Abstract
NaeI is a remarkable type II restriction endonuclease. It must bind two recognition sequences to cleave DNA, forms a covalent protein–DNA intermediate, and is only 1 aa change away from topoisomerase and recombinase activity. The latter activities apparently derive from reactivation of a cryptic DNA ligase active site. Here, we demonstrate that NaeI has two protease-resistant domains, involving approximately the N-terminal and C-terminal halves of the protein, linked by a protease-accessible region of 30 aa. The domains were purified by cloning. The C-terminal domain was shown by gel mobility-shift assay to have approximately 8-fold lower DNA-binding ability than intact NaeI. Analytical ultracentrifugation showed this domain to be a monomer in solution. The N-terminal domain, which contains the catalytic region defined by random mutagenesis, did not bind DNA and was a mixture of different-sized complexes in solution implying that it mediates self-association. DNA greatly inhibited proteolysis of the linker region. The results identify the DNA-binding domain, imply that DNA cleavage and recognition are independent and separable, and lead us to speculate about a cleft-like structure for NaeI.
Restriction endonuclease NaeI appears to have special mechanistic and evolutional importance. Showing its unusual heritage, NaeI must bind two DNA sequences to cleave DNA (1, 2). In addition, NaeI is the first restriction enzyme found to form a transient covalent intermediate with its newly cleaved substrate (3) much like the topoisomerases and recombinases. Finally, a single amino acid change, leucine to lysine at position 43 (L43K), radically transforms NaeI into a topoisomerase (3) and changes DNA recognition from double-stranded GCC/GGC to a preference for DNA containing unpaired bases (4). The L43K amino acid change is in a ligase-like active site that lies just upstream of the putative catalytic region (5). The ability of NaeI to act either as a restriction endonuclease or topoisomerase-recombinase depending on a single amino acid change is especially intriguing because NaeI has no detectable sequence homology with the topoisomerase or recombinase protein families. The unusual characteristics of NaeI suggest that it is either in the process of evolving from a topoisomerase to a restriction enzyme by abolishing its religation activity (3, 4) or may be able to serve both functions in vivo. NaeI may represent a missing link between some endonucleases and the topoisomerases and recombinases (4).
NaeI is a 317-aa polypeptide, has a molecular mass of 35 kDa (6), and forms a dimer in solution (Mr = 70) both in the absence and in the presence of cognate DNA (7). NaeI loops out DNA intervening distant NaeI DNA recognition sequences (8). This ability to loop out intervening sequences in DNA is highly unusual for a restriction enzyme but consistent with the requirement that NaeI bind two recognition sequences to activate DNA cleavage (1, 2).
Random mutagenesis shows that NaeI endonuclease has at least two functional regions: a region located within the N-terminal half of NaeI that mediates substrate binding and catalysis, and another region located within the C-terminal half of NaeI that binds DNA (5). We set out to map the protein-folding domains of NaeI for comparison with the regions defined by random mutagenesis, and to test whether the catalytic and DNA-binding functions could be separated by proteolysis. This knowledge is important because type II restriction endonucleases divide into subclasses in which sequence recognition and catalysis at the primary sequence level are either intertwined as in BamHI, EcoRI, EcoRV, and PvuII (reviewed in ref. 9) or independent as in FokI (10). Knowledge of these domains will enable comparison with the known domain structures of restriction endonucleases and other protein families. The comparisons may offer clues to how these protein families are related.
Proteolytic cleavage provides a direct probe of protein folding. Regions of proteins accessible to proteases generally occur in extended linker regions or loops between tightly folded domains that are exposed on the surface of the protein (11). The domains can usually be purified for further analysis. These methods have been used to identify the DNA-binding domains of several sequence-specific DNA-binding proteins (12–20). Here, we use these methods to map the domain organization of NaeI. NaeI protein was overexpressed by using a maltose-binding protein (MBP) fusion system. The purified fusion protein had similar activity to wild-type NaeI. Limited proteolysis of NaeI apoenzyme demonstrated two major domains resistant to proteolysis, encompassing almost the entire N-terminal and C-terminal halves of the protein, linked by a region relatively accessible to cleavage by both trypsin and chymotrypsin. The two protease-resistant domains were cloned, expressed as MBP fusions, and analyzed for DNA cleavage activity and specific DNA binding. The C-terminal domain was found to be a monomer in solution and bound DNA containing cognate recognition sequence with a KD 8-fold higher than that of NaeI. The N-terminal domain did not show DNA-binding ability and self-associated to give various- sized complexes in solution. The presence of DNA greatly reduced the susceptibility of the linker region to proteases. The change in susceptibility is the first indication of a protein conformational change associated with the switch from the inactive to active form of NaeI upon DNA binding (1).
MATERIALS AND METHODS
Materials.
Amylose resin and protein molecular weight markers were purchased from New England Biolabs. G-50 Sephadex, S-Sepharose, Q-Sepharose, and Phosphocellulose resin were purchased from Sigma. Escherichia coli strain CAA1 (F− e14− (mcrA−) lacY1 or D(lac)6 SupE44 galK2 galT22 mcrA rfbD1 mcrBa hsd(rk−mk+) M⋅MspI+) and plasmid pNEB786, containing the naeIR gene, were obtained from New England Biolabs. Bovine factor Xa was obtained from Haematologic Technologies (Burlington, VT).
Protein Purification.
The NaeIR gene and two NaeIR gene fragments were amplified from pNEB786 by PCR (21) and inserted into the plasmid vector pMALc2 (New England Biolabs) to give pMALc2:NaeI, pMALc2:NaeIΔC172, pMALc2:NaeIΔN168. The insertions were downstream from the malE gene, which encodes MBP and a factor Xa cleavage site so that either MBP∷NaeI or, after cleavage with factor Xa, NaeI proteins could be isolated. Plasmid pMALc2:NaeI was transformed into E. coli strain CAA1 and selected on Luria–Bertani plates containing ampicillin (100 μg/ml). Twelve-liter cultures were grown from single colonies with aeration at 25–30°C. MBP∷NaeI expression was induced with isopropyl β-d-thiogalactoside (0.5 mM), grown for an additional 6–8 hr, and harvested by centrifugation. Cell pellets were resuspended, washed in 0.9% NaCl, collected by centrifugation, and either processed or stored frozen. Cell pellets were resuspended in 4 volumes of column buffer (10 mM Tris⋅HCl (pH 7.4), 0.1 mM EDTA, 5% glycerol, 1 mM 2-mercaptoethanol, 50 mM NaCl). Phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM, to inhibit serine proteases. The resuspended cells were sonicated on ice (1 min per each 10 ml) and immediately centrifuged at 30,000 × g for 30 min to remove cellular debris.
MBP∷NaeI represented about 30% of total protein. Cell-free extract was passed over amylose resin equilibrated in column buffer plus 350 mM NaCl. Bound protein was eluted with 10 mM maltose in column buffer (fraction 2), which contained >70% of total protein as MBP∷NaeI. Fraction 2 was further purified by phosphocellulose chromatography using a NaCl gradient (50 mM–1 M NaCl in column buffer). Active fractions (225 mM NaCl) were pooled (fraction 3). For free NaeI protein, fraction 3 was digested with factor Xa (0.2% of total protein by weight) at 4°C for 48 hr (fraction 4), dialyzed in NaeI storage buffer (column buffer without glycerol), and purified by S-Sepharose column chromatography by using storage buffer and a NaCl gradient (50 mM–1 M). Active fractions were pooled (fraction 5) and a minor amount of uncleaved MBP∷NaeI protein was removed by using amylose resin (fraction 6). By these methods, NaeI was purified to >95%. MBP∷NaeI(1–145) and MBP∷NaeI(169–317) were purified by amylose resin chromatography. NaeI(169–317) was released from the fusion protein using factor Xa and purified to greater than 90% using phosphocellulose for further study.
Limited Proteolysis of NaeI.
Trypsin and chymotrypsin (Boehringer Mannheim) digestions were performed by adding at 37°C either 32 ng of trypsin or 1.6 μg of chymotrypsin in 16 μl of protease reaction buffer (10 mM Tris⋅HCl, pH 8.0/0.1 mM EDTA, pH 8.0/1 mM 2-mercaptoethanol/100 mM NaCl/5% glycerol/5 mM CaCl2) to 200 μl of protease reaction buffer containing 160 μg of purified NaeI. At various time points, 31-μl aliquots were removed and added to 0.3 μl of PMSF (100 mM in pure ethanol) to stop the reaction. SDS was added to 1%, the aliquots were heated to 100°C for 5 min, and reaction products were resolved by SDS/PAGE using 16% polyacrylamide gels. Gels were either stained with Coomassie brilliant blue-G250 or electroblotted onto poly(vinylidene difluoride) (PVDF) membrane for protein sequencing (22).
Amino Acid Sequence Analysis.
Polypeptide fragments were visualized as bands on PVDF membrane by staining with Ponceau S in water. Individual bands were cut from the membrane and subjected to N-terminal amino acid sequencing by the University of North Carolina Micro Protein Chemistry Facility using a Perkin–Elmer/ABI model 492 Precise instrument. The sequence of the first seven amino acid residues was determined for each fragment analyzed.
DNA Cleavage Activity Assays.
DNA cleavage activities of NaeI constructs were routinely monitored by examining their ability to cleave 0.5 μg of pBR322 as described elsewhere (3).
Gel Mobility-Shift Assays.
Radiolabeled single-stranded deoxyribooligonucleotide (oligonucleotide) was annealed to complementary oligonucleotide to give cognate and noncognate probe DNAs. Probes were incubated with protein in a 20-μl reaction volume containing 10 mM Tris⋅HCl (pH 8.0), 10 mM CaCl2, 20 mM NaCl, 10% glycerol, and BSA (0.1 mg/ml) for 20 min at room temperature. Cognate duplex DNA sequence was TTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCT; recognition sequence (underlined) was substituted by GAAGAA to give noncognate DNA. Reactions were immediately loaded onto 6 or 10% polyacrylamide nondenaturing gels and fractionated by electrophoresis as previously described (7). Results were analyzed by using a PhosphorImager (Molecular Dynamics).
Effect of DNA Binding on Proteolysis of NaeI.
To assess the effect of DNA binding on proteolysis of NaeI, 80 μg of NaeI in 100 μl of protease reaction buffer was incubated on ice for 30 min in the presence and absence of 107 μg of cognate DNA (TTGGGTGCCGGCAGGGTC). Protease digestion at 37°C was initiated by addition of 80 ng of trypsin or 800 ng of chymotrypsin. At timed intervals, 14 μl aliquots of reaction was mixed with 0.3 μl PMSF to terminate the reaction and fractionated by SDS/PAGE. To determine whether DNA was acting as a general protease inhibitor, this procedure was repeated using 80 μg of mammalian cell cycle protein p27 (23), which does not bind DNA, in place of NaeI.
Analytical Ultracentrifugation.
The oligomeric states of MBP-fusion proteins were determined by sedimentation velocity measurements in a Beckman XL-A analytical ultracentrifuge. Fusion proteins were dialyzed in centrifugation buffer containing 10 mM Tris (pH 7.4), 0.1 mM EDTA, 1 mM 2-mercaptoethanol, 100 mM NaCl, 10 mM maltose, and 5% glycerol. Maltose ensures against oligomerization of MBP (24). Samples (A280 = 1.5) were filtered (0.45 μm) and centrifuged (12,000 rpm) to eliminate particulates. Protein samples (400 μl) were centrifuged at 50,000 rpm. Sedimentation boundary was monitored by A280. Buffer density (1.0188 g/ml) was measured by specific gravity meter (DA-110M). Buffer viscosity (0.0114 poise) was determined from composition. Protein partial specific volumes were determined from amino acid composition (25). Molecular mass was derived from the Svedberg equation using the sedimentation coefficient (S) and diffusion coefficient (D) determined by the second boundary method (26), assuming globular spherical proteins with no hydration. S and D were corrected to standard state (25) using the axial program (Les Holladay, Alza Corp., Palo Alto, CA). Molecular mass was also determined by direct fit of sedimentation velocity profile (27).
RESULTS
Proteolysis of NaeI Endonuclease.
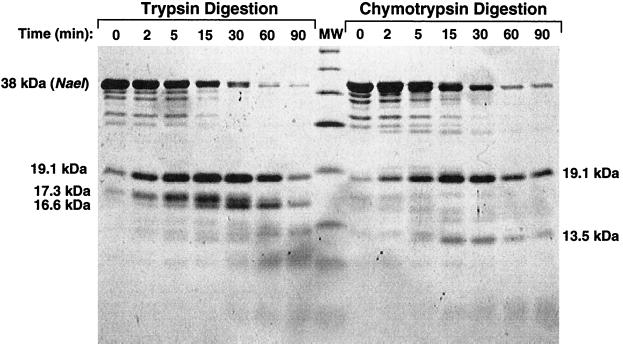
The absence of structural information for NaeI led us to map the domain structure of NaeI. By domains, we mean tightly folded regions of the protein resistant to proteolysis with trypsin and chymotrypsin. NaeI, purified as described in Materials and Methods, was incubated with limiting amounts of trypsin and chymotrypsin under conditions that digested the majority of full-length NaeI within 90 min (Fig. 1). Digestion with trypsin gave essentially three resistant fragments corresponding to molecular masses of 16.6, 17.3, and 19.1 kDa, which we refer to as fragments T16.6, T17.3, and T19.1 respectively. Digestion with chymotrypsin gave only two resistant fragments of approximately 13.5 (C13.5) and 19.1 (C19.1).
Figure 1.
Coomassie brilliant blue stained SDS-polyacrylamide gel showing pattern of polypeptide fragments produced by limited trypsin and chymotrypsin digestion of NaeI protein. The time of digestion is indicated along the tops of the lanes. The Mr of protease-resistant fragments analyzed in this study are indicated alongside the gel image and are based on the molecular weight (MW) markers glutamic dehydrogenase (55,561 Da), maltose-binding protein (42,710 Da), lactate dehydrogenase (36,487 Da), triosephosphate isomerase (26,625 Da), trypsin inhibitor (20,040–20,167 Da), lysozyme (14,313 Da), and aprotinin (6,517 Da). The digestion pattern at 0 min is caused by the amount of digestion taking place during mixing and sampling.
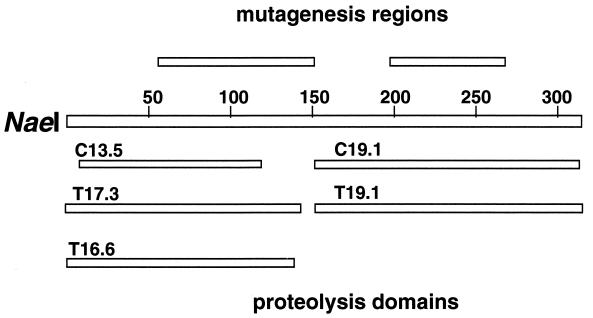
The N-terminal sequences of the stable fragments were determined by automated protein sequencing (Table 1). The protein fragment was mapped to the NaeI polypeptide by the N-terminal sequence of the fragment. Protein fragment length was estimated from the molecular mass of the fragment as determined by SDS/PAGE (Fig. 1). From the fragment’s N-terminal sequence and apparent molecular mass we mapped its location on the NaeI amino acid sequence as shown in Fig. 2. Based on this analysis, NaeI consists of two major protease-resistant domains linked by a region susceptible to protease cleavage. The protease-resistant domains reside within the C-terminal and N-terminal halves of the protein. To analyze the domains for DNA cleavage and binding, an attempt was made to separate the fragments by using ion-exchange chromatography (S-Sepharose, Q-Sepharose, and Phosphocellulose). The attempts failed to separate the two NaeI fragments and may indicate strong interactions between the two fragments. To overcome this problem, we cloned fragments of the gene, representing approximately the N-terminal (amino acids 1–145) and C-terminal (amino acids 169–317) domains, fused to MBP as described in Materials and Methods to give MBP∷NaeI(1–145) and MBP∷NaeI(169–317), respectively.
Table 1.
Polypeptide fragments of NaeI produced by limited proteolysis
| Fragment* | Sequence of N terminus† | Sequence position of polypeptide fragments‡ |
|---|---|---|
| T19.1 | LTPEGRA | 151 |
| T17.3 | MTELPLQ | 1 |
| T16.6 | MTELPLQ | 1 |
| C19.1 | LTPEGRA | 151 |
| C13.5 | QFAEPDD | 7 |
Fragments are named according to protease treatment (T, trypsin; C, chymotrypsin) and apparent molecular size of fragment (in kDa) based on relative mobility during SDS/PAGE (e.g., T19.1 = fragment of approximately 19.1 kDa produced by trypsin digestion).
Sequences shown were determined by automated amino acid sequencing of proteolytic reaction products as described in Materials and Methods.
The sequence position of each polypeptide fragment is given as the position of the first amino acid residue of the N-terminal end of the fragment relative to the amino acid sequence of NaeI.
Figure 2.
Map of the locations of the protease-resistant fragments relative to full-length NaeI polypeptide. Also shown is the location of the catalytic and DNA-binding regions mapped by random mutagenesis (5).
Proteins MBP∷NaeI(1–145), MBP∷NaeI(169–317), and MBP∷NaeI were purified >70% over amylose resin columns. DNA cleavage activities were examined by incubating up to 1 μg of each protein with pBR322. The specific activity of MBP∷NaeI was similar (within 2-fold) to that of native NaeI purified by other methods (28), indicating that MBP fusion has little effect on NaeI activity. Neither MBP∷NaeI(169–317) nor MBP∷NaeI(1–145) alone nor when combined showed detectable DNA cleavage activity.
DNA Binding.
Gel mobility-shift assays (29) were used as described in Materials and Methods to determine whether MBP∷NaeI(1–145) and MBP∷NaeI(169–317) (100 μM each) could bind DNA. MBP∷NaeI(169–317) showed strong binding affinity for DNA with NaeI cognate recognition sequence. The fraction of DNA bound was similar to that of the full-length NaeI fusion protein. No DNA-binding activity (specific or nonspecific) was detected for MBP∷NaeI(1–145) at protein concentrations of up to 1.1 μM (data not shown).
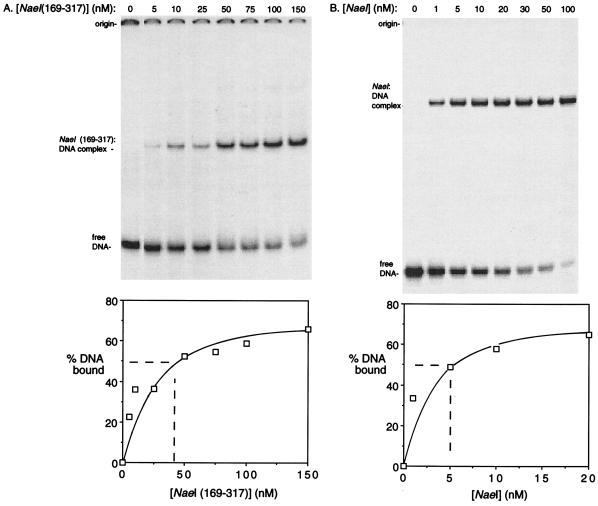
The ability of NaeI(169–317) to bind DNA was quantitated and compared with that of NaeI after isolating the two polypeptides free of MBP as described in Materials and Methods. Apparent DNA-binding coefficients (KD), or concentration at which 50% of the DNA was bound by protein, for NaeI and NaeI(169–317) binding to a 36-bp cognate DNA were approximately 5 nM for NaeI and 40 nM for NaeI(169–317) (Fig. 3).
Figure 3.
Determination of apparent KD for NaeI and NaeI(169–317) binding to DNA using gel mobility-shift assay. DNA probe was cognate 36-mer double-stranded DNA (0.2 nM). The protein concentrations used in each reaction are shown above each lane. The reaction conditions are described in Materials and Methods. The band intensities as a function of protein concentration were quantitated by densitometry and are plotted in the graphs at the bottom.
Effect of DNA Binding on Proteolysis of NaeI.
To gain insight into the domains that either interact with DNA in the intact NaeI molecule or undergo a conformational change upon binding DNA, we compared the trypsin and chymotrypsin digestion pattern in the presence and absence of DNA. We looked for regions of NaeI made either more sensitive or more resistant by DNA binding. The DNA was shown not to be a general inhibitor of either protease by incubation with mammalian cell cycle protein p27 (23) that does not bind DNA. Protein p27 was found to be equally susceptible to protease cleavage whether or not the DNA fragment was added to the reaction (results not shown).
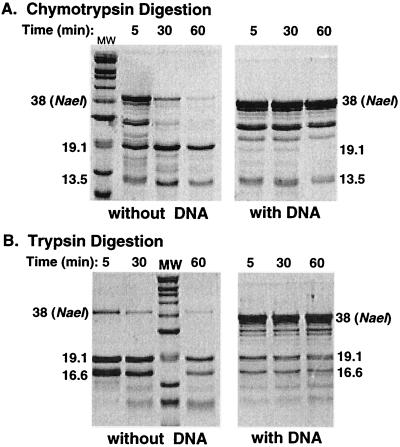
The protease digestion pattern of NaeI was determined in the presence and absence of cognate DNA as described in Materials and Methods. The digestion reactions were analyzed by SDS/PAGE (Fig. 4). Digestion with both trypsin and chymotrypsin was greatly attenuated by NaeI interaction with DNA, and the chymotrypsin digestion pattern was altered as well. Whereas the 13.5-kDa band was formed to a similar extent whether or not DNA was present, formation of the prominent 19.1-kDa band (corresponding to the C-terminal domain) was inhibited when DNA was included in the reaction. This indicates that the C terminus of the linker region is less accessible after DNA binding than its N terminus. This conclusion is consistent with the complete loss of cleavage by trypsin because trypsin only cleaves near the C terminus of the linker region (see Fig. 2).
Figure 4.
Coomassie brilliant blue stained SDS-polyacrylamide gel showing pattern of polypeptide fragments produced by limited chymotrypsin (A) and trypsin (B) digestion of NaeI protein in the presence or absence of DNA. Digestion times are shown above the lanes. The Mr of protease-resistant fragments are indicated alongside the gel image and are based on the molecular weight (MW) markers described in the legend to Fig. 2.
Self-Association.
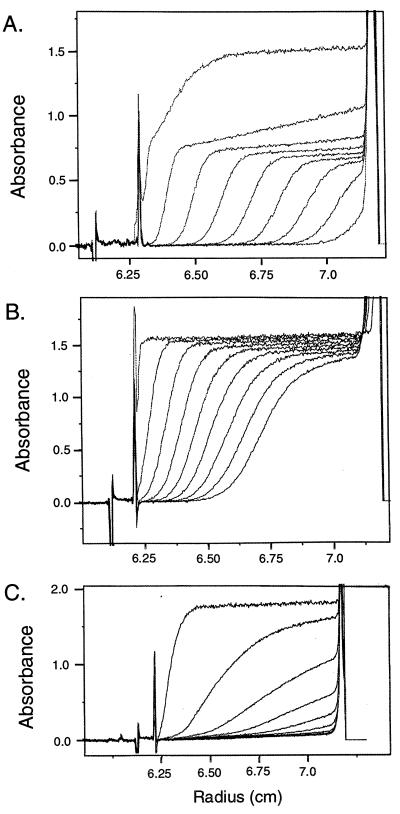
NaeI self-associates to form a dimer in solution (7). Analytical ultracentrifugation was used to determine the abilities of the NaeI domains to self-associate. From the determination of sedimentation and diffusion coefficients, the apparent molecular masses of MPB∷NaeI, MPB∷NaeI(1–145), and MPB∷NaeI(169–317) were calculated assuming spherical globular proteins with no hydration. Sedimentation of the two domains and wild-type protein fused to MBP are shown in Fig. 5. MBP∷NaeI centrifugation shows the presence of two major species (Fig. 5A). One, a rapidly sedimenting complex, is apparently a mixture of various-sized complexes of NaeI. The other major species showed a sedimentation coefficient of 5.125 × 10−13 sec, diffusion coefficient of 3.46 × 10−7 cm2/sec, and molecular mass of 147 kDa as determined by direct fitting of the sedimentation velocity profile (27). Similar values were obtained by other methods as described in Materials and Methods. The molecular mass determined is approximately two times the molecular mass of the monomeric MBP∷NaeI protein (78 kDa) determined from its amino acid composition. Thus MBP∷NaeI, like NaeI (7) is a dimer in solution, and also gave a rapidly sedimenting mixture of higher-ordered complexes that could be resuspended by mixing and recentrifuged to give the same sedimentation pattern. MBP∷NaeI(169–317) showed the presence of a single species (Fig. 5B) with sedimentation coefficient of 3.151 × 10−13 sec, diffusion coefficient of 6.53 × 10−7 cm2/sec, and molecular mass of 47 kDa. The apparent molecular mass value of 47 kDa is less than that determined from its amino acid composition (59 kDa); we infer that MBP∷NaeI(168–317) is a monomer in solution. MBP∷NaeI(1–145) showed a pattern corresponding to that expected for a mixture of higher-ordered complexes similar to that seen in the preparation of MBP∷NaeI (Fig. 5C).
Figure 5.
Analytical ultracentrifuge determination of molecular weight of maltose-binding protein fusions with NaeI, and its N-terminal and C-terminal domains. Centrifugation was at 50,000 rpm; A280 boundary profiles are shown at 5, 10, 15, 20, 25, 30, 35, 40, and 45 min (from left to right). x-axis shows the radius of the sample cell (from left to right is the direction of centrifugal force); y-axis shows the A280 of the solution, measured through the sample cell. (A) MBP∷NaeI; (B) MBP∷NaeI(169–317); (C) MPB∷NaeI(1–145).
DISCUSSION
Domain Organization of NaeI.
Limited digestion with trypsin and chymotrypsin reveals that NaeI is composed of at least two major domains that include amino acids 1–120 and 151–317. These two domains are linked by a short protease-sensitive region that includes amino acids 121–150. The boundaries of the protease-resistant domains approximately parallel (Fig. 2) the two functional regions defined by random mutagenesis (5). Cloning and characterization of the domains demonstrated that only the C-terminal domain detectably binds DNA. Thus, the separable N-terminal and C-terminal domains appear to have independent functions. Mutagenic studies imply that the N-terminal domain mediates catalysis: The L43K substitution gives NaeI topoisomerase activity (3) and T60I and E70K substitutions drastically reduce cleavage activity without reducing DNA binding (5). Mutagenic studies and the above DNA-binding results demonstrate that the C-terminal domain mediates sequence-specific DNA binding. Although it is clear from these studies that the C-terminal domain is mainly responsible for DNA recognition, it is not possible to fully rule out the presence in this domain of amino acids that contribute to catalytic function. No essential catalytic amino acids were located in this region by random mutagenesis, but the level of mutagenesis was not saturating.
The apparent independence of the catalytic and DNA-binding domains at the gene level is different from the type II restriction endonucleases for which information is available. For example, recognition and catalytic function are intertwined for BamHI, EcoRI, EcoRV, and PvuII (reviewed in ref. 9)—enzymes that recognize and cleave within a specific sequence. On the other hand, FokI binds a specific recognition sequence but cleaves nearby the sequence and has separable recognition and catalytic domains (although not in the same order as NaeI) (10). NaeI, therefore, may be unusual because it cleaves within its recognition sequence, yet appears to have separable functions. The domain structure of NaeI suggests it may be more related to the IIs type endonucleases like FokI than the type II enzymes like EcoRI.
DNA Binding.
DNAs encoding the two protease-resistant NaeI domains were cloned and overexpressed in E. coli. Isolation of the peptides and analysis of their DNA-binding abilities demonstrate that only the C-terminal domain has detectable DNA-binding activity in solution. Measurements of apparent KD for DNA binding show that the C-terminal domain accounts for approximately an eighth of the DNA-binding ability of intact NaeI. The C-terminal domain binds, however, as a monomer. Cooperative binding as a dimer in the wild-type protein could provide the missing binding energy.
The same DNA-binding measurements indicate that the N-terminal domain does not detectably bind DNA. Care must be taken, however, in concluding that the N-terminal domain has no DNA-binding capability. Mutagenesis studies (5) showed that some mutations in the N-terminal region modified DNA binding. These effects could be caused by disruptions to protein conformation that interfere with DNA binding to the C-terminal domain. Also likely, weak stabilizing interactions with DNA coming from the N-terminal domain, which must interact with DNA to catalyze cleavage in the native enzyme, may be undetectable when the N-terminal domain is separated from the major DNA-binding C-terminal domain.
Self-Association.
How NaeI dimerizes is important because none of the known protein dimerization motifs are obvious from the NaeI amino acid sequence. Analytical ultracentrifugation demonstrated that the isolated N-terminal domain is a mix of various-sized complexes. The ability of the N-terminal domain of NaeI to form various-sized complexes in solution implies that this domain mediates dimerization. Consistent with this conclusion, the C-terminal domain was shown to be a monomer in solution.
It is possible that self-association of the N-terminal domain contributes to the lack of DNA binding by this domain. We constructed other mutants with changes in the N-terminal domain that also form various-sized complexes in solution. The mutants bind DNA only if the C-terminal domain is intact (J.D.C. and M.D.T., unpublished results). The results show that formation of the various-sized complexes does not block NaeI accessibility to DNA.
Effects of DNA Binding on Proteolysis of NaeI.
Proteolysis of NaeI was strongly inhibited by DNA. The inhibition can, in principle, be explained by either of two models. In the first model, DNA directly binding to the protease-sensitive linker region blocks access of the protease and thus inhibits cleavage. In the second model, binding of DNA to another region of the protein induces a conformational shift in NaeI that reduces access or sensitivity of the linker region to cleavage. The linker region of NaeI (amino acids 121–150) has a concentration of positive charges (5 of 12 residues) located toward its C terminus. The involvement of this region in DNA direct binding could explain why the addition of DNA more effectively inhibits cleavage of the C terminus of the linker region than it does the N terminus.
A Low-Resolution Model.
Proteolysis demonstrates that NaeI has two tightly folded domains linked by a hinge region. The results imply independence of DNA binding and cleavage at the gene sequence level and show large effects of DNA on proteolysis of the hinge region. The requirement that both domains interact with DNA for sequence-specific cleavage leads us to speculate that docking of DNA in a crevice formed by these domains rearranges the linker region and makes it less accessible to protease. This low-resolution structure is consistent with the recently reported structure for FokI in which recognition and catalysis reside in separate domains (10); FokI has become a target for attempts to design new cleavage specificities based on domain swapping between proteins. Perhaps NaeI will also be useful for this purpose. In FokI, the cleavage domain is folded away from the DNA by protein–protein interactions with the recognition domain (10). Apparently, this provides a mechanism for avoiding DNA cleavage until interaction with cognate recognition sequence when DNA interactions cause the cleavage domain to swing over to the DNA for cleavage (10, 30). Such a mechanism could also apply to NaeI with its separate cleavage and recognition domains apparently linked by a switch that is toggled by DNA binding.
Acknowledgments
We thank Dr. Christian Lombardo for assistance with the analytical ultracentrifuge. This work was supported in part by a grant from the National Institutes of Health (GM 52123) and an institutional National Research Service Award (5-T32-CA09156).
References
- 1.Conrad M, Topal M D. Proc Natl Acad Sci USA. 1989;86:9707–9711. doi: 10.1073/pnas.86.24.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang C C, Topal M D. Biochemistry. 1992;31:9657–9664. doi: 10.1021/bi00155a019. [DOI] [PubMed] [Google Scholar]
- 3.Jo K, Topal M D. Science. 1995;267:1817–1820. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- 4.Jo K, Topal M D. Nucleic Acids Res. 1996;21:4171–4175. doi: 10.1093/nar/24.21.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtz J K, Topal M D. J Biol Chem. 1994;269:27286–27290. [PubMed] [Google Scholar]
- 6.Taron C H, Van Cott E M, Wilson G G, Moran L S, Slatko B E, Hornstra L J, Benner J S, Kucera R B, Guthrie E P. Gene. 1995;155:19–25. doi: 10.1016/0378-1119(94)00806-4. [DOI] [PubMed] [Google Scholar]
- 7.Baxter B K, Topal M D. Biochemistry. 1993;32:8291–8298. doi: 10.1021/bi00083a033. [DOI] [PubMed] [Google Scholar]
- 8.Topal M D, Thresher R J, Conrad M, Griffith J. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X, Balendiran K, Schildkraut I, Anderson J E. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wah D A, Hirsch J A, Dorner L F, Schildkraut I, Aggarwal A K. Nature (London) 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 11.Price N C, Johnson C M. In: Proteolytic Enzymes: A Practical Approach. Beynon R J, Bond J S, editors. Oxford: IRL; 1989. pp. 163–180. [Google Scholar]
- 12.Abdel-Meguid S S, Grindley N D F, Templeton N S, Steitz T A. Proc Natl Acad Sci USA. 1984;81:2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith D R, Jackson I J, Brown D D. Cell. 1984;37:645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- 14.Marzouki N, Camier S, Ruet A, Moenne A, Sentenac A. Nature (London) 1986;323:176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- 15.Huet J, Sentenac A. Proc Natl Acad Sci USA. 1987;84:3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama C, Teplow D B, Harshey R M. Proc Natl Acad Sci USA. 1987;84:1809–1813. doi: 10.1073/pnas.84.7.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vargas L M, Pargellis C A, Hasan N M, Bushman E W, Landy A. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 18.Boulanger P A, L’Etoile N D, Berk A J. Nucleic Acids Res. 1989;17:7761–7770. doi: 10.1093/nar/17.19.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan H, Clary D, Sadowski P D. J Biol Chem. 1991;266:11347–11354. [PubMed] [Google Scholar]
- 20.Adams G M, Blumenthal R M. Biochemistry. 1997;36:8284–8292. doi: 10.1021/bi961885n. [DOI] [PubMed] [Google Scholar]
- 21.Clackson T, Gussow D, Jones P T. In: PCR—A Practical Approach. Quirke P, Taylor G R, editors. New York: Oxford Univ. Press; 1991. pp. 187–214. [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl D, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 23.Toyoshima H, Hunter T. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 24.Richarme G. Biochem Biophys Res Commun. 1982;105:476–481. doi: 10.1016/0006-291x(82)91459-0. [DOI] [PubMed] [Google Scholar]
- 25.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 26.Goldberg R J. J Phys Chem. 1953;57:194–202. [Google Scholar]
- 27.Philo J S. In: Modern Analytical Ultracentrifugation. Schuster T M, Laue T M, editors. Basel: Birkhauser; 1994. pp. 157–170. [Google Scholar]
- 28.Yang C C, Baxter K B, Topal M D. Biochemistry. 1994;33:14918–14925. doi: 10.1021/bi00253a031. [DOI] [PubMed] [Google Scholar]
- 29.Fried M G. Electrophoresis. 1989;10:366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- 30.Waugh D S, Sauer R T. J Biol Chem. 1994;269:12298–12303. [PubMed] [Google Scholar]