Abstract
Protein splicing involves the self-catalyzed excision of protein splicing elements, or inteins, from flanking polypeptide sequences, or exteins, leading to the formation of new proteins in which the exteins are linked directly by a peptide bond. To study the enzymology of this interesting process we have expressed and purified N- and C-terminal segments of the Mycobacterium tuberculosis RecA intein, each ≈100 amino acids long, fused to appropriate exteins. These fragments were reconstituted into a functional protein splicing element by renaturation from 6 M urea. When renaturation was carried out in the absence of thiols, the reconstituted splicing element accumulated as an inactive disulfide-linked complex of the two intein fragments, which could be induced to undergo protein splicing by reduction of the disulfide bond. This provided a useful tool for separately investigating the requirements for the reconstitution of the intein fragments to yield a functional protein splicing element and for the protein splicing process per se. For example, the pH dependence of these processes was quite different, with reconstitution being most efficient at pH 8.5 and splicing most rapid at pH 7.0. The availability of such an in vitro protein splicing system opens the way for the exploration of intein structure and the unusual enzymology of protein splicing. In addition, this trans-splicing system is a potential protein ligase that can link any two polypeptides fused to the N- and C-terminal intein segments.
Protein splicing is an unusual process by which the flow of information from a gene to its protein product is modulated posttranslationally so as to yield two functionally unrelated proteins. It involves the precise, self-catalyzed excision of an intervening polypeptide sequence, the intein, from an inactive precursor protein with the concomitant joining of the flanking sequences, the exteins, to produce a new functional protein (Fig. 1). All information and catalytic groups required for protein splicing reside in the intein and the two flanking amino acids. With the elucidation of the chemical mechanism of protein splicing (for review see refs. 1 and 2), it has become clear that inteins constitute a class of highly unusual enzymes: (i) they catalyze three mechanistically distinct reactions, two of which, acyl rearrangement of a peptide bond adjacent to cysteine or serine (3, 4) and cyclization of asparagine coupled to peptide bond cleavage (5, 6), have also been found to occur naturally in polypeptides, but only under extreme conditions or at very slow rates; (ii) they act on amino acid residues at their own N and C termini, so that the intein enzymes are also their own substrate, analogous to the role of catalytic RNA in the self-splicing of group I introns (7); and (iii) they catalyze a transesterification reaction between their N and C termini, and their catalytic center therefore comprises both extremities of the polypeptide chain, a situation that is rarely encountered in conventional enzymes and suggests an unusual protein structure. Even greater complexity is added by the fact that most mature inteins are also highly specific DNA homing endonucleases (8).
Figure 1.
Schematic illustration of the protein splicing process. The heavy lines, solid or hatched, symbolize polypeptide chains and the light dotted lines represent chain–chain interactions that stabilize the separate domains involved in protein splicing and homing endonuclease activity. The reactive nucleophilic groups at the splice junctions that participate in protein splicing are shown as thiols, as in the M. tuberculosis RecA intein, but could also be hydroxyls.
An important advance in unraveling the complexities of intein function has been the recent discovery that the homing endonuclease domain of the intein, which accounts for about one-half of its amino acid sequence, can be deleted without affecting the process of protein splicing (9, 10), suggesting that the active centers for protein splicing and homing endonuclease activity reside in separate structural domains, as illustrated schematically in Fig. 1. This made possible the design not only of minimal inteins but also of trans-splicing systems in which N-terminal and C-terminal fragments of the intein were coexpressed as separate polypeptides fused to exteins and found to undergo protein splicing in vivo (10). In this paper, we describe the separate expression of such intein fragments followed by purification and reconstitution to yield a complex that can undergo efficient protein splicing in vitro. The availability of such an in vitro protein splicing system opens the way for the exploration of intein structure and the unusual enzymology of protein splicing.
MATERIALS AND METHODS
Plasmid Constructs.
The plasmids used in this work were all derivatives of plasmid pMU1B, in which the coding sequence of the Mycobacterium tuberculosis RecA intein (U) was inserted in-frame between the coding sequences for Escherichia coli maltose binding protein (MBP) and the Bacillus circulans chitin binding domain (10). The construction of plasmid pMU2s/sΔ6 was described earlier (10). This plasmid encodes a chimeric protein (MUNΔ) consisting of an N-terminal MBP fused to the 105 N-terminal amino acids of U (UNΔ), followed by the C-terminal sequence Arg-Gly-Glu-Phe. Plasmid pTrcUH4 was constructed by inserting a 0.46-kb PstI-HindIII fragment from plasmid pMU2H (10) into the PstI and HindIII sites of the polylinker region of pTrcHis2A (Invitrogen). As a result, the inducible trc promoter of pTrcUH4 drives the expression of a fusion protein (UCΔH) with the N-terminal sequence formyl-Met-Asp-Pro-Ser-Ser-Arg-Ser, followed by the 107 C-terminal amino acids of U fused to a 49-amino acid polypeptide with a C-terminal hexahistidine sequence (His-tag).
Protein Expression and Affinity Purification.
E. coli DH5α (F− deoR recA1 endA1 supE44 hsdR17 (rK-mK+) relA1 thi-1 gyrA96 λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169) carrying pMU2s/sΔ6 or pTrcUH4 was grown at 37°C with shaking in Luria–Bertani medium with 100 μg/ml ampicillin overnight. The overnight culture (100 ml) was added to 10 l of Luria–Bertani medium in a New Brunswick Scientific MagnaFerm fermentor and grown at 37°C with agitation to a culture density (A600) of 0.5. Isopropyl β-d-thiogalactoside was then added to a final concentration of 0.4 mM to induce the synthesis of the intein fragment and growth was continued for 7 hr at 25°C (A600 = 1.0–1.2). The cells were harvested in a Sorvall RC5 centrifuge at 6,000 × g with a continuous flow collection apparatus and stored frozen at −70°C.
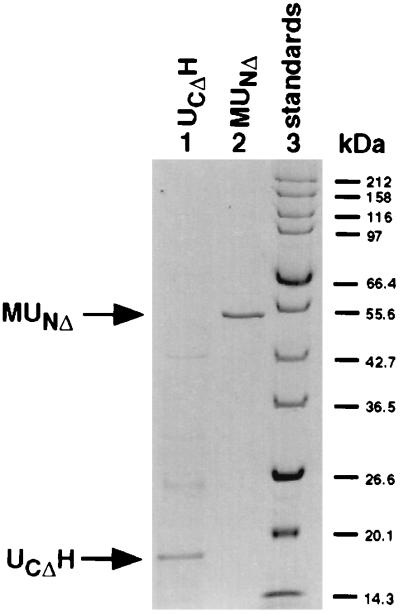
All subsequent steps were performed at 4°C. Cells were disrupted by passage through a French pressure cell after resuspension in 5 ml of buffer C (20 mM Tris⋅HCl, pH 7.5/0.1 M NaCl) for MUNΔ or 10 ml of buffer N (20 mM Tris⋅HCl, pH 7.5/0.5 M NaCl/1 mM EDTA, pH 7.5/8 M urea) for UCΔH, then centrifuged at 17,000 × g for 30 min. The MUNΔ supernatant was passed through a 0.5-ml amylose column (New England Biolabs), preequilibrated with buffer C. The column was washed with 15 ml of buffer C to elute unbound protein. All proteins containing the MBP domain were eluted with 1 ml of buffer C supplemented with 10 mM maltose. The UCΔH supernatant was passed batchwise through a Talon metal affinity resin spin column (CLONTECH), preequilibrated with 2 × 1 ml of buffer N. The column was washed with 2 × 1 ml of buffer N supplemented with 10 mM imidazole and eluted with 1 ml buffer N plus 100 mM imidazole. When analyzed by SDS/PAGE, purified MUNΔ and UCΔH each yielded one major band with an electrophoretic mobility consistent with its predicted molecular mass, 55 and 18 kDa, respectively (Fig. 2).
Figure 2.
Purification of the fusion proteins MUNΔ and UCΔH. MUNΔ and UCΔH were isolated as described in Materials and Methods. Approximately 5 μg UCΔH (lane 1), 0.2 μg MUNΔ (lane 2), and broad-range protein marker (New England Biolabs, lane 3) were subjected to SDS/PAGE by using the standard protocol (13). The gel was stained with Coomassie blue.
Reconstitution and Splicing Procedure.
MUNΔ and UCΔH were mixed in a volume ratio of 1:3, giving a final urea concentration of 6 M, and incubated at 4°C for 1 hr. The mixture was transferred to SpectraPor 8000 MWCO dialysis tubing (Fisher Scientific) and dialyzed against buffer N for 20 min, then against three changes of buffer O (20 mM Tris⋅HCl, pH 7.5/0.5 M NaCl/1 mM EDTA, pH 7.5) for 20 min each. Because self-aggregation of UCΔH competes with its interaction with MUNΔ as the urea concentration is lowered, MUNΔ and UCΔH were used in a 1:5 molar ratio so as to compensate for the loss of some UCΔH. After removal from the dialysis tubing, the mixture was kept at 4°C. Splicing was induced by the addition of DL-1,4-dithiothreitol (DTT) to 1.25 mM and incubation at 25°C. The splicing reaction was terminated by adding 1 volume New England Biolabs 3× SDS sample buffer to 2 volumes sample.
Assay of Reconstitution and Protein Splicing.
Reconstitution of MUNΔ and UCΔH and protein splicing were assayed by Western blot analysis. SDS/PAGE used precast 10–20% gradient Tris⋅glycine gels (Bio-Rad or Owl Scientific, Woburn, MA) and prestained protein markers (New England Biolabs). Gels were blotted onto nitrocellulose membranes (Hoefer Pharmacia Biotech) at 36 V overnight. The blot was first incubated for 1 hr in blocking buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/5% skim milk powder/0.25% gelatin/0.1% BSA), then with 0.2 μg/ml monoclonal mouse anti-Arg-Gly-Ser-His-His-His-His antibody (Qiagen, Chatsworth, CA) in dilution buffer (10 mM Hepes, pH 7.4/0.5 M NaCl/0.2% Tween-20/1% BSA) for 40 min, followed by washing for 4 × 15 min in wash buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.05% Tween-20). The blot was then incubated for 40 min in dilution buffer containing 0.3 μg/ml goat anti-mouse IgG conjugated with alkaline-phosphatase (Pierce) and washed again for 4 × 15 min in wash buffer. Immobilized alkaline phosphatase activity was detected by using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate tablets (Sigma). The reaction was allowed to proceed for ≈5 min and was terminated by washing the blots with water and incubation with 0.18 M trichloroacetic acid for 5 min. The Western blots were stored in water, scanned with a Supervista S12 scanner (Umax Data Systems), and analyzed densitometrically by using the National Institutes of Health image 1.60 program.
Protein Determination.
Protein concentration was estimated by either the Bradford (11) or the bichinchoninic acid method (12), with BSA as a standard, using the reagents and protocols supplied by Pierce, except that in the bichinchoninic acid assay the sample was not diluted before being added to the assay reagent. Because neither of these color tests give absolute protein concentrations, the color values obtained with MUNΔ and UCΔH were correlated to the absorbance of these proteins at 280 nm in buffer N. The molar absorption coefficients for MUNΔ and UCΔH, 83,190 and 9,650 M−1 cm−1, respectively, were calculated from their amino acid composition by using the DNAstar (Madison, WI) protean program. On a molar basis, the color yield at 595 nm in the Bradford test was found to be 17× greater for MUNΔ than for UCΔH.
RESULTS
Reconstitution of an in Vitro Trans-Splicing System.
Our observation that fusion proteins containing truncated N- and C-terminal fragments of the M. tuberculosis RecA intein (UNΔ and UCΔ, respectively) could undergo splicing when coexpressed in the same cell (10) suggested that trans-splicing should also be possible in vitro, provided that the intein fragments can be reconstituted to form a properly folded catalytic center for protein splicing. To test this possibility, we used an indicator system for protein splicing similar to that described earlier (10), which employed MBP (M) as the extein fused to the N terminus of the RecA intein and a polypeptide terminated by a His-tag (H) as the C-extein fused to the C terminus of the RecA intein. This choice of fusion proteins, termed MUNΔ and UCΔH, respectively, allowed purification of the N-terminal intein fragment by affinity chromatography on amylose resin and that of the C-terminal fragment by immobilized metal affinity chromatography. The intein fragments in these fusion proteins corresponded to the 105 N-terminal and the 107 C-terminal amino acids of the RecA intein plus four and seven additional amino acid residues, respectively, which were introduced in the course of engineering the translation termination and initiation sites. Separate E. coli cultures were transformed with the plasmids pMU2s/sΔ6 and pTrcUH4, which served as templates for the synthesis of MUNΔ and UCΔH, respectively. The purification of MUNΔ by amylose column chromatography followed standard procedures and yielded the expected 55.5-kDa component on SDS/PAGE (Fig. 2). Purification of UCΔH involved the solubilization of the expressed material with 8 M urea followed by metal affinity chromatography and elution with imidazole in 8 M urea, yielding a product with the expected molecular mass (18 kDa) (Fig. 2). Elution from the metal affinity resin in the absence of denaturant yielded no detectable product (data not shown), suggesting that UCΔH formed insoluble aggregates that remained trapped in the affinity column. To avoid such unproductive aggregate formation, our procedure for the reassociation of MUNΔ and UCΔH involved mixing the two components in 6 M urea, followed by dialysis at 4°C to remove urea and effect renaturation. The mixture was then diluted into appropriate buffers at 25°C and DTT was added to initiate the splicing of any productive complexes that may have formed during the renaturation step.
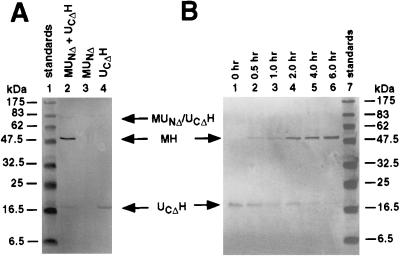
Protein splicing was assessed in terms of the amount of spliced exteins (MH) that could be detected upon SDS/PAGE of the incubation mixtures. Owing to the fact that UCΔH has a very low aromatic amino acid content and stains relatively weakly with Coomassie blue, the proteins separated by SDS/PAGE were usually blotted onto nitrocellulose membranes and visualized by Western blot analysis, using anti-His-tag antibody. The extent of protein splicing could be estimated by comparing the immunoreactivity of residual UCΔH precursor with that of the MH product, taking into consideration that the transfer of UCΔH to nitrocellulose membranes was somewhat less efficient than that of MH. As shown in Fig. 3A, spliced product was observed after renaturation of a mixture of MUNΔ and UCΔH, but not with MUNΔ or UCΔH alone. Examination of the time course of protein splicing, initiated by transferring the reconstitution mixtures to 25°C and adding DTT, showed that the reaction was nearly complete in 6 hr, with a half-time of 1–2 hr (Fig. 3B). This experiment also showed that, although a 5-fold excess of UCΔH was used in the reassociation reaction, the amount of UCΔH recovered after dialysis was roughly equivalent to the amount of spliced product formed subsequently. A possible explanation for the loss of UCΔH during the reconstitution procedure is that only UCΔH that formed a productive complex with MUNΔ could be recovered from the dialysis bag in soluble form.
Figure 3.
Protein splicing by reconstitution of fusion proteins containing N- and C-terminal fragments of the Mtu RecA intein. (A) Control experiment. Western blot analysis of MUNΔ and UCΔH (lane 2), MUNΔ alone (lane 3), and UCΔH alone (lane 4) carried through the renaturation and protein splicing protocols described in Materials and Methods. (B) Time course. Western blot analysis of reconstituted mixtures of MUNΔ and UCΔH allowed to undergo protein splicing for the times indicated. Western blot analysis was done as described in Materials and Methods using anti-His-tag antibody, which selectively reveal the unspliced precursor, UCΔH, and the spliced product, MH.
Separation of the Reconstitution and Splicing Steps.
Our standard conditions for reconstitution of the trans-splicing system from MUNΔ and UCΔH involved removal of urea by dialysis at 4°C in the absence of added thiols. When such a reconstitution mixture was subjected to SDS/PAGE by using the standard protocol (13), followed by Western blot analysis with anti-His-tag antibody, no spliced product could be observed and all of the His-tag could be accounted for as UCΔH (Fig. 4A, lane 3; see also Fig. 3B, lane 1). On the other hand, when SDS/PAGE was carried out without prior DTT-treatment of the samples, much if not all of the UCΔH migrated with an electrophoretic mobility corresponding to that predicted for an 80-kDa protein (Fig. 4A, lane 2; see also Fig. 4B). Upon incubation at 25°C of the reconstitution mixture in the presence of various concentrations of DTT, followed by SDS/PAGE and Western blot analysis, the slowly migrating component was seen to be converted to spliced product in a concentration-dependent manner (Fig. 4B). These observations suggested that, in the course of reconstitution in the absence of added thiols, MUNΔ and UCΔH formed a disulfide-linked adduct (predicted molecular mass, 73 kDa), which could either be dissipated into MUNΔ and UCΔH when exposed to DTT under denaturing conditions (Fig. 4A, lane 3) or undergo splicing to MH when incubated with DTT at 25°C and pH 7.5 (Fig. 4B). The somewhat lower than expected electrophoretic mobility of the putative disulfide-linked MUNΔ/UCΔH dimer may be a consequence of disulfide crosslinking and is reminiscent of the abnormally low electrophoretic mobility of the branched splicing intermediate observed earlier (14, 15).
Figure 4.
MUNΔ/UCΔH dimer formation in the course of reconstitution and the effect of DTT on its conversion to spliced product. (A) Dissociation of MUNΔ/UCΔH dimer formed during the reconstitution of MUNΔ and UCΔH to yield UCΔH when treated with DTT in SDS prior to electrophoresis. MUNΔ and UCΔH were reconstituted in the absence of thiols, subjected to SDS/PAGE without (lane 2) or with DTT (lane 3) in the sample buffer, and determined by Western blot analysis using anti-His-tag antibody. (B) Dependence of protein splicing on DTT concentration. Mixtures of MUNΔ and UCΔH, reconstituted in the absence of DTT, were supplemented with DTT at the concentrations indicated and incubated at 25°C and pH 7.5 for 6 hr, followed by SDS/PAGE without additional thiol in the sample buffer and Western blot analysis with anti-His-tag antibody.
The observation that the reconstituted MUNΔ/UCΔH complex could be trapped as a disulfide-linked dimer that can undergo protein splicing only after reduction by DTT provided a useful tool for separately investigating the requirements and properties of the reconstitution of the intein fragments to yield a functional trans-splicing system and of the protein splicing process per se.
Properties of the Reconstitution and Trans-Splicing Reactions.
The initial urea concentration for the reconstitution of the intein fragments was 6 M, needed to keep UCΔH in solution after elution from the metal affinity resin. However, significant reconstitution was seen even when MUNΔ was added to UCΔH after most urea had been removed by dialysis (data not shown). The standard reconstitution reaction was carried out in the absence of thiol reducing agents, but the addition of DTT or Tris(2-carboxyethyl)phosphine (TCEP) had no effect on the efficiency of subsequent protein splicing (data not shown), indicating that the disulfide-linked MUNΔ/UCΔH dimer is not an obligatory intermediate in the formation of a functional protein splicing precursor. When reconstitution was allowed to proceed at pH 6.5, 7.5, or 8.5 and measured in terms of the ability to undergo subsequent protein splicing at pH 7.5, the best yield of functional splicing precursor was obtained at pH 8.5 (Fig. 5).
Figure 5.
Dependence of protein splicing and intein reassociation on pH. Experiment 1 (• and ○): Mixtures of MUNΔ and UCΔH were reconstituted under standard conditions at pH 7.5 and diluted 2-fold into 200 mM buffers at the pH values indicated, by using either phosphate buffers (•) or Tris buffers (○). Protein splicing was initiated by the addition of DTT to 1.25 mM, was allowed to proceed at 25°C for 6 hr, and was assayed by Western blot analysis with anti-His-tag antibody. The relative amounts of MH were estimated by densitometry of the Western blot. Experiment 2 (▪ and □): Mixtures of MUNΔ and UCΔH were reconstituted at the pH values indicated, by using either phosphate (▪) or Tris buffers (□). Protein splicing was allowed to proceed under standard conditions for 6 hr and was assayed as in experiment 1. The densitometry values for reassociation were normalized to those for splicing by using the point for Tris⋅HCl, pH 7.5, as a common reference point.
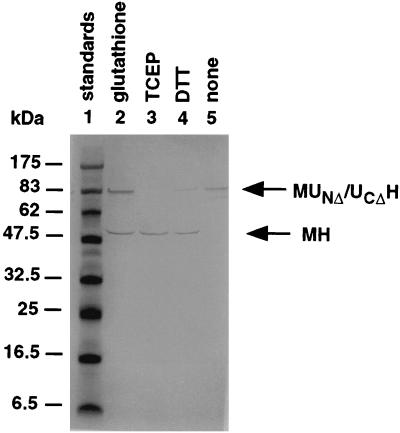
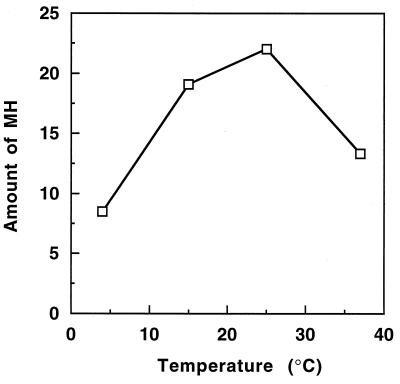
The optimal conditions for protein splicing were studied following activation of the disulfide-linked MUNΔ/UCΔH dimer with disulfide reducing agents. As shown in Fig. 6, either thiols or phosphines could serve as reducing agents, with TCEP being the most effective. Protein splicing was relatively slow at 4°C, the temperature at which reconstitution was carried out, and was optimal at 25°C, with some loss in splicing efficiency at 37°C or higher temperatures (Fig. 7). Optimal protein splicing was observed at pH 7.0, with very little splicing at pH 8.0 and above (Fig. 5). Low concentrations of urea were inhibitory to protein splicing, with ≈50% inhibition at 1 M urea and no detectable splicing above 2 M urea (data not shown).
Figure 6.
Effect of various reducing agents on the induction of protein splicing. Mixtures of MUNΔ and UCΔH, reconstituted in the absence of DTT, were supplemented with 1.25 mM glutathione (lane 2), 1.25 mM TCEP (lane 3), 1.25 mM DTT (lane 4), or no reducing agent (lane 5) and incubated at 25°C and pH 7.5 for 6 hr, followed by SDS/PAGE without additional reducing agent in the sample buffer and Western blot analysis with anti-His-tag antibody.
Figure 7.
Effect of temperature on protein splicing. Mixtures of MUNΔ and UCΔH were reconstituted under standard conditions at 4°C, supplemented with 1.25 mM DTT to induce splicing and incubated for 6 hr at the temperatures indicated. Protein splicing was assayed by Western blot analysis with anti-His-tag antibody, and the relative amounts of MH were estimated by densitometry of the Western blot.
DISCUSSION
This paper describes an in vitro experimental system for studying the enzymology of protein splicing and the structural organization of inteins. Several in vitro protein splicing systems have already been developed. The first was the classical work of Xu and coworkers (14), which involved the Psp pol1 intein from an extreme thermophile, which could be expressed as an unspliced fusion protein at low temperature in E. coli, purified and induced to undergo splicing by raising the temperature. This experimental system was instrumental in elucidating the mechanism of the protein splicing reaction (15–17), but may not lend itself as well to structural studies at ambient temperature because the structure of the Psp pol1 intein evolved to function optimally at ≈100°C. Another in vitro protein splicing system was based on a slow-splicing mutant form of the mesophilic Sce VMA intein (18), but the replacement of two of its catalytic amino acid residues does not make it an ideal subject for the study of intein catalysis and structure. A third in vitro splicing system, also involving the Sce VMA intein, was developed by Anraku and coworkers (19) and is based on the fact that expression of fusion proteins containing this intein in E. coli led to the accumulation of unspliced precursor as inclusion bodies, which could be solubilized in 6 M guanidine hydrochloride and made to undergo protein splicing by dialysis. Preliminary mechanistic studies by using this experimental system demonstrated that protein splicing is an intramolecular process, which is resistant to protease inhibitors (20); however, a serious limitation of these studies is that the refolding process and protein splicing were not clearly separated.
Our experimental system has several attributes that make it eminently suitable for studying both mechanistic and structural aspects of protein splicing. It involves the reconstitution of the protein splicing active center from two complementary fractions that correspond to N- and C-terminal fragments of the intein, UNΔ and UCΔ, respectively, to yield a functional protein splicing element. The intein fragments were ≈100 amino acids long, and together they comprised less than half the sequence of the 454-amino acid Mtu RecA intein, with the entire homing endonuclease domain deleted. An important feature of our system is that the reconstitution of the intein fragments into a functional protein splicing element can be separated from the splicing process itself by carrying out the reconstitution reaction under conditions of temperature, pH, and reductant levels at which protein splicing is suboptimal or does not occur. Especially striking was the observation that, in the absence of disulfide reducing agents, the reconstituted intein accumulated as a stable, disulfide-linked dimer, which had to be reduced by thiols or phosphines before it could undergo protein splicing (Figs. 4 and 6). In addition, at pH 8.5, which is optimal for reconstitution, protein splicing was almost completely inhibited (Fig. 5), and at 4°C, the temperature at which reconstitution was routinely carried out, protein splicing occurred at only 40% of the optimal rate (Fig. 7).
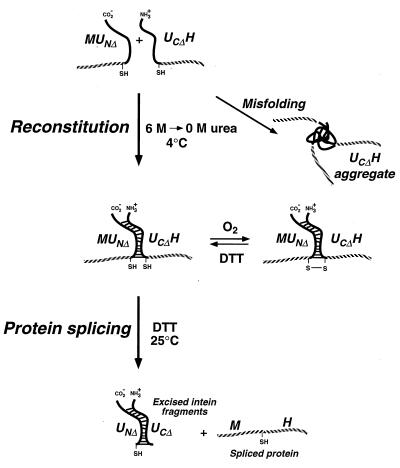
Although we have not yet studied the reconstitution of N- and C-terminal fragments of the Mtu RecA intein in detail, our preliminary results provide some interesting structural insights. When synthesized in E. coli, MUNΔ behaved like a typical soluble protein and could readily be purified by affinity chromatography on amylose resin. In contrast, synthesis of UCΔH in E. coli led to the formation of inclusion bodies, which could be solubilized with 8 M urea and bound to a metal chelate affinity resin but could be successfully eluted from the resin only in the presence of the denaturant, suggesting that even purified UCΔH has a tendency to precipitate. On the other hand, when urea was removed from UCΔH by dialysis in the presence of MUNΔ, a functional self-splicing precursor protein was obtained (Fig. 4A). This suggested that UCΔH alone has a tendency to misfold to yield nonfunctional aggregates but that the presence of MUNΔ prevents its misfolding. When MUNΔ and UCΔH were reconstituted in the absence of disulfide reducing agents, essentially all of the soluble UCΔH migrated with an electrophoretic mobility consistent with that expected of a disulfide-linked MUNΔ/UCΔH dimer. The disulfide bond must involve the cysteine residues at the splice junctions, which are the only thiols in MUNΔ and UCΔH, and the high efficiency at which a disulfide-linked MUNΔ/UCΔH dimer was formed by air oxidation suggest that these cysteines are situated in close proximity. This conclusion is consistent with the fact that the second step in protein splicing involves transesterification between these cysteines (17) and also with the crystal structures of the Sce VMA intein (21) and the homologous hedgehog autoprocessing domain (22). However, it should be noted that the reconstitution of functional inteins could also occur in the presence of DTT or TCEP, where disulfide bonds are not formed, and that the disulfide-linked dimer is therefore not an intermediate but a side-product in the reconstitution of the functional intein, as illustrated in Fig. 8.
Figure 8.
Diagram illustrating the reconstitution and splicing of fusion proteins containing N- and C-terminal fragments of the Mtu RecA intein.
Upon reassociation of MUNΔ and UCΔH in the absence of added thiols, protein splicing could be initiated by adding DTT or TCEP to reduce the disulfide-linked MUNΔ/UCΔH dimer, raising the temperature to 25°C, and—if necessary—adjusting the pH to 7.0. Whereas the reassociation of MUNΔ and UCΔH occurred most efficiently at high pH values, protein splicing was most rapid at pH 7.0 (Fig. 5). In that respect, the splicing of the Mtu RecA intein resembled that of the Psp pol1 intein, which also spliced poorly at high pH, with optimal splicing at ≈pH 6.0 (14). At 25°C and pH 7.5, the half-time of splicing of the reconstituted Mtu RecA intein was 1–2 hr, similar to that of the Psp pol1 intein (t1/2 = 2 hr) at 37°C and pH 7.5 (14). The splicing rate of the reconstituted Mtu RecA intein declined at higher temperatures (Fig. 7), perhaps owing to instability of the noncovalent MUNΔ/UCΔH complex. The sensitivity of the reconstituted complex to perturbation was also manifested in the inhibition of protein splicing by relatively low concentrations of urea (1–2 M).
Besides opening the way for detailed in vitro studies of protein splicing, the experimental system described in this paper is different in that protein splicing occurs in trans. The feasibility of trans protein splicing had already been demonstrated by our earlier in vivo experiments (10), but the availability of an in vitro trans-splicing system provides an opportunity for investigating the architecture of the intein catalytic site by studying the interaction of the N- and C-terminal intein fragments by biophysical and chemical means. Moreover, our results show that the N- and C-terminal intein fragments essentially constitute a polypeptide ligase system that allows the in vitro ligation of any two proteins fused to such fragments, which may find interesting applications in the field of protein engineering.
Acknowledgments
We thank Kaori Shingledecker for her help and advice. This paper is dedicated to Donald G. Comb, a pioneer in the protein splicing field, on the occasion of his 70th birthday. This work was supported by National Institute of General Medical Sciences Grant R01 GM55875 (H.P.) and by a Howard Hughes Medical Institute Predoctoral Fellowship (K.V.M.).
ABBREVIATIONS
- DTT
DL-1,4-dithiothreitol
- His-tag
hexahistidine sequence
- H
a 49-amino acid peptide with a C-terminal His-tag
- MBP or M
E. coli maltose-binding protein
- MH
spliced exteins
- MUNΔ
chimeric protein consisting of an N-terminal MBP fused to UNΔ
- TCEP
Tris(2-carboxyethyl)phosphine
- U
M. tuberculosis RecA intein
- UNΔ
the 105 N-terminal amino acids of U, followed by the C-terminal sequence Arg-Gly-Glu-Phe-COOH
- UCΔ
formyl-Met-Asp-Pro-Ser-Ser-Arg-Ser, followed by the 107 C-terminal amino acids of U
- UCΔH
chimeric protein consisting of N-terminal UCΔ fused to a 49-amino acid polypeptide with a C-terminal His-tag
References
- 1.Perler F, Xu M Q, Paulus H. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 2.Shao Y, Kent S B H. Chem Biol. 1997;4:187–194. doi: 10.1016/s1074-5521(97)90287-8. [DOI] [PubMed] [Google Scholar]
- 3.Elliott D F. Biochem J. 1952;50:542–550. doi: 10.1042/bj0500542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakakibara S, Shin K H, Hess G P. J Am Chem Soc. 1962;84:4921–4928. [Google Scholar]
- 5.Geiger T, Clarke S. J Biol Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 6.Voorter C E M, de Haard-Hoekman W A, Van den Oetelaar P J M, Bloemendal H, de Jong W W. J Biol Chem. 1988;263:19020–19023. [PubMed] [Google Scholar]
- 7.Cech T R. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 8.Gimble F S, Thorner J. Nature (London) 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- 9.Chong S, Xu M Q. J Biol Chem. 1997;272:15587–15590. doi: 10.1074/jbc.272.25.15587. [DOI] [PubMed] [Google Scholar]
- 10.Shingledecker K, Jiang S Q, Paulus H. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- 11.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Povenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Xu M-Q, Southworth M W, Mersha F B, Hornstra L J, Perler F B. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 15.Xu M-Q, Comb D G, Paulus H, Noren C J, Shao Y, Perler F B. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Xu M-Q, Paulus H. Biochemistry. 1995;34:10844–10850. doi: 10.1021/bi00034a017. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Xu M-Q, Paulus H. Biochemistry. 1996;35:3810–3815. doi: 10.1021/bi952592h. [DOI] [PubMed] [Google Scholar]
- 18.Chong S, Shao Y, Paulus H, Benner J, Perler F B, Xu M Q. J Biol Chem. 1996;271:22159–22168. doi: 10.1074/jbc.271.36.22159. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki M, Makino S-i, Matsuzawa H, Satow Y, Ohya Y, Anraku Y. Biochem Biophys Res Commun. 1996;222:827–832. doi: 10.1006/bbrc.1996.0826. [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki M, Satow Y, Ohya Y, Anraku Y. FEBS Lett. 1997;412:518–520. doi: 10.1016/s0014-5793(97)00850-8. [DOI] [PubMed] [Google Scholar]
- 21.Duan X, Gimble F S, Quiocho F A. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 22.Hall T M T, Porter J A, Young K E, Koonin E V, Beachy P E, Leahy D J. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]