Abstract
An abnormal pattern of DNA methylation occurs at specific genes in almost all neoplasms. The lack of high-throughput methods with high specificity and sensitivity to detect changes in DNA methylation has limited its application for clinical profiling. Here we overcome this limitation and present an improved method to identify methylated genes genome-wide by hybridizing a CpG island microarray with amplicons obtained by the methylated CpG island amplification technique (MCAM). We validated this method in three cancer cell lines and 15 primary colorectal tumors, resulting in the discovery of hundreds of new methylated genes in cancer. The sensitivity and specificity of the method to detect hypermethylated loci were 88% and 96%, respectively, according to validation by bisulfite-PCR. Unsupervised hierarchical clustering segregated the tumors into the expected subgroups based on CpG island methylator phenotype classification. In summary, MCAM is a suitable technique to discover methylated genes and to profile methylation changes in clinical samples in a high-throughput fashion.
In cancer, loss of expression of selected genes happens by either genetic mutation or epigenetic silencing. One of the main causes of epigenetic silencing in cancer is DNA methylation of cytosines in CG-rich regions (CpG islands) close to gene promoters. This happens by the enzymatic addition of methyl groups to CpG dinucleotides in an orchestrated reaction that involves DNA methyltransferases, methyl-binding domain proteins, and histone deacetylases (Herman and Baylin 2003; Laird 2005). While normal patterns of DNA methylation are important for genomic imprinting, X-chromosome inactivation, and to repress mobilization of repetitive elements, aberrant DNA methylation in cancer is associated with silencing of tumor-suppressor genes and genes involved in invasion, angiogenesis, and apoptosis (Sugimura and Ushijima 2000; Toyota and Issa 2005).
Much of the knowledge about aberrant DNA methylation in cancer came from genome-wide investigations. The use of techniques that test DNA methylation in an unbiased way, such as MCA (methylated CpG island amplification) and RLGS (restriction landmark genomic scanning), revealed that DNA methylation in cancer happens in a tissue-specific pattern (Costello et al. 2000) and also revealed the existence of coordinated hypermethylation of multiple genes in subsets of samples, a process termed CpG island methylator phenotype (Toyota et al. 1999a). Although efficient, both methods lack the high throughput required to study large sample collections for clinical and/or epidemiological purposes. Microarray chips containing promoter sequences filled this gap. Several platforms are available, with variation in genome representation and probe size (oligonucleotides, short DNA fragments, or BACs).
A major issue in methylation microarrays is the protocol for target preparation. Most of the published methods aim at selective enrichment for the methylated fraction of the genome, using either methylation-sensitive restriction enzymes (Yan et al. 2001; Lippman et al. 2004) or antibodies that recognize 5-methyl cytidine (Weber et al. 2005; Rauch et al. 2006). There are major problems with each published method. Methods that rely on frequent sites (HpaII/MspI) result in a high genome fraction to amplify (high complexity), which limits PCR efficiency and ends up favoring non-CpG island DNA. The sensitivity of antibody-based methods is undetermined and possibly low (in our experience). Finally, there are limited validation data of microarray results by a gold-standard technique (bisulfite-PCR).
In the present study, we introduce a new technique to detect DNA methylation in cancer by combining the simple and reliable MCA method with a CpG island microarray (MCAM, for methylated CpG island amplification microarray). This technique simultaneously reduces complexity and increases specificity by targeting methylated CpG islands before amplification. We find that MCAM provides reproducible results with a high validation rate, and demonstrate the use of MCAM to identify new methylated genes and altered molecular pathways in cancer cell lines, and also to classify clinical samples into distinctive clinical subgroups, for example, the ones determined by CIMP (CpG island methylator phenotype) in colorectal carcinomas.
Results
MCAM overview and procedure
In order to identify methylated targets in cancer, we generated PCR amplicons from three different cancer cell lines using the methylated CpG island amplification (MCA) protocol (Toyota et al. 1999b). Control amplicons were obtained from normal peripheral blood lymphocytes. MCA products from cancer cell lines were labeled with Cy5 dye, and Cy3 was used to label the control sample. Equimolar amounts of tumor and normal labeled amplicons were cohybridized to a microarray chip containing 12,192 CpG-island clones from the Sanger Institute (Heisler et al. 2005). Duplicate experiments were performed for each cell line, and they were averaged for data analysis. Done this way, hypermethylated genes in cancer were visualized as red spots in the microarray, and normalized log2 ratio values of ≥1.3 (equivalent to ∼2.5-fold tumor/normal signal intensity) were used as the cutoff for hypermethylation. This log2 ratio value was later supported by our validation experiments as the best cutoff to achieve optimal sensitivity and specificity. A schematic view of the method and a partial MCAM image are presented in Figure 1.
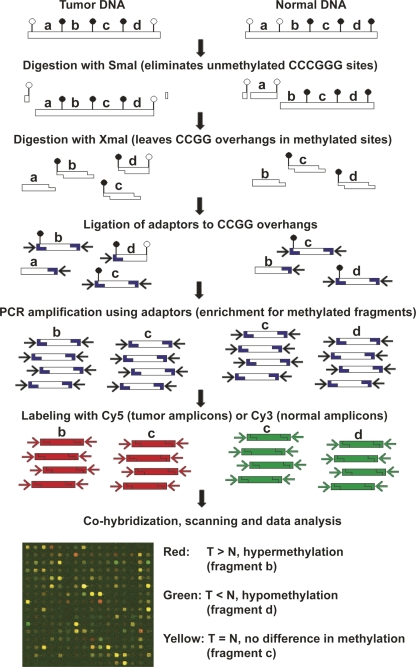
Figure 1.
Schematic diagram of the MCAM method. Enrichment for methylated DNA and reduction of genome complexity is achieved by serial digestion with SmaI (methylation sensitive) and XmaI (methylation insensitive) restriction enzymes, followed by ligation of adaptors and PCR amplification. The resulting amplicons, representative of the methylated fraction of tumor and normal cells, are labeled and cohybridized in a microarray platform. Image acquisition and data analysis allow identification of methylated and nonmethylated genes by comparing intensity values of Cy5 and Cy3 dyes for each pair of tumor and control samples.
By comparing probe intensity for each cancer cell line to normal controls, we can rapidly visualize the ones with significantly higher intensity in tumors (Fig. 2A). A total of 811 hypermethylated loci were identified for the three studied cell lines, representing colon tumors (RKO), leukemia (Raji), and melanoma (C8161). Several genes were methylated in more than one cell line, and after this redundancy was filtered, we found that 460 unique probes were hypermethylated. The actual number of investigated loci is inferior to the total number of probes in the microarray, since we excluded 4465 putative repetitive elements identified by Cot-1 hybridization. Also, the generation of MCA amplicons depends on the presence of two SmaI sites in relatively close proximity (no more than 1–2 kb apart), which occurs in ∼80% of the promoter CpG islands as calculated from in silico digestion of the human genome. In consequence, we estimate that 6180 probes are being investigated under our experimental conditions. Based on this estimation, we calculate the percent of methylated CpG islands in RKO, Raji, and C8161, respectively, as 4.7%, 5.5%, and 3.0%, or ∼4.4% on average, consistent with prior data (Smiraglia et al. 2001). The comparison of methylated probes between these three cell lines revealed that, on average, 26% of the methylated loci are exclusive to one cell line (Fig. 2B). This finding reinforces the fact that a large fraction of aberrant methylation in cancer occurs in a tissue-specific pattern (Costello et al. 2000), and also that the list of methylated genes in cancer is still largely incomplete.
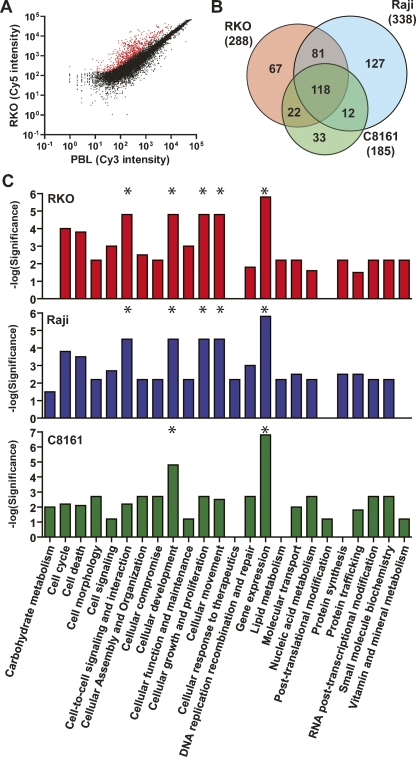
Figure 2.
Detection of DNA methylation by MCAM. (A) Scatterplot of Cy5 versus Cy3 intensity values for each probe showing the segregation of methylated (red spots) from unmethylated (black spots) in the colon cancer cell line RKO compared to normal peripheral blood lymphocytes. (B) Venn diagram representing the overlap and differences in methylated probes for the cancer cell lines RKO, Raji, and C8161. Note that a large number of loci are exclusively methylated in each individual cell line. (C) Comparison of significant representation of altered functional categories in cancer cell lines inferred by the presence of methylated genes network. The most significant categories in each cell line are labeled with asterisks (*). Note that “Gene expression” function is affected in all three cell lines, which is because of a large fraction of methylated transcription factors among all identified genes by MCAM.
To further evaluate whether tissue-specific methylation in normal samples biased the results in cancer cell lines, we hybridized normal peripheral blood lymphocytes (used in our experiments as a “universal control”) versus normal melanocytes (the theoretical optimal control to C8161). A small number of loci were differentially methylated between these tissues (31 loci were hypermethylated in normal peripheral blood lymphocytes, and 20 loci were methylated in normal melanocytes). The comparison experiment of peripheral blood lymphocytes versus peripheral blood lymphocytes showed no difference in methylation between samples, supporting a low rate of technical artifacts. These numbers are in agreement with our previous experience that tissue-specific methylation differences in normal tissues are rare and support our findings that differences in cancer cell lines represent cancer-relevant methylation rather than normal tissue-specific methylation differences. Similarly, Weber et al. (2005) reported that the use of either normal fibroblasts or normal colon mucosa as control samples when profiling methylation of the colon cancer cell line SW48 using mDIP resulted in virtually identical results.
Identification of new methylated genes and altered pathways in cancer using MCAM
The 460 probes found to be hypermethylated correspond to 376 genome loci according to the available annotation for this microarray platform, the remaining still requiring sequencing to confirm their origin. Among the annotated loci, 192 were representative of CpG island promoters. A comprehensive list of probes and their correspondence to known genes and CpG islands is presented as Supplemental Table 1. Since the significance of hypermethylation of non-promoter, non-CpG islands is still uncertain, we concentrate our analysis on those 192 genomic loci that more likely represent genes silenced by DNA methylation in the studied cell lines. Some loci correspond to two genes (bidirectional promoter), and others were represented more than once (replicated probes), resulting in 167 unique genes. Some of the known targets of DNA methylation in cancer were found among these genes, for example, BARHL1, CRABP1, GBX2, and HAND1 in solid tumors (Kaneda et al. 2002; Furuta et al. 2006; Ogino et al. 2006), which reinforce the use of MCAM to study aberrant hypermethylation in cancer. Of special interest are the genes that may play an important role in cancer and were found to be methylated for the first time, justifying the use of methylation arrays for gene discovery. For example, the candidate tumor-suppressor gene DUSP4, which maps to a chromosomal region of frequent LOH in breast cancer (Venter et al. 2005), and BTG2, described as a p53-inducible antiproliferative gene that was found to be down-regulated in breast and renal carcinomas (Kawakubo et al. 2004; Struckmann et al. 2004), were found to be methylated in our study.
Similarly to gene expression profiles, methylation data can be used to identify biological pathways and gene networks disrupted in cancer. To do such an analysis, the networks and functional analyses of genes found to be methylated in each one of the studied cell lines were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems; http://www.ingenuity.com), and the results are summarized in Figure 2C. The pattern of molecular and biological function categories with methylated genes was fairly similar between different cell lines, particularly the predominance of methylated genes involved in gene expression due to a large fraction of transcription factors found to be methylated by MCAM. Also of interest are the partners of methylated genes, and the network analysis identified some known cancer targets. Genes of special relevance are NOTCH1 and MYC in colon, TP53 and WT1 in leukemia, and TNF in melanoma (see Supplemental Table 2 for a comprehensive view).
Specificity, sensitivity, and reproducibility of MCAM
To validate MCAM results, we selected 15 genes (seven predicted hypermethylated and eight predicted unmethylated) and analyzed them by bisulfite-PCR. Figure 3A illustrates an example of one of these genes, HAND1, which codes for a basic helix–loop–helix transcription factor. The microarray probe fragment is completely included in the expected MCA amplicon, and a bisulfite-PCR and pyrosequencing assay was designed to investigate the methylation status of this gene close to the transcription start site. All four CpG sites tested were methylated in RKO but not in peripheral blood DNA, as predicted by MCAM (Fig. 3B), and the final methylation data are presented as the average methylation of the studied CpG sites. The other tested genes were BARHL1, INTS6, GDNF, DIS3, LHX9, NKX2-3, PAX2, POLR2J, TAF11, TFAP2C, TOP2A, RAD51, RSHL1, and SIN3A. Methylation of these genes was determined in one to three cell lines for which MCAM data were available (Fig. 3C). We found that 15/16 (93.8%) MCAM probes indicating positive methylation were also methylated by bisulfite-PCR. Conversely, among 17 probes indicating lack of methylation, 16 (94.1%) were unmethylated by MCAM. Considering bisulfite-PCR as gold-standard, these data yield a sensitivity of 93.8% and a specificity of 94.1% for MCAM. The MCAM log2 ratio and actual methylation density measured by bisulfite-PCR for each gene are included as Supplemental Table 3. Difference in CG content makes certain sequences more difficult to amplify in MCA, and also decreases hybridization efficiency, which may explain the presence of false-positive and false-negative data in MCAM. The low rate of false-positive data is very encouraging regarding the use of MCAM in large-scale investigations.
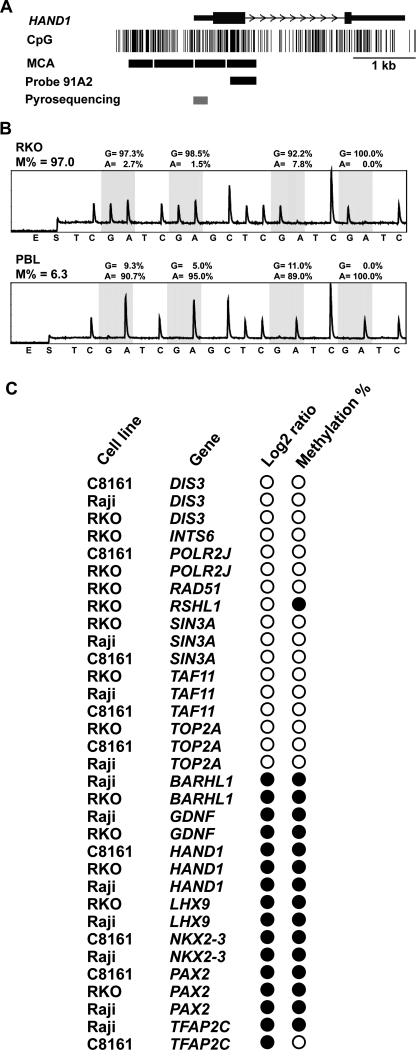
Figure 3.
Validation of methylation status of selected genes in cancer cell lines. (A) Representation of one identified methylated gene by MCAM. One of several possible MCA fragments (delimited by CCCGGG sequences) close to the transcription start site of the HAND1 gene overlaps with the DNA probe 91A2, allowing the investigation of this fragment in the microarray platform. Note that the remaining MCA fragments do not overlap additional probes, being therefore not investigated in this system but potentially investigated in other arrays with different probe collections. (Gray bar) The promoter region investigated by bisulfite PCR followed by pyrosequencing. (B) Representative pyrograms for HAND1. Four CpG sites close to the transcription start site were pyrosequenced, and a consistent pattern of high levels of methylation was observed for all of them in RKO, while in peripheral blood lymphocyte the methylation values were low, consistent with the MCAM results. The methylation density is presented in the top of each pyrogram as the averaged methylation of the four studied CpG sites. (C) Graphic representation of log2 ratio and methylation analysis by bisulfite-PCR results for selected genes for validation of MCAM. (Black circles) Hypermethylation in tumor (as determined as log2 ratio ≥ 1.3 in MCAM and methylation density >10% by bisulfite-PCR followed by pyrosequencing); (white circles) lack of methylation. Note the high concordance of methylation results between techniques.
Another level of validation required before applying MCAM to clinical and epidemiological studies is to determine the reproducibility of the method. The correlation between duplicate MCAM experiments was statistically significant (P < 0.0001), with Pearson r values of 0.90, 0.92, and 0.89 for RKO, Raji, and C8161, respectively (Fig. 4A). Most of the nonconcordant methylation occurred for probes in the immediate log2 ratio values close to the cutoff (Fig. 4B), with concordance increasing to 100% at log2 ratio ≥ 2.6. However, this increased reproducibility comes at the expense of lower sensitivity.
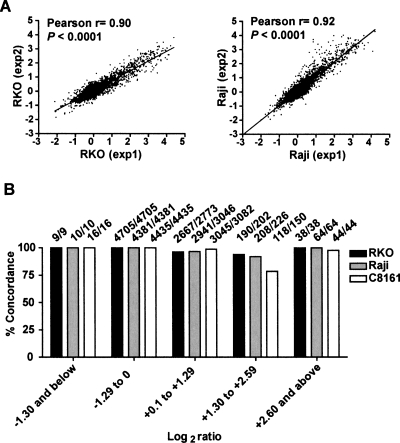
Figure 4.
Reproducibility of MCAM experiments. (A) Overall correlation between independent replicates for cancer cell lines RKO (colon) and Raji (leukemia). (B) General concordance between data in replicate experiments (both methylation and lack of methylation) per log2 ratio quantiles. The actual number of concordant loci is presented (above each bar).
Finally, we performed MCAM to the colorectal cancer cell line SW48 in order to generate a direct comparison to another microarray-based methylation analysis, the antibody-based mDIP technique described by Weber et al. (2005). We found a better Pearson’s correlation among duplicates in our study (0.91 for duplicate MCAM experiments compared to 0.79 for mDIP). Additionally, MCAM identified more than two times more hypermethylated promoter CpG islands in SW48 (Supplemental Table 4). In summary, MCAM compares favorably to mDIP and possibly to other antibody-based methods previously described.
Classification of colorectal carcinomas into clinical subgroups using MCAM
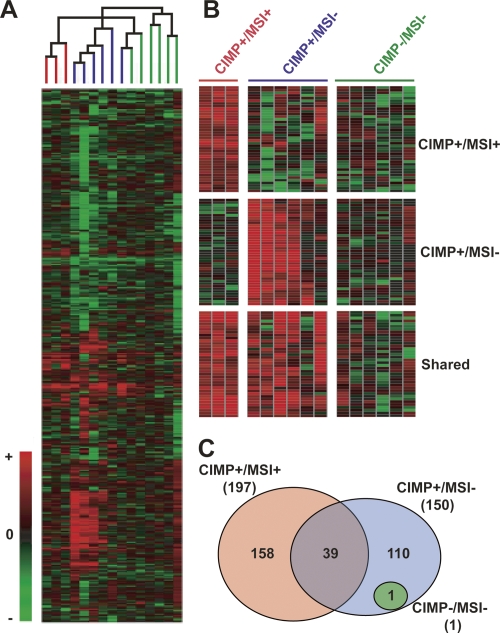
Once specificity, sensitivity, and reproducibility were determined, we applied MCAM to study clinical samples. Based on DNA availability, we selected 15 colorectal carcinoma samples to analyze by MCAM. The control sample used for each tumor was its corresponding normal-appearing adjacent mucosa. These colorectal carcinoma samples were previously investigated in our laboratory regarding their CpG island methylation phenotype (CIMP) and microsatellite instability (MSI) status (Toyota et al. 2000). MCAM on these primary colorectal cancers revealed an average of 300 hypermethylated probes (range: 29–878) (see Supplemental Fig. 7 for individual samples), corresponding to an average of 5.5% methylated probes. It was not possible to perform duplicate experiments for the colorectal tumor samples because of limited DNA availability. However, the high correlation between duplicated cell line experiments supports the idea that the variability in methylation observed among samples is unlikely to result from technical artifacts. Furthermore, to minimize the lack of duplicate experiments, the subsequent analyses of these samples were done for CIMP/MSI groups rather than to individual samples. We used unsupervised hierarchical clustering to analyze the data. The resulting cluster analysis based on MCAM satisfactorily resembles the expected CIMP/MSI groups (Fig. 5A). Except for one CIMP+/MSI− sample, all other samples were directed into their expected groups, resulting in 94% concordance between global and gene-specific CIMP/MSI classification. This result demonstrates the successful application of MCAM to clinical investigation and reinforces the existence of colorectal carcinomas with extensive epigenetic instability. Based only on this result, it is hard to discern whether the sample clustering generate from MCAM data or the predefined CIMP classification is more accurate. Since CIMP is better defined when age-related genes are removed from the analysis, and also since MSI cannot be determined using MCAM, we found it more appropriate to use the predefined CIMP/MSI classification to select genes that discriminate these groups. At P < 0.05, 40 probes were found to have a differential methylation status in CIMP+/MSI+ compared to CIMP+/MSI− and CIMP−/MSI− cases. Figure 5B represents probe clusters that are methylated mostly in CIMP+/MSI+, CIMP+/MSI− and the ones shared by these two groups but not CIMP−/MSI−. A complete list of methylated probes and corresponding genes is presented in Supplemental Table 5. Finally, we compared the different CIMP/MSI groups using probes that were methylated at the same time in at least 60% of the samples in each group (Fig. 5C). As expected, little or no concordance in methylation was observed in CIMP−/MSI− cases, while a high degree of concordance was seen in the other groups. Surprisingly, only a few probes (∼13%) were commonly methylated in CIMP+/MSI+ and CIMP+/MSI− cases.
Figure 5.
Global methylation analysis of colorectal carcinomas by MCAM. (A) Cluster analysis of MCAM data in primary colorectal carcinomas. A total of 4588 probe data was used for unsupervised hierarchical clustering analysis of 15 sample pairs (tumor compared to normal appearing mucosa), resulting in a classification that matched the known CIMP/MSI status of these samples. The terminal branches are color-coded to represent the CIMP/MSI status of the tumor sample: (red) CIMP+/MSI+; (blue) CIMP+/MSI−; (green) CIMP−/MSI−. (B) Probe clusters, which classifies CIMP+/MSI+ and CIMP+/MSI− samples apart from each other, and CIMP+ in general (shared group) from CIMP− samples. (C) Venn diagram of concordant probe methylation (at least 60% of samples methylated) for each CIMP/MSI group. The total number of methylated probes in each group is shown in parenthesis.
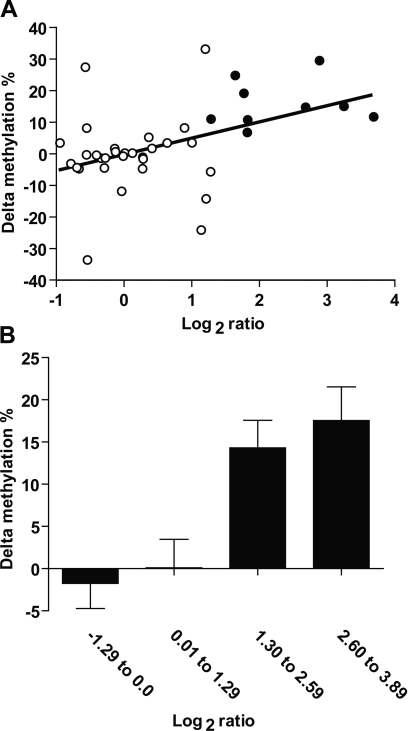
Finally, we selected four genes for validation in this colorectal carcinoma series, two predicted to be similarly methylated in normal and tumor (INTS6, RAD51) and two predicted to be hypermethylated in cancer compared to normal (BARHL1, RSHL1) by MCAM. We used bisulfite-pyrosequencing to measure the methylation of each gene in 11 normal adjacent/tumor pairs. Differently from normal peripheral blood lymphocytes (where methylation density for all tested genes was <5%), normal colon mucosa adjacent to tumor showed significant levels of methylation (>20%) in some cases. Since the log2 ratio is a relative value, we found it more accurate to compare the MCAM data with another relative value, the absolute difference in methylation among tumor and normal DNA methylation (i.e., methylation in tumor minus methylation in adjacent normal or delta methylation). As shown in Figures 6A, there was a positive correlation between log2 ratios and bisulfite-pyrosequencing data (presented in more detail in Supplemental Fig. 8); however, the positive predictive value was lower than that observed for cancer cell lines (90% compared to 93.8% in cancer cell lines) (Supplemental Fig. 9), which may be related to heterogeneous methylation, contamination by normal tissues, and lower DNA quality. Encouragingly, the averaged delta methylation significantly increases per log2 ratio quartile (Fig. 6B, P < 0.001), suggesting that higher log2 ratio values can indicate larger differences in methylation density between tumor and normal samples.
Figure 6.
Validation of methylation status of selected genes in primary colorectal carcinomas. (A) Correlation between delta methylation (methylation in tumor minus methylation in adjacent normal) and log2 ratio for four genes investigated by bisulfite-PCR and pyrosequencing. (White circles) Genes with lower methylation in tumor compared to normal samples (log2 ratio < 1.3) according to MCAM; (black circles) hypermethylated genes according this technique. (B) Average delta methylation per log2 ratio. (Bar graphic) The average delta methylation of samples in the same log2 ratio quartile window. Note that the difference in methylation between tested (colon tumor) and control (normal colon) increases with log2 ratio deviation from zero, revealing the semiquantitative nature of the MCAM measurements.
Discussion
We developed a new microarray strategy to study DNA methylation, named MCAM. Our method applies methylated CpG island amplification (MCA) to a microarray platform, and in the present study we used an array containing 12,192 CpG island clones from the Sanger Institute (Cross et al. 1994; Heisler et al. 2005). However, this method can be applied to other array platforms, such as oligonucleotide, tiling, and custom-made arrays with larger gene representation. Our method compares favorably to other published methylation microarray protocols because it provides reproducible results with a high validation rate and demonstrated use in clinical samples to cluster cases into distinct molecular groups. A recent method published by Rauch et al. (2006) using methyl-binding domain proteins has proved specific to detect new methylated genes in lung cancer, although the reproducibility of the method and sensitivity was not described. The same issues apply to methods based on anti-5 methyl cytidine antibodies (Weber et al. 2005). In our experience, the sensitivity of methods to isolate DNA using 5-mC antibodies is low, which limits its application in genome-wide studies. Indeed, it has been estimated that the SW48 colon cancer cell line has only 26 genes hypermethylated in cancer (Weber et al. 2005), a gross underestimation of the real number based on our studies. The use of methylation-sensitive enzymes with frequent cutting sites, for example, HpaII/MspI (Hatada et al. 2006), results in a high genome fraction to amplify. In such circumstance of high complexity, PCR efficiency is compromised and likely favors amplification of non-CpG-island DNA. Indeed, several reports using HpaII/MspI-derived libraries to identify novel methylation targets found a high frequency of hypomethylated compared to hypermethylated fragments in cancer and cloned mostly non-CpG-island DNA (Gonzalgo et al. 1997; Estecio et al. 2006). In comparison, our MCAM technique simultaneously reduces complexity and increases specificity by targeting methylated CpG islands before amplification.
A large fraction of hypermethylated genes in cancer is still unknown, and one of the uses for MCAM would be to identify such genes. Several of the genes we identified here in cancer cell lines were previously not known to be methylated in cancer, and some of them have potential biological functions that can participate in tumor formation or promotion. It is interesting to note that pathway analysis of genes hypermethylated in cancer leads to processes important in development/differentiation, reinforcing the interplay between tissue-specific expression and susceptibility to methylation. Separately, the top pathway affected in each cell line strikingly leads to key pathways in cancer formation—NOTCH1 for RKO, TP53 for RAJI, and TNF for melanoma. There is a remarkable concordance of functions affected in cancer by gene mutation (Sjoblom et al. 2006) and those affected by gene methylation, with a predominance of genes in signal transduction and transcriptional regulation. These stress the potential value of methylation microarrays in understanding cancer biology. However, there are limitations to our analyses in that (1) only three cell lines were analyzed; (2) this microarray platform has limited gene representation (indeed, several genes known to be methylated in cancer are missing, e.g., CDKN2A and RASSF1); and (3) MCA theoretically only covers 80% of CpG islands. The report of pathways and functional categories enriched for methylated genes in these cell lines by no means exhausts further investigations, although the relevance of our findings are reinforced by previous studies of these cell lines. For example, Raji is a well-known model for resistance to apoptosis induced by various stimuli (Kawabata et al. 1999; Kern et al. 2004). Additional studies on more cell lines and more extensive CpG island arrays should very revealing vis-à-vis cancer biology.
Important questions in cancer epigenetics can be clarified by investigating large collections of genes in an unbiased way. The CIMP concept has been controversial, but a recent study confirmed that CIMP-positive (CIMP+) cases exist and are associated with selected genetic alterations, like BRAF mutations (Weisenberger et al. 2006). Our present study adds to previous ones (Toyota et al. 2000; Shen et al. 2003) to further confirm the existence of CIMP using an unbiased approach and suggests specific markers for each CIMP+ group. In this study, we identified nearly 200 hypermethylated genes that can specifically classify CIMP+/MSI+ and CIMP+/MSI− cases apart from others, and a more detailed analysis of these genes may help us to understand the origin of such epigenetic dysregulation. An important aspect of CIMP+ cases is the association with microsatellite instability (MSI) in a portion of cases, due to MLH1 hypermethylation. This CIMP+ subgroup, named CIMP+/MSI+, also presents more concordant hypermethylation than CIMP+/MSI− cases, suggesting an underlying genetic defect that boosts hypermethylation.
In conclusion, we have developed MCAM, a new unbiased, high-throughput technique to detect DNA methylation in cancer. MCAM is highly specific and sensitive; it will be useful to detect new methylation markers in cancer and may be extended to clinical studies. Also, our results validate the presence of CIMP in colorectal carcinomas and identify a collection of genes that defines this phenotype.
Methods
Samples
Fifteen matched pairs of tumor and apparently normal adjacent colon specimens were obtained from patients treated at Johns Hopkins University (Baltimore, MD). This study was approved by the Ethics Committee of Johns Hopkins University, and informed consent was obtained from all participants. CpG island methylation phenotype (CIMP) and microsatellite analysis were previously determined for these samples (Toyota et al. 2000). Cancer cell lines RKO, SW48 (both colon), Raji (leukemia), and C8161 (melanoma) were obtained from ATCC and cultured using standard methods. Peripheral blood lymphocytes were collected from normal donors, and normal melanocytes were obtained from foreskin of newborn infants. DNA from patients, subjects, and cell lines was extracted using standard phenol–chloroform extraction methods.
Methylated CpG island amplication (MCA)
The procedure was performed according to Toyota et al. (1999b). In brief, 5 μg of DNA was digested with 100 units of SmaI for 16 h (all restriction enzymes were from New England Biolabs), followed by digestion with 20 units of XmaI for 6 h. DNA fragments were then precipitated with ethanol. RXMA PCR adaptors were prepared by incubation of the oligonucleotides RXMA24 (5′-AGCACTCTCCAGCCTCTCACCGAC-3′) and RXMA12 (5′-CCGGGTCGGTGA-3′) for 2 min at 65°C, followed by cooling to room temperature for 1 h. DNA (0.5 μg) was ligated to 0.5 nmol of RXMA adaptor using T4 DNA ligase (New England Biolabs). PCR was performed using 3 μL each of the ligation mix as a template in a 100-μL volume containing 100 pmol of RXMA24, 5 units of Taq DNA polymerase, 67 mM Tris-HCl (pH 8.8), 4 mM MgCl2, 16 mM NH4(SO4)2, and 10 μg/mL BSA. The reaction mixture was incubated for 5 min at 72°C and for 3 min at 95°C. Samples were then subjected to 20 cycles of amplification consisting of 1 min at 95°C and 3 min at 72°C in a thermal cycler. The final extension time was 10 min. Ten microliters of PCR product was resolved in 1.5% agarose gel and visualized under UV after ethidium bromide staining. Successful MCA reactions result in amplicon smear ranging from 300 bp to 3 kb, with most amplicons at 1 kb. PCR was purified using QIAGEN PCR purification to remove unincorporated nucleotides.
CpG island microarray
Incorporation of amino-allyl dUTP (aa-dUTP; Sigma) into 600 ng each of tumor DNA and normal DNA was conducted using the Bioprime DNA-labeling system protocol (Life Technologies) (Weinmann et al. 2002). Cy5 and Cy3 fluorescent dyes were coupled to aa-dUTP-labeled tumor and normal DNA, respectively, and cohybridized to the HCGI12K-Human CpG 12K Array (Microarray Center, University Health Network, Toronto, Canada). Microarray protocols including the hybridization and post-hybridization procedures are according to protocols developed by DeRisi et al. (1996), except for washing steps, which were extended to 10 min each to decrease background. Hybridized slides were scanned with the GenePix 4000A scanner (Axon), and the acquired images were analyzed with the software GenePix Pro 3.0. Two-step global lowess normalization was done using the background-subtracted median intensity of each spot, and the resultant log2 ratios were averaged from duplicate experiments (except for primary tumors, where only one array experiment was available per sample). Based on published expression microarray methods, CpG island tags having a Cy5/Cy3 log2 ratio ≥ 1.3 (∼2.5-fold enrichment) were chosen as hypermethylation-specific signals. This log2 ratio value was further supported by our validation experiments as the best cutoff to achieve optimal sensitivity and specificity.
Pyrosequencing PCR
Validation of the methylation status of 15 genes (BARHL1, INTS6, GDNF, HAND1, DIS3, LHX9, NKX2-3, PAX2, POLR2J, TAF11, TFAP2C, TOP2A, RAD51, RSHL1, and SIN3A) was performed using pyrosequencing- or COBRA- (combined bisulfite restriction analysis) based methylation analysis. The primer sequences are available in Supplemental Table 6. For each gene, a 50-μL PCR was carried out in 60 mM Tris-HCl (pH 8.5), 15 mM ammonium sulfate, 2 mM MgCl2, 10% DMSO, 1 mM dNTP mix, 1 unit of Taq polymerase, 5 pmol of the forward primer, 2 pmol of the reverse primer, and ∼50 ng of bisulfite-treated genomic DNA. For pyrosequencing only, 4.5 pmol of biotinylated universal primer (5′-GGGACACCGCTGATCGTATA-3′) was added to the PCR reactions. The forward primer has a 20-bp linker sequence on the 5′-end that is recognized by a biotin-labeled primer so that the final PCR product can be purified using Sepharose beads. PCR cycling conditions were 30 sec at 95°C, 30 sec at 50°–57°C, and 30 sec at 72°C for 50 cycles. The biotinylated PCR product was purified and made single-stranded to act as a template in a pyrosequencing reaction as recommended by the manufacturer using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc.). In brief, the PCR product was bound to Streptavidin-Sepharose HP (Amersham Biosciences), and the Sepharose beads containing the immobilized PCR product were purified, washed, denatured using a 0.2 M NaOH solution, and washed again. Then, 0.3 μM pyrosequencing primer (sequence-specific to each gene) was annealed to the purified single-stranded PCR product, and pyrosequencing was performed using the PSQ HS 96 Pyrosequencing System (Pyrosequencing, Inc.). Methylation quantification was performed using the provided software. The methylation degree of each gene was computed as the average of two to six CpG sites. Alternatively, for COBRA assays, the PCR products were digested with the restriction enzyme TaqI, followed by electrophoresis on 6% polyacrylamide gels. Gels were stained with ethidium bromide, imaged, and quantitated in a Bio-Rad Geldoc 2000 imager (Bio-Rad). The methylation density for each sample was computed as a ratio of the density of the digested band to the density of all bands in a given lane.
Statistical analysis
Unsupervised hierarchical clustering of primary colorectal tumors was done using the program GeneSpring (Agilent Technologies, Inc.) with calculation for distance using absolute correlation and complete linkage clustering. Gene classifiers of each CIMP/MSI group were selected based on the significance of the differences observed between means of individual spots using the two-sided Student’s t-test. A P-value of <0.05 was considered statistically significant. The Pearson product-moment correlation coefficient was used to compare log2 ratios between duplicate experiments. Statistical analyses were carried out with the Statistica software package (StatSoft). Gene network and pathway analysis was done using Ingenuity Pathways Analysis (Ingenuity Systems; http://www.ingenuity.com). The network genes associated with biological functions and/or diseases in the Ingenuity Pathways Knowledge Base were considered for the analysis. The Fischer’s exact test was used to calculate a P-value determining the probability that each biological function and/or disease assigned to that network is due to chance alone.
Acknowledgments
This work was supported in part by National Institutes of Health grants P50CA100632, RO1CA098006 and R33CA89837.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6417007
References
- Costello J.F., Fruhwald M.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Fruhwald M.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Rush L.J., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Robertson G.P., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Gao X., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Wright F.A., Feramisco J.D., Peltomaki P., Lang J.C., Feramisco J.D., Peltomaki P., Lang J.C., Peltomaki P., Lang J.C., Lang J.C., et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Cross S.H., Charlton J.A., Nan X., Bird A.P., Charlton J.A., Nan X., Bird A.P., Nan X., Bird A.P., Bird A.P. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- DeRisi J., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Ray M., Chen Y., Su Y.A., Trent J.M., Chen Y., Su Y.A., Trent J.M., Su Y.A., Trent J.M., Trent J.M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Estecio M.R., Youssef E.M., Rahal P., Fukuyama E.E., Gois-Filho J.F., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Youssef E.M., Rahal P., Fukuyama E.E., Gois-Filho J.F., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Rahal P., Fukuyama E.E., Gois-Filho J.F., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Fukuyama E.E., Gois-Filho J.F., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Gois-Filho J.F., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Maniglia J.V., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Goloni-Bertollo E.M., Issa J.P., Tajara E.H., Issa J.P., Tajara E.H., Tajara E.H. LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene. 2006;25:5018–5026. doi: 10.1038/sj.onc.1209509. [DOI] [PubMed] [Google Scholar]
- Furuta J., Nobeyama Y., Umebayashi Y., Otsuka F., Kikuchi K., Ushijima T., Nobeyama Y., Umebayashi Y., Otsuka F., Kikuchi K., Ushijima T., Umebayashi Y., Otsuka F., Kikuchi K., Ushijima T., Otsuka F., Kikuchi K., Ushijima T., Kikuchi K., Ushijima T., Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- Gonzalgo M.L., Liang G., Spruck C.H., Zingg J.M., Rideo W.M., Jones P.A., Liang G., Spruck C.H., Zingg J.M., Rideo W.M., Jones P.A., Spruck C.H., Zingg J.M., Rideo W.M., Jones P.A., Zingg J.M., Rideo W.M., Jones P.A., Rideo W.M., Jones P.A., Jones P.A. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 1997;57:594–599. [PubMed] [Google Scholar]
- Hatada I., Fukasawa M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Fukasawa M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yamanaka S., Endo C., Sakurada A., Sato M., Endo C., Sakurada A., Sato M., Sakurada A., Sato M., Sato M., et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- Heisler L.E., Torti D., Boutros P.C., Watson J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Torti D., Boutros P.C., Watson J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Boutros P.C., Watson J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Watson J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Chan C., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Winegarden N., Takahashi M., Yau P., Huang T.H., Farnham P.J., Takahashi M., Yau P., Huang T.H., Farnham P.J., Yau P., Huang T.H., Farnham P.J., Huang T.H., Farnham P.J., Farnham P.J., et al. CpG island microarray probe sequences derived from a physical library are representative of CpG islands annotated on the human genome. Nucleic Acids Res. 2005;33:2952–2961. doi: 10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J.G., Baylin S.B., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Kaneda A., Kaminishi M., Yanagihara K., Sugimura T., Ushijima T., Kaminishi M., Yanagihara K., Sugimura T., Ushijima T., Yanagihara K., Sugimura T., Ushijima T., Sugimura T., Ushijima T., Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645–6650. [PubMed] [Google Scholar]
- Kawabata Y., Hirokawa M., Kitabayashi A., Horiuchi T., Kuroki J., Miura A.B., Hirokawa M., Kitabayashi A., Horiuchi T., Kuroki J., Miura A.B., Kitabayashi A., Horiuchi T., Kuroki J., Miura A.B., Horiuchi T., Kuroki J., Miura A.B., Kuroki J., Miura A.B., Miura A.B. Defective apoptotic signal transduction pathway downstream of caspase-3 in human B-lymphoma cells: A novel mechanism of nuclear apoptosis resistance. Blood. 1999;94:3523–3530. [PubMed] [Google Scholar]
- Kawakubo H., Carey J.L., Brachtel E., Gupta V., Green J.E., Walden P.D., Maheswaran S., Carey J.L., Brachtel E., Gupta V., Green J.E., Walden P.D., Maheswaran S., Brachtel E., Gupta V., Green J.E., Walden P.D., Maheswaran S., Gupta V., Green J.E., Walden P.D., Maheswaran S., Green J.E., Walden P.D., Maheswaran S., Walden P.D., Maheswaran S., Maheswaran S. Expression of the NF-kappaB-responsive gene BTG2 is aberrantly regulated in breast cancer. Oncogene. 2004;23:8310–8319. doi: 10.1038/sj.onc.1208008. [DOI] [PubMed] [Google Scholar]
- Kern C., Cornuel J.F., Billard C., Tang R., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Cornuel J.F., Billard C., Tang R., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Billard C., Tang R., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Tang R., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Simonin P.Y., Feldblum S., Feldblum S., et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood. 2004;103:679–688. doi: 10.1182/blood-2003-02-0540. [DOI] [PubMed] [Google Scholar]
- Laird P.W. Cancer epigenetics. Hum. Mol. Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Lavine K., Mittal V., May B., Kasschau K.D., Mittal V., May B., Kasschau K.D., May B., Kasschau K.D., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Ogino S., Cantor M., Kawasaki T., Brahmandam M., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Cantor M., Kawasaki T., Brahmandam M., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Kawasaki T., Brahmandam M., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Brahmandam M., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Kirkner G.J., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Weisenberger D.J., Campan M., Laird P.W., Loda M., Fuchs C.S., Campan M., Laird P.W., Loda M., Fuchs C.S., Laird P.W., Loda M., Fuchs C.S., Loda M., Fuchs C.S., Fuchs C.S. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T., Li H., Wu X., Pfeifer G.P., Li H., Wu X., Pfeifer G.P., Wu X., Pfeifer G.P., Pfeifer G.P. MIRA—Assisted Microarray Analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- Shen L., Kondo Y., Hamilton S.R., Rashid A., Issa J.P., Kondo Y., Hamilton S.R., Rashid A., Issa J.P., Hamilton S.R., Rashid A., Issa J.P., Rashid A., Issa J.P., Issa J.P. P14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology. 2003;124:626–633. doi: 10.1053/gast.2003.50102. [DOI] [PubMed] [Google Scholar]
- Sjoblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Mandelker D., Leary R.J., Ptak J., Silliman N., Leary R.J., Ptak J., Silliman N., Ptak J., Silliman N., Silliman N., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smiraglia D.J., Rush L.J., Fruhwald M.C., Dai Z., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Rush L.J., Fruhwald M.C., Dai Z., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Fruhwald M.C., Dai Z., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Dai Z., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Held W.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Costello J.F., Lang J.C., Eng C., Li B., Wright F.A., Lang J.C., Eng C., Li B., Wright F.A., Eng C., Li B., Wright F.A., Li B., Wright F.A., Wright F.A., et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum. Mol. Genet. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- Struckmann K., Schraml P., Simon R., Elmenhorst K., Mirlacher M., Kononen J., Moch H., Schraml P., Simon R., Elmenhorst K., Mirlacher M., Kononen J., Moch H., Simon R., Elmenhorst K., Mirlacher M., Kononen J., Moch H., Elmenhorst K., Mirlacher M., Kononen J., Moch H., Mirlacher M., Kononen J., Moch H., Kononen J., Moch H., Moch H. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res. 2004;64:1632–1638. doi: 10.1158/0008-5472.can-03-1687. [DOI] [PubMed] [Google Scholar]
- Sugimura T., Ushijima T., Ushijima T. Genetic and epigenetic alterations in carcinogenesis. Mutat. Res. 2000;462:235–246. doi: 10.1016/s1383-5742(00)00005-3. [DOI] [PubMed] [Google Scholar]
- Toyota M., Issa J.P., Issa J.P. Epigenetic changes in solid and hematopoietic tumors. Semin. Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P., Herman J.G., Baylin S.B., Issa J.P., Baylin S.B., Issa J.P., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. 1999a;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M., Ho C., Ahuja N., Jair K.W., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P., Ho C., Ahuja N., Jair K.W., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P., Ahuja N., Jair K.W., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P., Jair K.W., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P., Li Q., Ohe-Toyota M., Baylin S.B., Issa J.P., Ohe-Toyota M., Baylin S.B., Issa J.P., Baylin S.B., Issa J.P., Issa J.P. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999b;59:2307–2312. [PubMed] [Google Scholar]
- Toyota M., Ohe-Toyota M., Ahuja N., Issa J.P., Ohe-Toyota M., Ahuja N., Issa J.P., Ahuja N., Issa J.P., Issa J.P. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc. Natl. Acad. Sci. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter D.J., Ramus S.J., Hammet F.M., de Silva M., Hutchins A.M., Petrovic V., Price G., Armes J.E., Ramus S.J., Hammet F.M., de Silva M., Hutchins A.M., Petrovic V., Price G., Armes J.E., Hammet F.M., de Silva M., Hutchins A.M., Petrovic V., Price G., Armes J.E., de Silva M., Hutchins A.M., Petrovic V., Price G., Armes J.E., Hutchins A.M., Petrovic V., Price G., Armes J.E., Petrovic V., Price G., Armes J.E., Price G., Armes J.E., Armes J.E. Complex CGH alterations on chromosome arm 8p at candidate tumor suppressor gene loci in breast cancer cell lines. Cancer Genet. Cytogenet. 2005;160:134–140. doi: 10.1016/j.cancergencyto.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Haase M., Lam W.L., Schubeler D., Lam W.L., Schubeler D., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Yan P.S., Oberley M.J., Huang T.H., Farnham P.J., Yan P.S., Oberley M.J., Huang T.H., Farnham P.J., Oberley M.J., Huang T.H., Farnham P.J., Huang T.H., Farnham P.J., Farnham P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Kang G.H., Widschwendter M., Weener D., Buchanan D., Widschwendter M., Weener D., Buchanan D., Weener D., Buchanan D., Buchanan D., et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Yan P.S., Chen C.M., Shi H., Rahmatpanah F., Wei S.H., Caldwell C.W., Huang T.H., Chen C.M., Shi H., Rahmatpanah F., Wei S.H., Caldwell C.W., Huang T.H., Shi H., Rahmatpanah F., Wei S.H., Caldwell C.W., Huang T.H., Rahmatpanah F., Wei S.H., Caldwell C.W., Huang T.H., Wei S.H., Caldwell C.W., Huang T.H., Caldwell C.W., Huang T.H., Huang T.H. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]