G2252, G2253, A2439, A2451, U2506, U2584, U2585 | This paper |
E. coli 70S ribosomes or 50S large ribosomal subunits.
Refers to relative reactivity of individual 23S rRNA bases to chemical modification.
Binding of the P site substrates to 70S ribosomes increases accessibility of A2602 to modification with dimethyl sulfate.
Refers to 3′-terminal fragments generated by RNase T1 hydrolysis of corresponding N-acyl-aminoacyl-tRNAs.
Binding of tRNA fragments to 70S ribosomes was performed in the fragment reaction conditions.
Protection of G2251 was reported in ref. 22.
Binding of biotinylated N-Ac-Tyr-tRNABiotTyr to 50S ribosomal subunits was performed in the fragment reaction conditions.
The complex of N-Ac-Tyr-tRNABiotTyr with the large ribosomal subunit can be captured on the avidin resin (Fig. 2B). In agreement with filter-binding experiments, the amount of ribosomal subunits retained on the column increased approximately 2.5-fold when N-Ac-Tyr-tRNABiotTyr was bound in the presence of sparsomycin. Significantly less ribosomal material was retained when nonbiotinylated N-Ac-Tyr-tRNATyr was used.
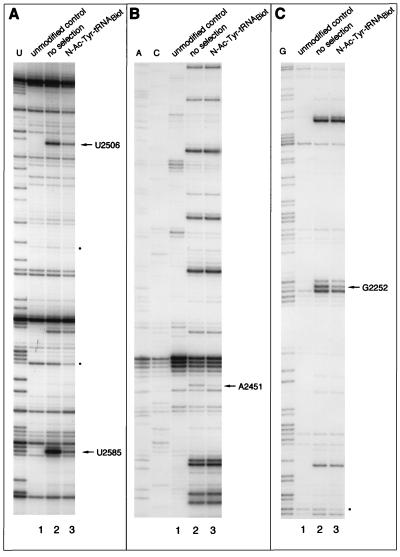
To define 23S rRNA residues that participate in tRNA binding, E. coli large ribosomal subunits were randomly modified with the chemical reagents specific to RNA bases, dimethyl sulfate, kethoxal, or carbodiimide (14, 17) and allowed to form a complex with N-Ac-Tyr-tRNABiotTyr in the presence of sparsomycin. The resulting 50S subunit–tRNA complexes were trapped on the avidin resin, and captured 50S subunits were eluted with SDS/EDTA-containing buffer. Comparison of the 23S rRNA modification pattern in the eluted subunits with that of unselected subunits revealed four residues—G2252, A2451, U2506, and U2585—that reproducibly were found undermodified in the 50S subunits that were able to form a complex with N-Ac-Tyr-tRNABiotTyr (Fig. 3 and Table 2). Except for these four residues in domain V, modification of the remaining 23S rRNA accessible positions did not interfere with tRNA binding. When the 50S subunit–tRNA complex was formed (with a low yield) in the absence of sparsomycin, selection against modification of the same four nucleotides was observed (not shown). We concluded that G2252, A2451, U2506, and U2585 most probably form direct contacts with the peptidyl-tRNA bound in the P site of the large ribosomal subunit.
Figure 3.
Analysis of base modifications of 23S rRNA in randomly modified 50S ribosomal subunits before and after selection on the avidin resin. (A) Modification with 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide. (B) Modification with dimethyl sulfate. (C) Modification with kethoxal. rRNA was extracted from the 50S subunits, and modifications were localized by primer extension. Lanes: 1, 23S rRNA from the unmodified 50S subunits; 2, modified, unselected 50S subunits; 3, modified subunits complexed in the presence of sparsomycin with N-Ac-Tyr-tRNA and captured on the avidin resin. Dideoxy sequencing lanes corresponding to RNA bases U, A, C, and G are shown. Dots show the reverse transcriptase “strong stop” bands whose intensity is decreased in the selected samples.
Table 2.
Selection against modification of 23S rRNA bases in the large ribosomal subunits complexed with biotinylated tRNA substrates and captured on the avidin resin*
| Position | N-Ac-Tyr-tRNABiot† | N-Ac-Tyr-tRNABiot‡ (puromycin elution) |
|---|---|---|
| G2252 | ++ | + |
| A2451 | +++ | + |
| U2506 | ++ | + |
| U2585 | +++ | + |
| U2555 | − | + |
Band intensities were measured by scanning gels on phophorimager and, after subtraction of background, normalized to intensities of control bands. The Table summarizes the results of four independent experiments. +++, Strong selection (>75%) against base modification; ++, considerable selection (50–75%); +, moderate selection (25-50%); −, no selection.
N-Ac-Tyr-tRNABiotTyr was bound to 50S ribosomal subunits in the fragment reaction conditions in the presence of 0.1 mM sparsomycin, and the complexes were captured on the avidin resin and eluted with SDS/EDTA-containing buffer. 23S rRNA was extracted and subjected to primer-extension analysis.
Same as †, but binding was performed in the absence of sparsomycin, and captured ribosomal subunits were eluted from the avidin complex by puromycin treatment.
Close examination of the gels showed that some reverse transcriptase “strong stops” were reproducibly weaker in the resin-selected samples compared with unselected 50S subunits (marked by dots in Figs. 3 and 4C). The “strong stops” may result either from the reverse transcriptase pausing “in front” of tight RNA structures or from RNase cuts present in a small proportion of the 23S rRNA molecules (14, 17). Selection against such “stops” upon formation of the complex with tRNA suggests that either the rRNA cuts or local rRNA conformations in a subpopulation of 50S subunits prevents formation of a stable complex with tRNA.
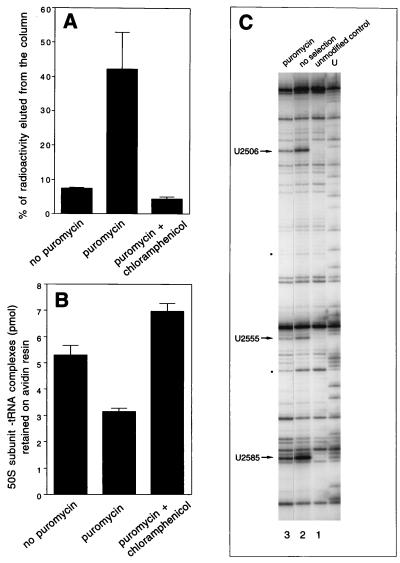
Figure 4.
(A) Puromycin-dependent release of radioactivity from the N-Ac-[3H]Tyr-tRNABiotTyr complexed with the large ribosomal subunit and captured on the avidin resin. Biotinylated N-Ac-[3H]Tyr-tRNA was incubated with 50S subunits and captured on the resin in the absence of sparsomycin (see Materials and Methods). The resin was saturated at 0°C with the FR/M buffer containing either no puromycin, 0.2 mM puromycin, or 0.2 mM puromycin and 8 mM chloramphenicol. After 10 min of incubation, eluted radioactivity was counted and plotted as percentage of the total radioactive tRNA captured on the column. (B) Puromycin-dependent release of large subunits from the complexes with tRNA captured on the avidin resin. 32P-labeled 50S subunits were complexed with N-Ac-Tyr-tRNABiotTyr, captured on the avidin resin, and incubated 10 min at 0°C in the FR/M buffer containing either no puromycin, 0.2 mM puromycin, or 0.2 mM puromycin and 8 mM chloramphenicol. The column was washed with the FR/M buffer, and 50S subunits remaining on the column after this treatment were eluted and counted. (C) Analysis of base modifications of 23S rRNA in randomly modified 50S ribosomal subunits eluted by puromycin from the complex with N-Ac-Tyr-tRNABiotTyr captured on avidin resin. Lanes: 1, rRNA from the unmodified 50S subunits; 2, unselected 50S subunits modified with cyclohexyl-3-(2-morpholinoethyl)carbodiimide; 3, rRNA from modified subunits complexed with N-Ac-Tyr-tRNABiotTyr, captured on the avidin resin, and eluted by puromycin-containing buffer. U-specific dideoxy sequencing lane is shown. Dots indicate the reverse transcriptase “strong stop” bands whose intensity is decreased in the selected samples.
Puromycin Reaction.
Having established 23S rRNA positions that are essential for P site binding of the donor substrate of the PT reaction we attempted identification of residues important for interaction of the acceptor substrate in the ribosomal A site. Puromycin is a structural and functional analog of the 3′ terminus of aminoacyl tRNA. It binds to the A site of the large ribosomal subunit and serves as an acceptor substrate in the peptidyl transfer reaction. During transpeptidation, the peptidyl (or N-acetyl-aminoacyl) moiety is transferred from the P site-bound tRNA to puromycin. The large ribosomal subunit, complexed to biotinylated N-Ac-[3H]Tyr-tRNABiotTyr and captured on the avidin resin in the fragment reaction conditions, retained its ability to catalyze the puromycin reaction. When N-Ac-[3H]Tyr-tRNABiotTyr was complexed to 50S subunits and trapped on the avidin column, more than 1/3 of resin-bound radioactivity could be eluted by puromycin treatment (Fig. 4A). PT inhibitors chloramphenicol (Fig. 4A) and sparsomycin (not shown) prevented puromycin-dependent release of radioactivity, confirming that it was the result of transpeptidation. The radioactivity remaining bound to the resin after puromycin treatment likely corresponded to avidin-bound N-Ac-[3H]Tyr-tRNABiotTyr not complexed with 50S subunits.
Deacylated tRNA, formed as a result of transpeptidation, binds to the large ribosomal subunit in a more labile mode than the initial N-acetyl-aminoacyl-tRNA (18, 19). Therefore, transfer of N-acetyl-tyrosyl moiety from N-Ac-Tyr-tRNABiotTyr to puromycin leads to partial dissociation of the 50S subunit–tRNA complex. When 32P-labeled large ribosomal subunits with P site-bound N-Ac-Tyr-tRNABiotTyr were captured on the avidin resin and incubated with puromycin, a significant amount of the bound material was eluted from the column (Fig. 4B). Similar to the puromycin-dependent release of radioactive amino acid from the P site-bound tRNA (Fig. 4A), elution of 50S subunits from the avidin-trapped complex was prevented by PT inhibitors chloramphenicol (Fig. 4B) and sparsomycin (not shown), demonstrating dependence of the subunit release on catalysis of the puromycin reaction. (Note, some “leakage” of the ribosomal material from the avidin-bound complexes was observed even in the absence of puromycin. This leakage was decreased in the presence of PT inhibitors, suggesting that it resulted from the transfer of a peptidyl residue to methanol catalyzed by the PT center of the large ribosomal subunit.)
Puromycin-dependent release of 50S subunits from the trapped complexes allowed us to identify 23S rRNA residues essential for puromycin reaction. N-Ac-Tyr-tRNABiotTyr was bound in the absence of sparsomycin to the P site of randomly modified large ribosomal subunits; the complexes were trapped on the avidin resin and then treated with puromycin on the column, resulting in release of 50S ribosomal subunits from puromycin-reactive complexes. Analysis of modified 23S rRNA residues in puromycin-eluted material confirmed the result of the previous experiment, because we again observed selection against modification of the four nucleotides essential for the P site binding of N-Ac-Tyr-tRNABiotTyr (G2252, A2451, U2506, and U2585). However, one more residue, U2555, was reproducibly found undermodified in the puromycin-eluted large subunits compared with unselected material (Fig. 4C and Table 2). Because modification of U2555 did not hinder binding of peptidyl tRNA (see Fig. 3A), but prevented puromycin-dependent release of ribosomal material, we concluded that presence of unmodified U2555 is essential for the puromycin reaction.
DISCUSSION
P Site.
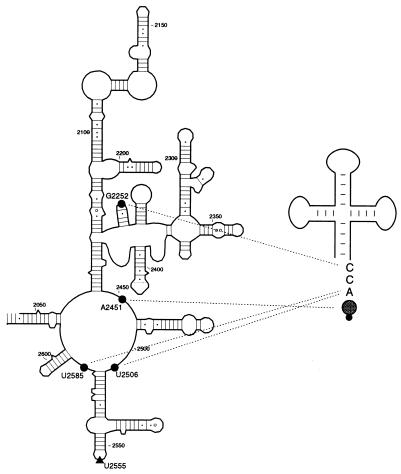
Modification of four nucleotides in 23S rRNA—G2252, A2451, U2506, and U2585—interferes with binding of peptidyl-tRNA to the P site of the large ribosomal subunit (Fig. 5). This finding agrees with previous affinity labeling and RNA footprinting results that implicated these four residues, among others, as being in the vicinity of the tRNA 3′ terminus (2–4). Importantly, however, the modification-interference results show that not all the 23S rRNA residues, whose accessibility to chemical modification is affected by tRNA, are essential for tRNA binding (compare Tables 1 and 2). Thus, peptidyl-tRNA bound to the P site of 50S subunits protects eight positions in 23S rRNA from chemical modification (Table 1). However, modification of half of these positions (G2251, G2253, A2439, and U2584) does not notably inhibit tRNA binding (Table 2). Three such nucleotides (G2251, G2253, and U2584) are located next to positions where a strong selection was observed in modification-interference experiments suggesting that their protection is a result of tRNA binding to the neighboring RNA base. Alternatively, binding of tRNA may induce a conformational transition in the ribosome, resulting in an altered accessibility of several rRNA bases. Similar to our results, only a subset of nucleotides protected by tRNA in 16S rRNA appeared to be important for the binding of tRNA to the small ribosomal subunit (8).
Figure 5.
Summary of the positions in domain V of 23S rRNA whose modification interfere with binding of N-acetyl-aminoacyl-tRNA in the P site (•) or with puromycin reaction (▴). Possible interactions of individual P site residues with the CCA-end and aminoacyl moiety of N-acetyl-aminoacyl-tRNA are shown by dotted lines.
Functional interactions between tRNA and PT are confined to the universal CCA terminus of tRNA (9, 10). Hence, rRNA residues G2252, A2451, U2506, and U2585, identified in this work as important for P site tRNA binding, are likely to form contacts either with the CCA end or the aminoacyl moiety of N-Ac-Tyr-tRNA. The available experimental data support this model. Mutations of G2252 abolish PT activity by preventing tRNA binding (7, 20). The binding is restored by compensatory mutations at C74 of the tRNA CCA end (7), suggesting a Watson–Crick interaction between G2252 in 23S rRNA and C74 in the P site-bound tRNA.
The 3′ terminal A76 of tRNA apparently interacts with U2506 and U2585 because protection of U2506 and U2585 from chemical modification is lost upon removal of the 3′ terminal adenosine from the P site-bound tRNA. However, though mutations of either U2506 or U2585 severely diminish PT activity, apparently by preventing binding of peptidyl tRNA to the ribosome, they could not be compensated by mutations in tRNA (21, 22), arguing that interactions other than Watson–Crick base pairing may occur between U2506 and/or U2585 in 23S rRNA and A76 in tRNA. Formation of a Hoogsteen base pair between the 3′ terminal adenosine of tRNA and U2585 of 23S rRNA was proposed previously (21). However, coherently similar behavior of U2585 and U2506 in the RNA footprinting and modification-interference experiments appear to be more consistent with a triple nucleotide interaction involving these two rRNA positions and A76 of tRNA.
Modification of A2451 interfered with binding of N-Ac-Tyr-tRNABiotTyr (Fig. 3B) but did not affect binding of deacylated tRNA to the P site of the large ribosomal subunit (data not shown). This agrees with the footprinting data because protection of A2451 by aminoacyl derivatives of tRNA bound to the P and A sites of the 70S ribosome depends on the presence of the 2′,3′ linked aminoacyl group (2). Thus, modification interference and footprinting data place A2451 in contact with the aminoacyl moiety of N-acetyl-aminoacyl-tRNA. Interestingly, modification of A2439, whose protection, similar to A2451, depended on the presence of the N-acetyl-aminoacyl group at the tRNA 3′ end, did not interfere with the binding of N-Ac-Tyr-tRNABiotTyr to the P site, which suggests that A2439 plays a less important role than A2451 in tRNA binding.
Our data did not reveal interactions involving the universally conserved C75, a penultimate nucleotide at the tRNA 3′ end. Though it was proposed (23) that G2581 or U2580 form a part of the C75 binding site, there is no strong evidence implicating these positions (or any other in 23S rRNA) in a direct interaction with C75. Furthermore, modification of U2580 did not interfere with binding of tRNA in the modification-interference assay (Fig. 3A), contradicting its involvement in C75 binding, whereas G2581 was inaccessible for modification with kethoxal in the 50S subunits. It is possible that C75 interacts with rRNA residues that are not modified by chemical reagents or with a sugar–phosphate backbone or ribosomal protein; these types of contacts would not be revealed by base-specific modifications.
Summarizing, we can conclude that the main functional interactions between 23S rRNA and peptidyl-tRNA in the P site of the large ribosomal subunit, as revealed by modification-interference experiments, are apparently limited to a Watson–Crick base pair between G2252 and C74, a noncanonical (possibly, triple-nucleotide) interaction of U2506 and U2585 with A76, and a contact between A2451 and the aminoacyl moiety of peptidyl-tRNA (Fig. 5). Because rRNA residues essential for tRNA binding participate in different types of interactions with functional groups at the 3′ end of peptidyl-tRNA, they contribute differently to the free energy of tRNA binding. Thus, contacts formed by G2252 with C74 of tRNA or a noncanonical interaction between U2506/2585 and A76 are crucial for tRNA binding, such that mutations at corresponding rRNA positions prevent tRNA binding and are lethal for the cell (7, 20, 22, 24). In contrast, though the contact between A2451 and the aminoacyl moiety of peptidyl-tRNA is important for P site binding, it is not crucial. Mutations at A2451 can be tolerated by the ribosome (25) and, as RNA footprinting and direct-binding measurements show, deacylated tRNA, which apparently does not interact with A2451, can still bind to the P site of the large ribosomal subunit, though probably with a lower affinity than peptidyl tRNA (2, 19).
Puromycin Reaction.
Five 23S rRNA positions were found undermodified in the subunits released from the resin-trapped complex as the result of the peptidyl transfer from the P site-bound N-Ac-Tyr-tRNABiotTyr to the A site-bound puromycin. Four of these residues (G2252, A2451, U2506, and U2585) corresponded to the positions essential for P site binding of N-acetyl-aminoacyl-tRNA. Modification at the fifth position, U2555, apparently prevented the puromycin reaction. Electrophilic derivatives of puromycin could be crosslinked to the sequence GU(U/C)CG in 23S rRNA, with the 2553–2557 GUUCG sequence being the most likely site of incorporation (26, 27). 4-Thio-dT-C-puromycin could be crosslinked, with a high yield, to G2553 in 23S rRNA (28). U2555 is one of the positions protected by aminoacyl-tRNA bound to the A site of 70S ribosomes; removal of the tRNA 3′ terminal adenosine eliminates the protection (2). Because puromycin resembles, structurally and functionally, the aminoacylated 3′ terminal adenosine of aminoacyl tRNA, we propose that U2555 may directly interact with puromycin and, correspondingly, with the 3′ terminal aminoacyl-adenosine of the A site-bound tRNA. Substitutions of U2555 by A or G (viable only when a low copy number of mutated 23S rRNA genes were present) affected the efficiency of nonsense suppression and frame shifting, both processes that are related to tRNA binding in the A site (29). Interestingly, substitution of U2555 by C did not have much effect either on in vitro PT activity or on cell viability (29, 30), suggesting that the C2 carbonyl group, common between uracyl and cytosine bases, may be responsible for interaction with A site-bound tRNA.
We would like to note, however, that modification interference data are also consistent with U2555 being directly involved in the catalysis of the transpeptidation reaction.
Acknowledgments
We thank Tanel Tenson for helpful discussions and Patricia Kloss for help in preparing the manuscript. This work was supported by National Institutes of Health Grant GM53762.
ABBREVIATIONS
- N-Ac-Tyr-tRNABiotTyr
biotinylated N-acetyl-Tyr-tRNATyr
- PT
peptidyl transferase
- FR
fragment reaction
References
- 1.Noller H F. J Bacteriol. 1993;175:5297–5300. doi: 10.1128/jb.175.17.5297-5300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moazed D, Noller H F. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D, Noller H F. Proc Natl Acad Sci USA. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steiner G, Kuechler E, Barta A. EMBO J. 1988;7:3949–3955. doi: 10.1002/j.1460-2075.1988.tb03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wower J, Hixson S S, Zimmermann R A. Proc Natl Acad Sci USA. 1989;86:5232–5236. doi: 10.1073/pnas.86.14.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P, Stade K, Osswald M, Brimacombe R. Nucleic Acids Res. 1993;21:887–896. doi: 10.1093/nar/21.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samaha R R, Green R, Noller H F. Nature (London) 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 8.Von Ahsen U, Noller H F. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- 9.Monro R E, Marcker K A. J Mol Biol. 1967;25:347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- 10.Monro R E, Cerna J, Marcker K A. Proc Natl Acad Sci USA. 1968;61:1042–1049. doi: 10.1073/pnas.61.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rappoport S, Lapidot Y. Methods Enzymol. 1974;29:685–688. doi: 10.1016/0076-6879(74)29060-8. [DOI] [PubMed] [Google Scholar]
- 12.Spiegel S, Wilchek M. J Immunol. 1981;127:572–575. [PubMed] [Google Scholar]
- 13.Spedding G. In: Ribosomes and Protein Synthesis, A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 1–29. [Google Scholar]
- 14.Stern S, Moazed D, Noller H F. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 15.Kasai H, Ohashi Z, Harada F, Nishimura S, Oppenheimer N J, Crain P F, Liehr J G, von Minden D L, McCloskey J A. Biochemistry. 1975;14:4198–4209. doi: 10.1021/bi00690a008. [DOI] [PubMed] [Google Scholar]
- 16.Monro R E, Celma M L, Vazquez D. Nature (London) 1969;222:356–358. doi: 10.1038/222356a0. [DOI] [PubMed] [Google Scholar]
- 17.Christiansen J, Egebjerg J, Larsen N, Garrett R A. In: Ribosomes and Protein Synthesis; A Practical Approach. Spedding G, editor. Oxford: Oxford Univ. Press; 1990. pp. 229–252. [Google Scholar]
- 18.Rheinberger H-J, Sternbach H, Nierhaus K H. Proc Natl Acad Sci USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parfenov D V, Saminsky E M. Mol Biol. 1985;19:1130–1136. [Google Scholar]
- 20.Lieberman K R, Dahlberg A E. J Biol Chem. 1994;269:16163–16169. [PubMed] [Google Scholar]
- 21.Porse B T, Thi-Ngoc H P, Garrett R A. J Mol Biol. 1996;264:472–483. doi: 10.1006/jmbi.1996.0655. [DOI] [PubMed] [Google Scholar]
- 22.Green R, Samaha R R, Noller H F. J Mol Biol. 1997;266:40–50. doi: 10.1006/jmbi.1996.0780. [DOI] [PubMed] [Google Scholar]
- 23.Spahn C M T, Schäfer M A, Krayevsky A A, Nierhaus K H. J Biol Chem. 1996;271:32857–32862. doi: 10.1074/jbc.271.51.32857. [DOI] [PubMed] [Google Scholar]
- 24.Gregory S T, Lieberman K R, Dahlberg A E. Nucleic Acids Res. 1994;22:279–284. doi: 10.1093/nar/22.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearsey S E, Craig I W. Nature (London) 1981;290:607–608. doi: 10.1038/290607a0. [DOI] [PubMed] [Google Scholar]
- 26.Eckermann D J, Symons R H. Eur J Biochem. 1978;82:225–234. doi: 10.1111/j.1432-1033.1978.tb12015.x. [DOI] [PubMed] [Google Scholar]
- 27.Branlant C, Krol A, Machatt M A, Pouyet J, Ebel J P, Edwards K, Kossel H. Nucleic Acids Res. 1981;9:4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green, R., Switzer, C. & Noller, H. F. (1998) Science, in press. [DOI] [PubMed]
- 29.O’Connor M, Dahlberg A E. Proc Natl Acad Sci USA. 1993;90:9214–9218. doi: 10.1073/pnas.90.19.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porse B T, Garrett R A. J Mol Biol. 1995;249:1–10. doi: 10.1006/jmbi.1995.0276. [DOI] [PubMed] [Google Scholar]