Abstract
NF-κB is activated by various stimuli including inflammatory cytokines and stresses. A key step in the activation of NF-κB is the phosphorylation of its inhibitors, IκBs, by an IκB kinase (IKK) complex. Recently, two closely related kinases, designated IKKα and IKKβ, have been identified to be the components of the IKK complex that phosphorylate critical serine residues of IκBs for degradation. A previously identified NF-κB-inducing kinase (NIK), which mediates NF-κB activation by TNFα and IL-1, has been demonstrated to activate IKKα. Previous studies showed that mitogen-activated protein kinase/ERK kinase kinase-1 (MEKK1), which constitutes the c-Jun N-terminal kinase/stress-activated protein kinase pathway, also activates NF-κB by an undefined mechanism. Here, we show that overexpression of MEKK1 preferentially stimulates the kinase activity of IKKβ, which resulted in phosphorylation of IκBs. Moreover, a catalytically inactive mutant of IKKβ blocked the MEKK1-induced NF-κB activation. By contrast, overexpression of NIK stimulates kinase activities of both IKKα and IKKβ comparably, suggesting a qualitative difference between NIK- and MEKK1-mediated NF-κB activation pathways. Collectively, these results indicate that NIK and MEKK1 independently activate the IKK complex and that the kinase activities of IKKα and IKKβ are differentially regulated by two upstream kinases, NIK and MEKK1, which are responsive to distinct stimuli.
Exposure of cells to certain cytokines [e.g., tumor necrosis factor (TNF) and interleukin (IL)-1] or environmental stresses (e.g., UV and γ irradiation) leads to activation of the transcription factors NF-κB and c-Jun (1–4). NF-κB is composed of hetero- or homodimers of Rel family proteins and is involved in the inflammatory response, cell adhesion, growth control, and cell death (2, 5, 6). In unstimulated cells, NF-κB is sequestrated in the cytoplasm as a complex with inhibitory proteins called IκBs (1). In the family of IκBs, the most important ones seem to be IκBα, IκBβ, and a recently cloned IκBɛ (1, 7). Various stimuli to activate NF-κB result in phosphorylation of two serines at the N terminus of IκBα and IκBβ and subsequent degradation of the IκBs, resulting in translocation of NF-κB into the nucleus and activation of target genes. The mutation of the two serine residues Ser-32 and Ser-36 in IκBα decreases phosphorylation and degradation of IκBα protein (8–11). IκBβ and IκBɛ also have the two conserved serine residues at the N terminus for signal-induced degradation (7, 12, 13). These results indicate that identification of the kinases responsible for the IκB phosphorylation is a critical step for understanding the mechanism of NF-κB activation. A previous study demonstrated that the IκB kinase (IKK) forms a large complex with a molecular mass of 700 kDa, and this complex could be activated by ubiquitination or mitogen-activated protein kinase/ERK kinase kinase-1 (MEKK1), a member of the MAP kinase kinase kinase (MAPKKK) family (14). However, the function of MEKK1 in TNF-mediated NF-κB activation still remains controversial (14–18).
The second pathway of stress responses is the c-Jun N-terminal kinase (JNK)/stress-activated protein kinase (SAPK) pathway (3, 4). MEKK1 has been implicated in this pathway, which activates MKK4 that in turn activates JNK/SAPK (19, 20). Then, the JNK/SAPK not only activates c-Jun that constitutes the transcription factor AP-1 but also contributes to apoptosis by an undefined mechanism (21).
Tumor necrosis factor receptor-associated factors (TRAFs) have emerged as signal-transducing molecules through members of the TNF-R superfamily and IL-1R1 (22–35). TRAFs, except for TRAF4, have been shown to directly or indirectly interact with cytoplasmic domains of the TNF-R superfamily molecules and IL-1R1. TRAF2, TRAF5, and TRAF6 mediate NF-κB activation by these receptors (27–36). NF-κB-inducing kinase (NIK) was first identified as a TRAF2-interacting protein and has structural homology to the MAPKKK family (37). Overexpression of NIK-activated NF-κB and a kinase inactive mutant of NIK blocked TNF-, IL-1-, and TRAFs-mediated NF-κB activation, suggesting that NIK is a common downstream mediator of NF-κB activation by TNF, IL-1, and TRAFs (18, 37). TRAF2, TRAF5, and TRAF6 also activate MEKK1, which in turn activates the JNK/SAPK pathway (17, 18). Collectively, these results demonstrate that two responses including NF-κB activation and JNK/SAPK activation diverge downstream of TRAFs.
Recently, three groups (38–42) have independently identified two subunits of the IKK complex, designated IKKα (or IKK-1) and IKKβ (IKK-2) by using a protein purification method or a yeast two-hybrid assay to clone interacting molecules of NIK. Human IKKα, a previously cloned serine-threonine kinase called CHUK (43), and human IKKβ are composed of an N-terminal serine-threonine kinase domain, a central leucine zipper domain, and a C-terminal helix-loop-helix domain. These two kinases show 52% identity at the amino acid level. Biochemical analysis demonstrated that IKKα and IKKβ independently phosphorylate both serine 32 and 36 in IκBα (38–42). Furthermore, overexpression of IKKα or IKKβ activated an NF-κB-dependent reporter and a kinase negative mutant of IKKα- or IKKβ-inhibited NF-κB activation by TNF or IL-1 (38–42). These results clearly demonstrated that the IKKα and IKKβ constitute the functional IKK complex. Although NIK has been shown to activate IKKα (39), the regulation of IKKα and IKKβ kinase activities is still largely unknown.
In the present study, we identified a murine homologue of human IKKβ, which is implicated in NF-κB activation by the TNF-R family members and TRAFs. We demonstrated that overexpression of MEKK1 preferentially stimulated the kinase activity of IKKβ, which resulted in phosphorylation of IκBs. By contrast, overexpression of NIK activated both IKKα and IKKβ comparably, indicating differential regulation of the IKK complex by NIK and MEKK1.
MATERIALS AND METHODS
Reagents and Cell Lines.
Anti-Flag mAb and anti-hemagglutinin (HA) mAb (12CA5) were purchased from Kodak International Biotechnology and Boehringer, respectively. The human embryonic kidney 293 cells were cultured in DMEM supplemented with 10% fetal bovine serum.
cDNA Cloning.
To identify an IKKα-related kinase, we searched an expressed sequence tag (EST) database in the National Center for Biotechnology Information (NCBI) DNA database and identified a cDNA clone (AA326115) showing high homology to IKKα. Then, we screened a murine spleen cDNA library (Stratagene) with a PCR fragment corresponding to the EST sequence. Several overlapping clones were obtained and sequenced using series of oligonucleotide primers by standard methods. A full-length IKKα cDNA was obtained by screening the same library by standard methods.
Expression Vectors.
Mammalian expression vectors encoding CD27 (C. Morimoto, Institute of Medical Science, University of Tokyo) (44), TRAF2 and CD30 (T. Watanabe, Institute of Medical Science, University of Tokyo) (30), CD40 (H. Kikutani, Research Institute for Microbial Disease, University of Osaka) (45), lymphotoxin-β receptor (LT-βR) (C. F. Ware, La Jolla Institute for Allergy and Immunology) (28), TRAF5 (28), TRAF6, NIK, and NIK-KM(KK429–430AA) (D. Wallach, Weizmann Institute of Science) (37), and MEKK1 and MEKK1-KM(K432M) (S. Ohno, Yokohama City University) (15) have been described previously. Expression vectors for Flag or HA epitope-tagged IKKα and IKKβ were constructed in-frame with DNA encoding an N-terminal Flag or HA epitope in pCR-3 (Stratagene). Expression plasmids encoding IKKα-KM(K44A) and IKKβ-KM(K44A) were generated by using a mutagenesis kit (Stratagene) according to the manufacturer’s instruction.
pGEX-IκBα(1–100), pGEX-IκBβ(1–120), and pGEX-IκBɛ(1–61) were constructed by subcloning the RT-PCR products encoding corresponding amino acids into pGEX-4T vector (Pharmacia). pGEX-IκBα(1–100) (S32A, S36A; designated as 1–100AA), pGEX-IκBβ(1–120) (S19A, S23A; designated as 1–120AA), and pGEX-IκBɛ(1–61) (S18A, S22A; designated as 1–61AA) were generated by using the mutagenesis kit. Expression and purification of the glutathione S-transferase (GST) fusion proteins were performed as described previously (46).
NF-κB-Dependent Reporter Assays.
293 cells (1 × 106) were plated in 35-mm dishes. On the following day, the cells were transfected with the indicated expression vectors using Lipofectamine (Promega). Every transfection included 50 ng of β-actin-β-gal (K. Yokota, NIH, Japan), β-actin promoter-driven β-galactosidase expression plasmid, for the normalization of transfection efficiency, together with 100 ng of the reporter plasmid and various amounts of each expression vector. Total DNA was kept constant by supplementation with pCR-3. The reporter plasmid, 3xκB-L, has three repeats of the NF-κB site upstream of a minimal thymidine kinase promoter and a luciferase gene in pGL-2 vector (Promega) (M. Kashiwada, NIH, Japan). After 24 h, the cells were harvested in PBS and lysed in a luciferase lysis buffer, LC-β (Piccagene, Toyo Ink, Tokyo). The lysates were assayed for luciferase and β-galactosidase activities using a luminometer (Berthold).
In Vitro Phosphorylation Assays.
293 cells (2 × 106) were plated in 60-mm dishes and transfected with various expression vectors using Lipofectamine. After 24–36 h, the cells were washed with ice-cold PBS and lysed for 30 min on ice in 1 ml of a lysis buffer containing 1% Nonidet P-40, 50 mM Hepes (pH 7.3), 150 mM NaCl, 2 mM EDTA, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM sodium orthovanadate, and 1 mM NaF. Nuclei were removed by centrifugation, and the supernatant was precleared with protein G-Sepharose (Pharmacia) for 1–2 h. The cleared lysates were incubated with anti-HA or anti-Flag mAb for 1 h at 4°C. After addition of 30 μl of protein G-Sepharose, the lysates were incubated for a further 1 h. The immunoprecipitates were washed three times with the lysis buffer and twice in a kinase buffer containing 20 mM Hepes (pH 7.3), 20 mM MgCl2, 20 mM MnCl2, 1 mM EDTA, 1 mM NaF, 0.1 mM sodium orthovanadate, and 1 mM DTT. The immunoprecipitates were then incubated with 1 μg of GST-IκBα(1–100), GST-IκBα(1–100AA), GST-IκBβ(1–120), GST-IκBβ(1–120AA), GST-IκBɛ(1–61), or GST-IκBɛ(1–61AA) and [γ-32P]ATP (10 μCi) in the kinase buffer for 20 min at 30°C. The reaction was stopped by addition of the Laemmli’s sample buffer. The eluted proteins were subjected to SDS/PAGE, and the autoradiograms were visualized on an image analyzer (Fujix, BAS2000). In all cases, expression of the transfected proteins was verified by immunoblotting of aliquots of the cell lysates as described previously (47). In some experiments, amounts of the GST-IκBs in the reaction mixtures were verified by Coomassie blue staining.
RESULTS AND DISCUSSION
cDNA Cloning and Expression of Murine IKKβ.
Recent identification of the first subunit of the IKK complex (IKKα) (38, 39) prompted us to search for IKKα-related kinases. We found a homologous sequence in the EST database and subsequently cloned a full-length cDNA from a murine spleen cDNA library. During preparation of this manuscript, a human kinase highly related to IKKα has been cloned and named IKKβ or IKK-2 (40–42). As our clone has the highest homology to human IKKβ (hIKKβ) (see below), it seems to be the murine IKKβ (mIKKβ).
The mIKKβ cDNA encodes 758 amino acids, which shows 92% and 50% identity to human IKKβ and murine IKKα in amino acid level, respectively (Fig. 1). mIKKβ is composed of an N-terminal serine threonine kinase domain, a leucine zipper domain, and a C-terminal helix-loop-helix domain, as is mIKKα (Fig. 1B). Northern blot analysis with the mIKKβ cDNA probe revealed a ubiquitous expression of a 4-kb transcript in various murine tissues (data not shown).
Figure 1.
Amino acid alignment of murine and human IKKα and IKKβ. (A) Comparison of murine and human IKKβ. The full-length amino acid sequences are shown and numbered. Dashes in the human sequence indicate residues identical to those in mice. (B) Comparison of murine IKKβ and IKKα. Identical residues are indicated by asterisks. The amino-terminal kinase domains are indicated by large boxes, and the conserved amino acids to form a leucine zipper are indicated by small boxes. The leucine zipper and the helix-loop-helix domains are indicated by boldface type.
A Kinase Inactive Mutant of IKKβ Blocks NF-κB Activation by Members of the TNF-R Superfamily and TRAFs.
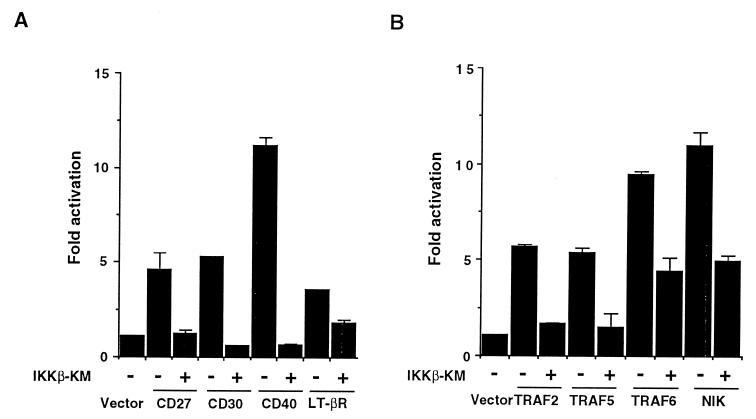
A kinase inactive mutant of IKKα or IKKβ blocked TNF- and IL-1-induced NF-κB activation (refs. 39, 41, and 42, and our unpublished results). To test whether IKKβ is also involved in NF-κB activation by other members of the TNF-R superfamily, we constructed a kinase inactive mutant of murine IKKβ (IKKβ-KM), in which a lysine at the ATP binding site in the kinase domain was substituted by an alanine. We transiently transfected 293 cells with expression vectors for members of the TNF-R superfamily along with a reporter plasmid, 3xκB-L. As shown in Fig. 2A, cotransfection of IKKβ-KM blocked CD27-, CD30-, CD40-, and LT-βR-induced reporter gene activation. NF-κB activation, induced by these receptors, TNF, and IL-1, is mediated by TRAF2, TRAF5, or TRAF6 and their interacting kinase NIK (refs. 18, 25, 28–32, 36, and 37, and our unpublished results). Cotransfection of IKKβ-KM also inhibited NF-κB-dependent reporter gene activity elicited by TRAF2, TRAF5, and TRAF6 (Fig. 2B). Furthermore, NF-κB activation by NIK was also inhibited by IKKβ-KM, indicating a critical contribution of IKKβ to the NIK-mediated NF-κB activation, as has been demonstrated for IKKα (39). Collectively, these results indicated that IKKβ is a common downstream kinase for NF-κB activation through members of the TNF-R superfamily and their signal transducers, TRAFs and NIK.
Figure 2.
A kinase inactive mutant of IKKβ (IKKβ-KM) blocks NF-κB activation by members of the TNF-R superfamily or TRAFs. (A) Effect of IKKβ-KM on CD27-, CD30-, CD40-, and LT-βR-induced NF-κB-dependent reporter activity. 293 cells were transiently transfected with 100 ng of 3xκB-L and 0.5 μg of expression vectors for CD27, CD30, CD40, or LT-βR along with or without 0.5 μg of IKKβ-KM. Total amount of the DNAs was kept constant by supplementation with pCR-3. The cells were harvested 24 h posttransfection. Luciferase activities were determined and normalized on the basis of β-galactosidase (β-gal) expression from cotransfected β-actin-β-gal (50 ng). The level of induction in luciferase activity was compared as a ration to cells transfected with the control vector. Data are shown as mean ± SEM of triplicated samples and represent one of three experiments with similar results. (B) Effect of IKKβ-KM on TRAFs- and NIK-induced NF-κB reporter activity. 293 cells were transfected with 0.5 μg of expression vectors for TRAF2, TRAF5, TRAF6, or NIK with or without 0.5 μg of IKKβ-KM along with 100 ng of 3xκB-L and 50 ng of β-actin-β-gal. Data were obtained and are represented as in A.
IKKβ Phosphorylates IκBɛ As Well As IκBα and IκBβ and Is Activated by NIK.
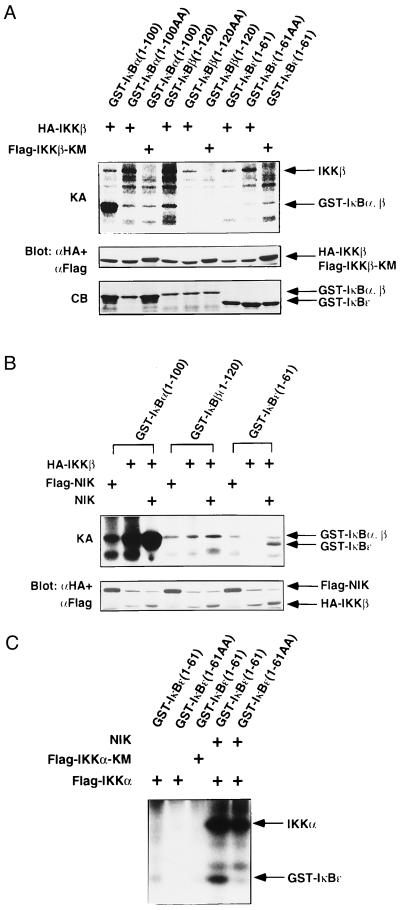
To characterize the kinase activity of IKKβ, HA-tagged IKKβ or Flag-tagged IKKβ-KM was transiently expressed in 293 cells. The immunoprecipitates with anti-HA or anti-Flag mAb were subjected to in vitro phosphorylation assays using GST fusion proteins of IκBα, β, or ɛ as substrates. As shown in Fig. 3A, IKKβ, but not IKKβ-KM, phosphorylated the wild-type GST-IκBα(1–100) and GST-IκBβ(1–120) but not their mutants, in which the two critical serines for phosphorylation (Ser-32 and Ser-36 for IκBα and Ser-19 and Ser-32 for IκBβ) were both replaced by alanines. The kinase activity of IKKβ to GST-IκBβ was consistently lower than that to GST-IκBα. IκBɛ is a newly identified member of the IκB family, and phosphorylation of Ser-18 and Ser-22 is required for degradation (7). It has not been determined whether IκBɛ is also phosphorylated by IKKα or IKKβ. Then, we also examined the phosphorylation of IκBɛ by IKKα or IKKβ. As shown in Fig. 3 A–C, neither IKKβ nor IKKα alone phosphorylated GST-IκBɛ(1–61).
Figure 3.
In vitro phosphorylation of IκBα, -β, and -ɛ by IKKβ and IKKα. (A) Specificity of IκBs phosphorylation by IKKβ. 293 cells were transiently transfected with expression vector for HA-IKKβ or Flag-IKKβ-KM. Twenty-four hours after transfection, IKKβ proteins were immunoprecipitated with anti-Flag or anti-HA mAb. The precipitates were incubated with GST-IκBα(1–100), GST-IκBα(1–100) (S32A, S36A; designated as 1–100AA), GST-IκBβ(1–120), GST-IκBβ(1–120) (S29A, S32A; designated as 1–120AA), GST-IκBɛ(1–61), or GST-IκBɛ(1–61) (S18A, S22A; designated as 1–61AA) and [γ-32P]ATP, resolved by SDS/PAGE, and analyzed by autoradiography. The kinase activity (KA) is indicated (Top). The amounts of IKKβ and IKKβ-KM were determined by immunoblotting with anti-Flag and anti-HA mAbs (Middle). The amounts of GST-fusion proteins were assessed by Coomassie Blue (CB) staining (Bottom). Right arrows mark the positions of each protein. (B) NIK enhances phosphorylation of IκBs by IKKβ. 293 cells were transiently transfected with expression vectors for HA-IKKβ, Flag-NIK, and/or NIK. HA-IKKβ or Flag-NIK was immunoprecipitated and incubated with GST-IκBα(1–100), GST-IκBβ(1–120), or GST-IκBɛ(1–61) in the presence of [γ-32P]ATP. The kinase activity (KA) is indicated (Upper). The amounts of HA-IKKβ and Flag-NIK were determined by immunoblotting with anti-Flag and anti-HA mAbs (Lower). The positions of each protein are indicated at the right. (C) NIK enhances phosphorylation of IκBɛ by IKKα. 293 cells were transiently transfected with expression vectors for FlagIKKα, Flag-IKKα-KM, and/or NIK. Flag-tagged proteins were immunoprecipitated, and in vitro phosphorylation of GST-IκBɛ(1–61) or GST-IκBɛ(1–61AA) was performed as in A. The positions of phosphorylated IKKα and GST-IκBɛ are indicated (Right).
A previous study demonstrated that NIK stimulates the kinase activity of IKKα to IκBα (39). To test the effect of NIK on the kinase activity of IKKβ, we transfected 293 cells with expression vectors encoding HA-IKKβ along with NIK. The expressed IKKβ was precipitated with anti-HA mAb and subjected to in vitro phosphorylation assays. The cotransfected NIK markedly enhanced the phosphorylation of GST-IκBα by IKKβ (Fig. 3B). Notably, the coexpression of NIK induced phosphorylation of GST-IκBɛ(1–61) by IKKβ (Fig. 3B). GST-IκBɛ(1–61) (S18A, S22A; designated as 1–61AA), in which both Ser-18 and Ser-22 were mutated to alanines, was not phosphorylated under the same conditions (data not shown). We next examined whether IKKα could also phosphorylate GST-IκBɛ(1–61) when coexpressed with NIK. As shown in Fig. 3C, NIK stimulated IKKα to phosphorylate GST-IκBɛ(1–61) but not GST-IκBɛ(1–61AA). These results indicated that NIK activates IKKβ as well as IKKα and that both IKKα and IKKβ can specifically phosphorylate the critical serine residues of IκBα, IκBβ, and IκBɛ for their degradation. The kinase activity of IKKs to IκBβ and IκBɛ seems to be weaker than that to IκBα, which could explain the slower kinetics of degradation of IκBβ and IκBɛ (7).
NF-κB Activation by MEKK1 Is Mediated by IKKα and IKKβ.
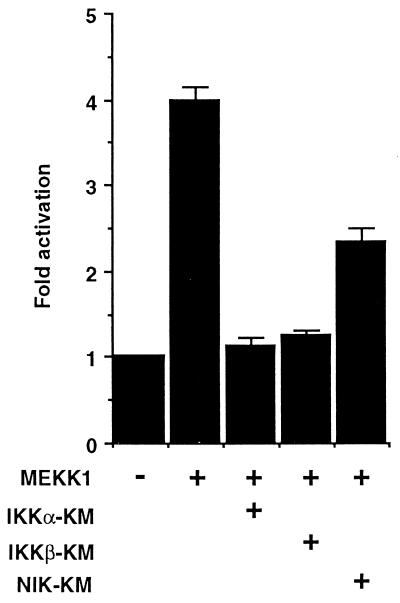
NF-κB activation by members of the TNF-R superfamily is mediated by TRAF2, -5, or -6 and their interacting kinase NIK (18, 27–37). On the other hand, these receptors and TRAFs also activate the JNK/SAPK pathway, which is mediated by MEKK1 (17, 18). MEKK1 has been also shown to be involved in TNF-induced NF-κB activation, but the precise mechanism of this pathway remains undefined (14–16). We then examined the contribution of IKKβ and IKKα to the MEKK1-mediated NF-κB activation. We first tested the effect of a catalytically inactive mutant of IKKα or IKKβ on the MEKK1-induced NF-κB activation using reporter assays (Fig. 4). Coexpressed IKKβ-KM or IKKα-KM inhibited the reporter gene activity elicited by MEKK1, indicating that both IKKα and IKKβ are involved in the MEKK1-mediated NF-κB activation. We further examined the contribution of NIK to MEKK1-mediated NF-κB activation. As also shown in Fig. 4, a kinase inactive mutant of NIK (NIK-KM) partially inhibited MEKK1-induced reporter gene activity, suggesting some contribution of NIK to this pathway.
Figure 4.
Effect of IKKβ-KM, IKKα-KM, or NIK-KM on MEKK1-induced NF-κB activation. 293 cells were transiently transfected with 100 ng of 3xκB-L and 0.5 μg each of the indicated expression vectors. The NF-κB reporter assays were performed as in Fig. 2. Data represent one of three experiments with similar results.
MEKK1 and NIK Differentially Activate IKKα and IKKβ.
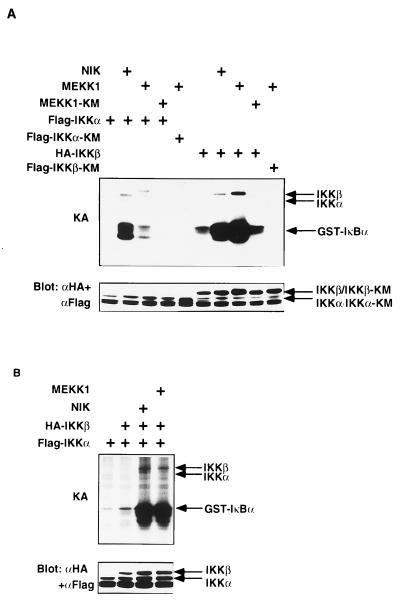
To further characterize the MEKK1-mediated NF-κB activation pathway, we examined the effect of MEKK1 on the kinase activity of IKKα and IKKβ. As shown in Fig. 5A, coexpression of MEKK1, but not MEKK1-KM, markedly enhanced the phosphorylation of GST-IκBα by IKKβ, whereas the IκBα phosphorylation by MEKK1-stimulated IKKα was marginal. In contrast, coexpression of NIK markedly enhanced the kinase activity of both IKKα and IKKβ, and comparable levels of IκBα phosphorylation were observed with NIK-activated IKKα and IKKβ. An apparently greater extent of activation of IKKα than IKKβ by NIK (16.5-fold versus 4.0-fold) is consistent with previous studies (39, 41). These results indicated that MEKK1 preferentially activates IKKβ, whereas NIK efficiently activates both IKKα and IKKβ to phosphorylate IκBα.
Figure 5.
Regulation of IKKα and IKKβ activities by MEKK1 and NIK. (A) Effect of NIK and MEKK1 on IκBα phosphorylation by IKKα or IKKβ. 293 cells were transiently transfected with Flag-IKKα, Flag-IKKα-KM, HA-IKKβ, or Flag-IKKβ-KM along with NIK, MEKK1, or MEKK1-KM. IKKα or IKKβ were immunoprecipitated, and in vitro phosphorylation of GST-IκBα(1–100) was performed as in Fig. 3. The kinase activity (KA) is indicated (Upper). The amounts of IKKα and IKKβ were determined by immunoblotting with anti-Flag and anti-HA mAbs (Lower). The positions of each protein are indicated (Right). (B) Both NIK and MEKK1 activate IKKαβ heterodimer. 293 cells were transiently transfected with expression vectors for Flag-IKKα and HA-IKKβ along with NIK or MEKK1. IKKα was precipitated with anti-Flag mAb, and in vitro phosphorylation of GST-IκBα was performed as in Fig. 3. The kinase activity (KA) is indicated (Upper). The amounts of IKKα and IKKβ were determined by immunoblotting with anti-Flag and anti-HA mAbs (Lower). The positions of each protein are indicated (Right).
Given that overexpressed IKKα or IKKβ most likely forms homodimers, we next examined the effect of NIK or MEKK1 on the kinase activity of IKKαβ heterodimer, which can normally exist in cells (40–42). To form the IKKαβ heterodimer, we transfected Flag-IKKα with or without HA-IKKβ. When IKKα alone was expressed, the immunoprecipitates with anti-Flag mAb phosphorylated GST-IκBα very weakly. In contrast, when Flag-IKKα and HA-IKKβ were coexpressed, phosphorylation of GST-IκBα by the anti-Flag immunoprecipitate was substantially enhanced (Fig. 5B), suggesting the coprecipitation of IKKβ with IKKα. In this condition, additional coexpression of either NIK or MEKK1 markedly enhanced the phosphorylation of GST-IκBα, indicating that both NIK and MEKK1 can activate the IKKαβ heterodimer comparably. Taken together, these results suggest that MEKK1 can activate the IKK complex as potently as NIK but in a different manner with a preferential activation of IKKβ. Because NIK has been demonstrated to interact with both IKKα and IKKβ directly (39, 41), the partial inhibition of MEKK1-induced reporter gene activity by NIK-KM (Fig. 4) seems to result from competitive inhibition of MEKK1-mediated activation of IKKβ by overexpressed NIK-KM, rather than a direct contribution of NIK as the downstream kinase of MEKK1. The less efficiency of MEKK1 compared with NIK to activate IKKα also supports this notion and suggests that MEKK1 can activate the IKK complex independently of NIK. In our preliminary experiments, MEKK1 appears not to interact with IKKα or IKKβ directly. However, MEKK1 has been identified to be a component of the large IKK component (42). Therefore, a putative downstream kinase of MEKK1 for IKKβ activation may be involved in the IKK complex, which remains to be identified in the future study.
In the present study, we characterized for the first time the molecular mechanism of the MEKK1-mediated NF-κB activation and found a qualitative difference in the MEKK1- and NIK-mediated NF-κB activation pathways. Our present data are consistent with previous findings that MEKK1 was present in the IKK complex (42) and exogenously added MEKK1 stimulated kinase activity of the IKK complex in vitro (14). Various stresses, including UV light, protein synthesis inhibitor, and hyperosmolarity shock, activate both NF-κB and JNK/SAPK pathways. Unlike the TNF case, JNK/SAPK activation by these stresses is not blocked by a dominant negative form of TRAF2, suggesting that this pathway is independent of TRAF and NIK (48). It has been known that these stresses can activate members of the MAPKKK family including MEKK1 (4). Because the activation of MEKK1 by these stresses is independent of TRAF, the presently revealed MEKK1-mediated IKK activation pathway could play a crucial role in NF-κB activation by these stresses. It remains to be determined whether pathways from all stimuli finally converge on IKKα and/or IKKβ, or stimulate other kinases such as p90rsk1 (49), for the phosphorylation of IκBs.
Acknowledgments
We thank Drs. Chikao Morimoto, Toshiki Watanabe, Hitoshi Kikutani, Carl F. Ware, David Wallach, Kyoko Yokota, Masaki Kashiwada, Shigeo Ohno, Eisuke Nishida, and Hisaya Akiba for gifts of plasmids. This work was supported by grants from the Ministry of Education, Science, and Culture, the Ministry of Health, Japan, and Uehara Memorial Foundation.
ABBREVIATIONS
- GST
glutathione S-transferase
- HA
hemagglutinin
- IKK
IκB kinase
- IL
interleukin
- JNK
c-Jun N-terminal kinase
- MAPKKK
MAP kinase kinase kinase
- MEKK1
mitogen-activated protein kinase/ERK kinase kinase-1
- NIK
NF-κB-inducing kinase
- SAPK
stress-activated protein kinase
- TNF
tumor necrosis factor
- TRAF
tumor necrosis factor receptor-associated factor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF026524).
References
- 1.Verma I M, Steveson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 4.Kyriakis J M, Avruch J. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin A S. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 7.Whiteside S T, Epinat J-C, Rice N R, Israel A. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteside T, Ernst M K, LeBail O, Laurent-Winter C, Rice N, Israel A. Mol Cell Biol. 1995;15:5339–5345. doi: 10.1128/mcb.15.10.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traencker E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlicht U. Science. 1995;267:1485–1491. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 12.DiDonato J A, Mercurio F, Rosette C, Wu-li J, Suyang H, Ghosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinsey T A, Brockman J A, Scherer D C, Al-Murrani S W, Green P L, Ballard D W. Mol Cell Biol. 1996;16:2083–2090. doi: 10.1128/mcb.16.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee F S, Hagler J, Chen Z J, Maniatis T. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 15.Hirano M, Osada S, Aoki T, Hirai S, Hosaka M, Inoue J-I, Ohno S. J Biol Chem. 1996;271:13234–13238. doi: 10.1074/jbc.271.22.13234. [DOI] [PubMed] [Google Scholar]
- 16.Meyer C F, Wang X, Chang C, Templeton D, Tan T-H. J Biol Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z-G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 18.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 20.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Nature (London) 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 21.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1996;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 22.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 23.Cheng G, Cleary A M, Ye Z-S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 24.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C F, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 25.Gedrich R W, Gilfillan M C, Duckett C S, Van Dongen J L, Thompson C B. J Biol Chem. 1996;271:12852–12858. doi: 10.1074/jbc.271.22.12852. [DOI] [PubMed] [Google Scholar]
- 26.Regnier C H, Tomasetto C, Moog-Lutz C, Chenard M-P, Wendling C, Basset P, Rio M-C. J Biol Chem. 1995;270:25715–25721. doi: 10.1074/jbc.270.43.25715. [DOI] [PubMed] [Google Scholar]
- 27.Hsu H, Shu H-B, Pan M-G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 28.Nakano H, Oshima H, Chung W, Williams-Abbot L, Ware C F, Yagita H, Okumura K. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 30.Aizawa S, Nakano H, Ishida T, Horie R, Nagai M, Ito K, Yagita H, Okumura K, Inoue J-I, Watanabe T. J Biol Chem. 1997;272:1–4. doi: 10.1074/jbc.272.4.2042. [DOI] [PubMed] [Google Scholar]
- 31.Ishida T, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, Yamamoto T, Inoue J-I. Proc Natl Acad Sci USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, Yamamoto T, Inoue J-I. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 33.Hsu H, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 34.Marsters S A, Ayres T M, Skubatch M, Gray C L, Rothe M, Askenazi A. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 35.Chinnaiyan A M, O’Rourke K, Yu G-L, Lyons R H, Garg M, Duan R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 36.Rothe M, Sarma V, Dixit V M, Goeddel D V. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 37.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 38.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 39.Regnier C H, Song H Y, Gao X, Goeddel D, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 40.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 41.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 42.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J W, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 43.Connelly M A, Marcu K B. Cell Mol Biol Res. 1995;41:537–549. [PubMed] [Google Scholar]
- 44.Kobata T, Jacquot S, Kozlowski S, Agematsu K, Schlossman S F, Morimoto C. Proc Natl Acad Sci USA. 1995;92:11249–11253. doi: 10.1073/pnas.92.24.11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inui S, Kaisho T, Kikutani H, Stamenkovic H, Seed B, Clark E A, Kishimoto T. Eur J Immunol. 1990;20:1747–1753. doi: 10.1002/eji.1830200819. [DOI] [PubMed] [Google Scholar]
- 46.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 47.Nakano H, Ohno H, Saito T. Mol Cell Biol. 1994;14:1213–1219. doi: 10.1128/mcb.14.2.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natoli G, Costanzo A, Ianni A, Templeton D J, Woodgett J R, Balsano C, Levrero M. Science. 1997;275:200–203. doi: 10.1126/science.275.5297.200. [DOI] [PubMed] [Google Scholar]
- 49.Schouten G J, Vertegaal A C O, Whiteside S T, Israel A, Toebes M, Dorsman J C, van der Eb A J, Zanteman A. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]