Abstract
While for two of three mammalian olfactory receptor families (OR and V2R) ortholog teleost families have been identified, the third family (V1R) has been thought to be represented by a single, closely linked gene pair. We identified four further V1R-like genes in every teleost species analyzed (Danio rerio, Gasterosteus aculeatus, Oryzias latipes, Tetraodon nigroviridis, Takifugu rubripes). In the phylogenetic analysis these ora genes (olfactory receptor class A-related) form a single clade, which includes the entire mammalian V1R superfamily. Homologies are much lower in paralogs than in orthologs, indicating that all six family members are evolutionarily much older than the speciation events in the teleost lineage analyzed here. These ora genes are under strong negative selection, as evidenced by very small dN/dS values in comparisons between orthologs. A pairwise configuration in the phylogenetic tree suggests the existence of three ancestral Ora subclades, one of which has been lost in amphibia, and a further one in mammals. Unexpectedly, two ora genes exhibit a highly conserved multi-exonic structure and four ora genes are organized in closely linked gene pairs across all fish species studied. All ora genes are expressed specifically in the olfactory epithelium of zebrafish, in sparse cells within the sensory surface, consistent with the expectation for olfactory receptors. The ora gene repertoire is highly conserved across teleosts, in striking contrast to the frequent species-specific expansions observed in tetrapod, especially mammalian V1Rs, possibly reflecting a major shift in gene regulation as well as gene function upon the transition to tetrapods.
The olfactory sense is one of the main tools that animals developed to make sense of their environment. Thousands of structurally diverse odor molecules perceived and discriminated by vertebrates supply them with a wide range of vital information, ranging from prey and predator localization to mating behavior.
In mammals odor molecules are detected by three olfactory receptor families that are expressed in olfactory sensory neurons: olfactory receptors (ORs), with ∼1000 genes in rodents (Buck and Axel 1991; Mombaerts 2004), and two types of vomeronasal receptors (V1Rs and V2Rs, respectively), with ∼100 genes in rodents (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997; Mombaerts 2004).
While several studies classified the V2R and OR receptors as evolutionary old families, with ∼50–150 members already present in several fish species (Hashiguchi and Nishida 2005; Niimura and Nei 2005), the V1R receptor family was considered a recent family that originated in a single V1R-like receptor gene in fish (Pfister and Rodriguez 2005) or rather a single gene pair (Shi and Zhang 2007). Although species-specific expansion and loss of genes and even whole subfamilies are recurrent themes in all three mammalian receptor families (Lane et al. 2004; Zhang et al. 2004; Grus et al. 2005), as well as in fish OR (Niimura and Nei 2005) and fish V2R-related gene families (Hashiguchi and Nishida 2005; Alioto and Ngai 2006), the V1R expansion from a single gene pair to over a hundred genes in some mammalian species appeared somewhat extreme. This prompted us to examine the genome of several fish species for the presence of hitherto overlooked V1R-like genes. We report here the identification and characterization of a novel family of six teleost V1R-like genes and suggest a new, consistent nomenclature for this family. These genes are highly conserved between five evolutionary distant teleost species, in stark contrast to the frequent gene gains and losses seen in the mammalian V1R family.
Results
Five novel V1R-like genes were identified in each of five teleost species
A recursive search strategy starting with all known V1R genes and using automatic ortholog annotation in combination with the TBLASTN algorithm (for details, see Methods) uncovered five novel genes in the zebrafish genome (Fig. 1). All orthologs of all five Danio rerio genes could be identified in four further fish species, Gasterosteus aculeatus, Oryzias latipes, Takifugu rubripes, and Tetraodon nigroviridis (three-spined stickleback, medaka, and two pufferfish, respectively). No pseudogenes were detected in any of the five teleost species. We propose to name these V1R-like genes ora (olfactory receptors related to class A, cf. Schiöth and Fredriksson 2005; www.gpcr.org/), not V1Rs, since that name refers to the vomeronasal system which fish do not possess—all the fish olfactory receptors are expressed in the main olfactory epithelium. The designation “ora” reflects both the olfactory-specific nature of these receptors as well as their phylogenetic position within the GPCR superfamily. Individual ora genes were numbered from 1 to 6, beginning with the first zebrafish ora gene to be identified (Pfister and Rodriguez 2005). The new nomenclature is shown in Supplemental Table 1. It mirrors the recent renaming of the fish V2R-like genes as OlfC genes (cf. Alioto and Ngai 2006). In the phylogenetic comparison with other teleost chemosensory receptor families (see below) ora genes emerge as a monophyletic group.
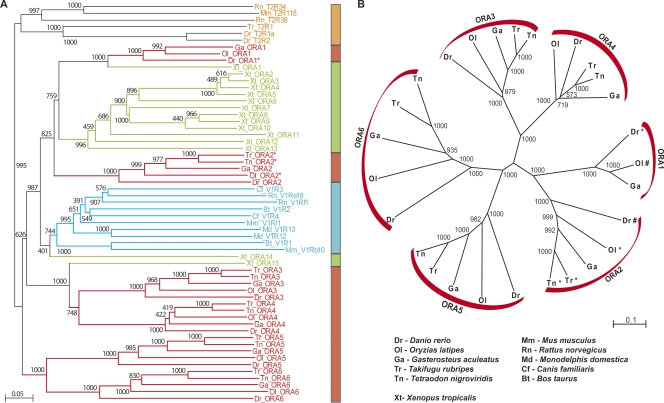
Figure 1.
Phylogenetic tree of the fish Ora family. (A) Twenty-eight fish Ora (red), 15 frog Ora (green) with some mammalian V1R (light blue) representatives and T2R (orange) as the closest relatives. (B) Twenty-eight fish ora genes. Trees were constructed using the NJ method. Bootstrap support (total 1000 replications) is indicated at the major nodes. Scale bar indicates the number of amino acid substitutions per site. *, see Pfister and Rodriguez 2005; #, see Shi and Zhang 2007. Ora, olfactory receptors of class A; T2R, putative taste receptors of type 2 (Ishimaru et al. 2005); V1R, vomeronasal type 1 receptors (Grus et al. 2005). The V1R receptors are a subset of V1Rs from all mammalian organisms annotated in the NCBI database (mouse, rat, human) and described in publications (opossum, cow, dog; Grus et al. 2005). The phylogenetic position of the full mammalian V1R set of annotated and published genes is identical (cf. Supplemental Fig. 1).
The ora genes form a single clade together with mammalian V1Rs
Using the T2Rs as an outgroup we compared the newly found genes to the most closely related chemosensory receptor families, the mammalian V1R genes (Fig. 1A). With respect to T2R, OR (Supplemental Fig. 1), and other chemosensory receptor gene families (T1R, OlfC; data not shown), all fish ora genes form a monophyletic clade, supporting their identification as a single family separate from the other chemosensory receptor families. The Ora clade includes all mammalian V1R receptors (Fig. 1A; Supplemental Fig. 1); thus, the Ora family can be considered paraphyletic, with the mammalian V1Rs originating as a single subclade within the Ora family. Both mammalian and fish taste receptors of the T2R family (Ishimaru et al. 2005) are phylogenetic neighbors, but nevertheless clearly segregate from the Ora family with high bootstrap values (Fig. 1A). Odorant receptor genes (ORs) are even more distant from the ora genes (Supplemental Fig. 1).
The six ora genes subdivide into three pairs, ora1–ora2, ora3–ora4, and ora5–ora6. In the phylogenetic tree these subclades are supported by maximal bootstrap values (Fig. 1B). Orthologs of the individual genes in all cases can be identified unambiguously, again with maximal bootstrap values (Fig. 1A). Conserved amino acids commonly are restricted to the orthologs of a single gene, but often a particular position is conserved in all orthologs of a gene pair (cf. Fig. 2 and Supplemental Fig. 2), consistent with the branchpoint pattern of the phylogenetic tree. Motifs conserved across two or all three gene pairs are comparatively rare, as detailed below.
Figure 2.
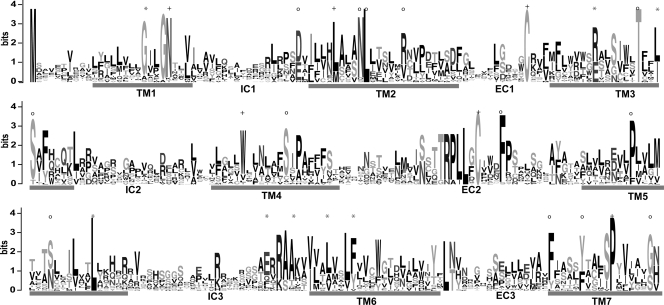
Conserved sequence motifs of the Ora family. Conservation of predicted amino acid sequence for the fish Ora repertoire is displayed as a sequence logo. In this representation, the relative frequency with which an amino acid appears at a given position is reflected by the height of its one-letter amino acid code in the logo, with the total height at a given position proportional to the level of sequence conservation. The regions corresponding to the transmembrane (TM) domains and the intracellular and intracellular domains (EC and IC) are numbered and indicated. Sequence alignments were manually edited (for details, see Methods). Of 14 motifs conserved in V1Rs (all of them single amino acids, identified by Rodriguez et al. 2002) eight are not conserved in ORs (cf. Niimura and Nei 2005) and consequently were chosen as analytical criterion here (*). (+) Residues conserved in ORs, some also in other GPCR families; (○) residues conserved in fish ora genes, but not in mammalian V1R genes.
Low overall similarity but high degree of conservation of motifs that are characteristic for mammalian V1Rs
The ora genes constitute a highly heterogeneous family, with homologies often as low as 15% and minimally 11% (Supplemental Table 2). We, therefore, analyzed the retention of characteristic sequence motifs in order to obtain a second line of evidence supporting the identification of ora genes both as a new family and as a V1R-related family.
Mammalian V1Rs are already quite divergent and comprise, e.g., in mice twelve distantly related subfamilies (Rodriguez et al. 2002). Consequently there are very few defining sequence motifs, all of them single amino acids, which are retained in nearly all family members. Most of these are highly conserved in the fish ora genes (Fig. 2; Supplemental Fig. 2), supporting the assignment as V1R-like genes. The degree of conservation is highest for the ora3–ora4 gene pair, slightly lower for the ora1–ora2 gene pair, and lowest (though clearly significant) for the ora5–ora6 gene pair, consistent with its larger distance from the mammalian V1Rs in the phylogenetic tree.
Beyond these motifs ora genes contain some general class A GPCR-specific motifs and 13 conserved amino acids, which are—with two exceptions—not conserved in either fish OR or T2R genes (Ishimaru et al. 2005; Niimura and Nei 2005), and thus distinguish the Ora family from both the odorant receptor and the taste receptor family (Fig. 2). Five of these motifs are conserved in all six ora genes, three in five of six genes, and five in two of three ora gene pairs (the differing gene pair is variantly ora1–ora2, ora3–ora4, or ora5–ora6). In mammalian V1Rs some of these motifs are weakly conserved, and others are not conserved at all, supporting the assignment of the fish ora genes as a family separate from, but related to, mammalian V1Rs.
ora genes precede teleost speciation
The presence of Ora orthologs in five distantly related fish species suggests the ancient evolutionary origin of this family. Orthologs are without exception closer related than paralogs (Figs. 1, 3; Supplemental Table 2). Thus, we conclude that all six Ora family members are evolutionarily older than the speciation events in the teleost lineage taken into account here. Moreover, for each ora gene the phylogenetic tree closely reflects the phylogenetic relationships of the species. The two pufferfish studied belong to the same family (Tetraodontidae) and their orthologs are related closer to each other than to those from any other species. The stickleback and medaka orthologs constitute their next neighbors, as pufferfish, stickleback, and medaka belong to three subdivisions of the same superorder Acanthopterygii (orders Tetraodontiformes, Gasterosteiformes, and Beloniformes, respectively). In all cases the zebrafish orthologs occupy the most distant position (zebrafish being the evolutionarily most distant fish in this comparison, as it is an Ostariophysi, not a Neoteleostei like the four other species). For zebrafish, Ora1 orthologs were found in stickleback and medaka, but not in the two pufferfish species. Since stickleback, medaka, and pufferfish belong to the same superorder Acanthopterygii, but zebrafish does not, this appears to be a case of gene loss in the pufferfish family, possibly related to the extreme reduction in genome size so characteristic of that family.
Figure 3.
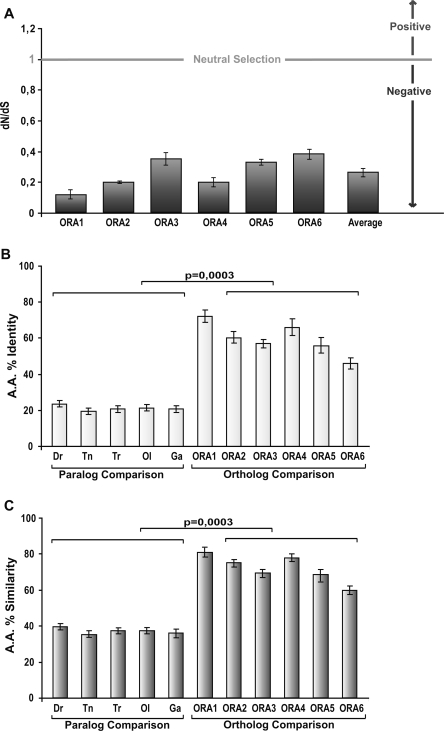
Identity, similarity, and conservation level of the ora genes. (A) dN/dS ratios of the six ora genes. For each gene, the dN/dS ratio was determined for all possible pairwise comparisons between orthologs and the mean value was plotted. (B) Amino acid percent identity is calculated for each gene comparisons between orthologs or species (paralog comparisons) by averaging the values of all possible pairwise comparisons inside the described group. (C) Amino acid % similarity is calculated for each gene or species by averaging the values of all possible pairwise comparisons inside the described group. (A–C) The bars correspond to the associated standard deviation as a measure of the variance within each group. (B,C) Differences between values for ortholog and paralog comparisons are highly significant.
The clear separation in three subclades (Fig. 1) suggests the presence of three ancestral genes, of which the corresponding phylogenetic level(s) remain to be elucidated.
Gene loss and gene gain upon transition to tetrapods
Mammalian V1R genes all belong to the Ora1–Ora2 clade, and all mammalian genes form a separate subtree within that clade (Fig. 1A; Supplemental Fig. 1; data not shown). These results are consistent with a loss of two clades (Ora3–Ora4 and Ora5–Ora6) somewhere after the teleost/tetrapod split, as well as a massive expansion of the remaining clade. Such a course of events is very reminiscent of the evolution within the OR gene family, where nearly all subfamilies present in teleosts disappeared in mammals, and exactly one subfamily underwent a massive expansion (Niimura and Nei 2005). To clarify the time scale of the Ora evolution we have searched the genome of an amphibian tetrapod for ora genes. In Xenopus tropicalis we found 15 ora genes in total, which belong to two of the three fish clades (Fig. 1; Supplemental Fig. 1; Supplemental Table 1). Clade Ora5–Ora6 is missing, and clade Ora3–Ora4 is only represented by a single gene, Xtora15. However, clade Ora1–Ora2 contains a single ortholog of ora1, another isolated gene (Xtora14) closest to the nodal point of origin of this clade, as well as a large expansion of highly related genes (Xtora2–13), well within the range of such expansions in mammalian species. Thus, clade Ora1–Ora2 may have existed already as gene pair in the most recent common ancestor (MRCA) of teleosts and tetrapods. Due to their phylogenetic position it appears appropriate to name the Xenopus genes as oras, since they intermingle with fish oras, but not with mammalian V1Rs (Fig. 1; Supplemental Fig. 1). Numbering begins with Xtora1, because the ortholog assignment is unambiguous only in this case; following numbers are given according to the position in the phylogenetic tree (cf. Fig. 1). With respect to gene loss, Xenopus represents an intermediate stage, but with respect to gene gain it resembles the mammalian situation.
We hypothesize that the partial gene loss might be due to the loss of a fully aquatic lifestyle in amphibians, and that the gene expansion is related to the transition to a terrestrial environment, consistent with a major shift in function of ora genes during this transition.
Strong negative selection for ora genes, but no evidence for positive selection
To better understand both the high degree of intraspecies variability and the high degree of interspecies conservation of ora genes we analyzed the evolutionary constraints that are acting on this gene family.
Paralog homology is usually below 25% amino acid identity and often as low as 15%, with average values for each species close to 20% (Fig. 3B; Supplemental Table 2). Most of this divergence is due to radical amino acid changes, since average similarity of paralogs is still <40% for all species (Fig. 3C). Homology within paralog gene pairs is somewhat higher than between them, with ora5 vs. ora6 comparisons always resulting in lower values than those obtained for the other two gene pairs, ora1 vs. ora2 and ora3 vs. ora4.
Ortholog homologies are much higher and in fact identity of any ortholog pair is higher than that of any paralog pair in all possible pairwise comparisons bar one (Supplemental Table 1). The average identity in all ortholog comparisons is 60%, with mean values for the individual ora genes ranging between 46% and 72%, and average ortholog similarities go up from 60% to 81%, with an average value for all ora genes of 72% (Fig. 3).
These values appear large enough (cf. Wolfe and Sharp 1993) to allow analysis of nonsynonymous (dN) vs. synonymous (dS) substitutions as a means to calculate the selective pressures acting on the ora genes. When the number of dN equals the number of dS, the dN/dS ratio equals 1, which corresponds to neutral selection. If the number of nonsynonymous changes is higher than the number of synonymous changes, then dN/dS > 1, which indicates positive selection. On the other hand, if the number of synonymous (dS) changes is higher than the number of nonsynonymous changes, then dN/dS < 1 and we are in the presence of negative selection (Nei and Gojobori 1986).
To avoid distortion of the dN/dS ratio by beginning saturation of synonymous substitutions (Gojobori 1983) the dS values should not exceed a certain value, differently given as 2 or 3 (Mank et al. 2007). We, therefore, analyzed the dS values for each of the pairwise comparisons separately and verified that 49% of the dS values (26 values) are <1.0, 32% (17 values) are between 1.0 and 1.5, 15% (eight values) are between 1.5 and 2.0, and only two values are slightly >2.0—the highest being 2.2 (data not shown). Since nearly no dS values are >2 and the vast majority is even <1.5, we assume that saturation of the synonymous substitutions does not distort the overall dN/dS calculations.
We observe a very low average dN/dS ratio for comparisons between orthologs (0.25), with values for individual genes ranging between 0.11 (ora1) and 0.37 (ora6) (Fig. 3A). All values clearly indicate strong negative selection, i.e., the ora genes are slowly evolving genes. A low dN/dS value of the ora genes together with a high divergence between ora genes indicates a very ancient origin of this slowly evolving gene family. This is drastically different from the properties of the mammalian V1R family, which is characterized by fast evolution and consequently highly species-specific gene repertoires. Incidentally, this difference in evolution rates may be related to the difference in pseudogene frequency: High numbers of pseudogenes are present in the V1R family (Zhang et al. 2004; Grus et al. 2005) but none were detected in the Ora family.
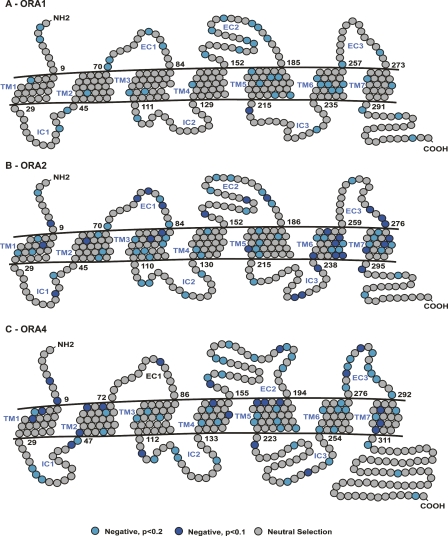
Since overall strong negative selection could mask positive selection at a few individual codon sites, we also analyzed the dN/dS ratio for each sequence position, using a manually optimized alignment of all orthologs for a particular ora gene. This analysis was performed separately for ora1, ora2, and ora4, the genes with the highest ortholog homologies. As expected from the summary dN/dS analysis, extended regions of the coding sequence show evidence for moderate to strong negative selection (Fig. 4). Moreover, no evidence for any positively selected site was found in any of these genes. A comparison between ora1, ora2, and ora4 shows rough similarity in the pattern of negative selection, although no specific motifs could be identified between genes (Fig. 4).
Figure 4.
Sites under positive and negative selection in ora coding sequences. A schematic representation of site-by-site selective pressure is shown on ora receptor sequences drawn based on the edited nucleotide alignment, which was generated from the corresponding amino acid alignment used for Fig. 3. SLAC analysis shows the probability of sites being under selective pressure (negative selection in light blue (P < 0.2) or blue (P < 0.1), neutral selection in gray, positive selection not observed even at P < 0.2 level. All orthologs of each gene were used for this analysis; the results for three genes are shown: (A) ora1, (B) ora2, (C) ora4.
Multiexonic structure of some ora genes is highly conserved between species
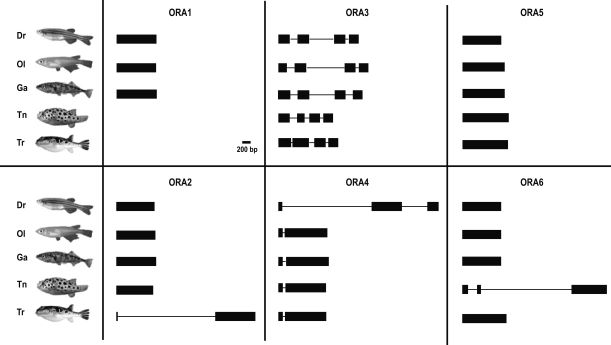
It is generally believed that all mammalian V1R genes possess a single exon structure (Dulac and Axel 1995; Saito et al. 1998; Rodriguez et al. 1999; Del Punta et al. 2000; Grus et al. 2005). It came, therefore, as quite a surprise that about half of the fish ora genes show a multi-exon structure in the coding region with two, three, or four exons (Fig. 5). A few introns occur only in one species: Tnora2 and Drora4 exhibit one additional intron, and Trora6 even two, compared to the exon structure in the remaining four species (Fig. 5). These isolated findings are best explained by late events in evolution, after teleost speciation. The ancestral genomic structure thus appears to be monoexonic for ora1, ora2, ora5, and ora6. This structure is maintained in the mammalian relatives of the Ora1–Ora2 clade (Dulac and Axel 1995; Grus et al. 2005).
Figure 5.
Genomic structure of the 28 fish ora genes. Predicted exon/intron structure is drawn to scale for all the ora genes: six zebrafish oras (Dr, Danio rerio), six medaka (Ol, Oryzias latipes), six stickleback (Ga, Gasterosteus aculeatus), five fugu (Tr, Takifugu rubripes), and five tetraodon (Tn, Tetraodon nigroviridis) oras. Exons are represented by the black filled rectangles and introns are represented by the black line connecting the exons.
In marked contrast, ora4 possesses two exons in four fish species (three in Danio rerio), and in all cases a small N-terminal exon is followed by a several-fold larger C-terminal exon. For ora3 four approximately equal-sized exons are predicted in all five fish species examined (Fig. 5). For the zebrafish representatives these exon predictions have been confirmed by sequencing all products of the RT-PCR analysis (Fig. 7A, see below; Supplemental Fig. 3). ora3 intron/exon borders are exactly conserved between all five teleost species, and the same holds true for ora4 (Supplemental Fig. 2). The sole intron/exon border in ora4 does not correspond to any intron/exon border in ora3. The striking conservation of particular intron/exon structures could indicate a role of noncoding elements in regulation of ora gene expression.
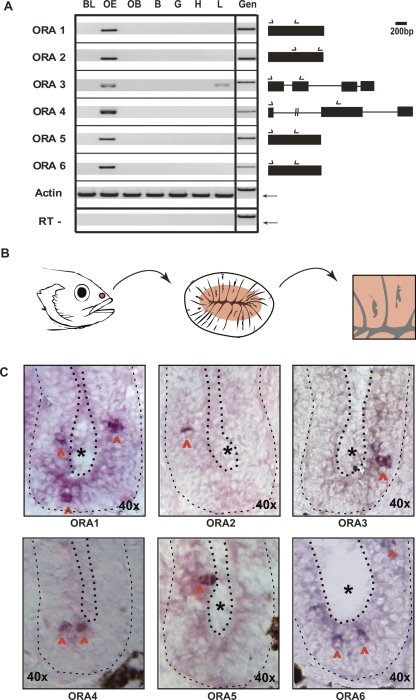
Figure 7.
Expression of ora transcripts in the zebrafish olfactory system. (A) Expression of ora mRNA detected by RT-PCR. PCR amplifications were performed by using gene-specific primers (arrows above the gene structure scheme). BL, barbels + lips; OE, olfactory epithelium; OB, olfactory bulb; B, brain; G, gills; H, heart; L, liver; Gen, genomic DNA. Actin, both plus and minus RT, and genomic DNA as template for all oras and actin were used as controls. The single actin band as well as the absence of actin amplification products in the “minus RT” condition confirm the absence of genomic DNA contamination. That genomic DNA, if present, would have generated a visible amplification product is shown in the lane labeled “Gen.” Gel sections shown all correspond to the 400–900 bp range, with exception of the much larger genomic product of ora4. The weak band with ora3 in liver cDNA might be due to minor ectopic expression, as has been reported for several olfactory receptor genes (e.g., Vanderhaeghen et al. 1993). Arrows in the actin rows point to the expected position for the cDNA product. (B) Schematic representation of the localization of the OE followed by a drawing of a horizontal section of OE (lamellae are cut perpendicular to their flat face) and finally an enlargement of two lamellae. The central rose-colored area in the lamellae indicates the location of the sensory neuroepithelium (cf. Weth et al. 1996), gray lines, basal lamina, gray jagged spots, lumen. (C) In situ hybridizations with ora1, ora2, ora3, ora4, ora5, and ora6 in horizontal sections of the OE, with antisense RNA probes. The area shown corresponds roughly to one half of the schematical representation in the right panel of B. The black asterisks indicate the lumen. Each half-lamella is enclosed by dashed lines, thicker in the apical region and thinner in the basal region adjoining the basal lamina. Red arrowheads point to the labeled neurons.
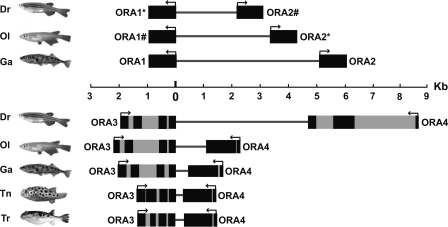
Four ora genes are arranged in closely linked gene pairs in head-to-head and tail-to-tail orientation
Between clades Ora1–2, Ora3–4, and Ora5–Ora6 there is no genomic linkage apparent. This holds true also for the gene pair ora5–ora6, as these two genes occur on different chromosomes or at least in different contigs in all species studied. However, the gene pairs ora1–ora2 and ora3–ora4 exhibit a striking pairwise arrangement with very short intergenic distance of a few kilobases, in the case of the pufferfish <1 kb (Fig. 6). The orientation is head-to-head (head means 5′ end) for the ora1–ora2 gene pair, and tail-to-tail (tail means 3′ end) for the ora3–ora4 gene pair. This pairwise arrangement and even the orientation are conserved without exception in all five teleost species examined (Fig. 6). Thus, a significance of this arrangement for regulation of expression may be assumed. This would result in an evolutionary constraint and consequently slower evolution than for the non-linked ora5, ora6 genes. Indeed, ortholog identities are always minimal for ora6, and nearly always minimal for ora5 compared to the remaining four genes (cf. Supplemental Table 2), consistent with an accelerated evolution of both ora5 and ora6.
Figure 6.
Genomic arrangement of the ora1–ora2 and ora3–ora4 gene pairs. Exons are represented by the black filled rectangles, introns are represented by the gray filled rectangles, and the thick line represents the intergenic distance between the two members of a gene pair. All elements are drawn to scale.
Specific expression of all ora genes in the olfactory epithelium
Any olfactory receptor is expected to be specifically expressed in olfactory receptor neurons situated in the olfactory epithelium. To test that prediction, we performed RT-PCR with seven different tissues and all zebrafish ora genes. All six genes were expressed specifically in the olfactory epithelium (Fig. 7A), supporting their assignment as olfactory receptors. None of the genes was expressed in the taste cell-containing tissues barbels and lips (Fig. 7A), confirming the segregation of the ora gene family from its closest phylogenetic neighbors, the T2R family of taste receptor genes. To analyze the expression of ora genes at the cellular level we performed in situ hybridization for all six ora genes. All ora genes are expressed in sparse cells confined to the sensory region of the zebrafish olfactory epithelium (Fig. 7B,C), consistent with their expression in olfactory receptor neurons.
Discussion
We have identified a novel olfactory receptor gene family in teleost fish, which we named ora for olfactory receptor genes related to class A GPCRs. The homologous mammalian V1R genes have not been formally assigned to any of the five major GPCR classes yet, but are most related to the class A or rhodopsin family of GPCRs (Schiöth and Fredriksson 2005; this study). All mammalian V1R genes belong to one of three clades within the Ora family. Previously a single V1R-related gene had been described in several teleost species (Pfister and Rodriguez 2005). The genes reported there correspond to ora1 (zebrafish) and ora2 (pufferfishes and medaka). The latest search for teleost V1R-related genes (Shi and Zhang 2007) yielded further orthologs of ora1 (medaka) and ora2 (zebrafish). ora3–ora6 presumably have been overlooked so far due to the low sequence identity between paralogs and their peculiar gene structure.
The specific expression of all Ora family members in the olfactory epithelium, and indeed in olfactory receptor neurons, as well as the relationship to mammalian V1R receptors support the identification of this novel family as olfactory receptors. Among olfactory receptor families the Ora family is unique for its small, rigidly maintained gene repertoire (no pseudogenes, no gene gains in five teleost species, only one gene loss in pufferfish), as well as the strict genomic arrangement in symmetrical gene pairs (for four of the six genes). Accordingly, we found extensive negative selection in the ora genes, but no evidence for positive selection, in striking contrast to the situation in the mammalian V1R family (Grus et al. 2005) and also the OR and V2R families (Niimura and Nei 2005; Alioto and Ngai 2006). While the function of the ora genes is currently unknown, these features of the family suggest a small set of evolutionarily rather conserved ligands. If these ligands are pheromones as in the related mammalian V1R family (Boschat et al. 2002), they may be expected to be less species-specific than is usual for pheromones. Among known fish pheromones are some reproductive hormones, which are additionally recognized by the olfactory system (Friedrich and Korsching 1998; Stacey et al. 2003). Such a double role engenders a double constraint, which could impede changes in the chemical structure of these pheromones during evolution. This hypothesis will be tested directly by analyzing the ligand binding properties of the novel ora gene family.
The evolutionary origin of the Ora family is not completely resolved. The presence of all six ora genes in zebrafish (an Ostariophysi, i.e., a rather primitive teleost) as well as in two more modern fish species (stickleback and medaka, both Neoteleostei) indicates their presence at least in Otocephala, while the existence of ora1, ora1–ora2, and ora3–ora4 orthologs in Xenopus would seem to suggest that the most recent common ancestor (MRCA) of tetrapods and teleosts already possessed the ora1–ora2 gene pair and at least one gene from the Ora3–Ora4 clade. Since in the phylogenetic analysis the Ora5–Ora6 clade appears to be at least as ancient as the other two clades, it may have been already present in the MRCA. In fact, the Ora family appears to be much more ancient than the actinopterygian/sarcopterygian split leading to teleosts and tetrapods, respectively, since a member of the Ora3–Ora4 clade is present already in the genome of a jawless vertebrate, Petromyzon marinus (L.R. Saraiva and S.I. Korsching, unpubl.). The phylogenetic tree would support two ancient large scale genome duplications to generate the three clades observed here from a single ancestral gene. These duplications could correspond to the two whole genome duplications 590 and 440 million years ago (Meyer and Schartl 1999), i.e., before the teleost and tetrapod lineages split, or, alternatively, one of these in combination with a later whole genome duplication, which occurred in the ray-finned fish lineage after the teleost/tetrapod split (cf. Hoegg et al. 2004).
The inverse orientation of the gene pair in the Ora1–Ora2 clade vs. the Ora3–Ora4 clade supports an independent origin of these two local duplication events. Thus the genesis of the three clades most likely preceded the genesis of the ora gene pairs, consistent with the phylogenetic tree. Whether the MRCA already contained both genes of the ora3–ora4 and the ora5–ora6 gene pairs cannot be decided with current data. Further evolutionary analysis with more ancestral species will be necessary to unequivocally resolve these issues.
In Xenopus the first example for the species-specific expansion of subfamilies so common in mammalian V1Rs (Grus et al. 2005) is seen, with 14 members in the Ora1–Ora2 clade, all but one within a single genomic cluster. It is assumed that such expansions are caused by recurrent gene duplications. With this pattern of gene gains Xenopus oras exhibit a feature common to all tetrapod olfactory receptor families and also to other teleost olfactory receptor families (Grus et al. 2005; Hashiguchi and Nishida 2005; Niimura and Nei 2005; Shi and Zhang 2007). The striking absence of any gene gains in teleost ora genes is consistent with the notion of a radical shift in the composition of the ligand repertoire during the teleost-tetrapod transition.
The genomic arrangement of the ora genes presented two unexpected findings. In contrast to all mammalian V1R genes several instances of multiexonic organization are found for the ora genes. A few occur only in one species and these isolated findings are best explained by late events in evolution, after teleost speciation. The ancestral genomic structure thus appears to be monoexonic for ora1, ora2, ora5, and ora6. This structure is maintained in the tetrapod relatives of the Ora1–Ora2 clade, the amphibian and mammalian genes (Saito et al. 1998; Rodriguez et al. 1999; Del Punta et al. 2000). In stark contrast, ora4 possesses two exons and for ora3 four exons are predicted in all five fish species examined. This strict conservation of intron-exon structure may indicate the existence of regulatory elements within these introns. The size distribution of the exons is rather conserved between species and intron/exon borders are accurately maintained.
Interestingly, none of these four intron/exon borders are found in the monoexonic Xenopus Ora3–Ora4 clade member (Xtora15; data not shown) nor in the intronless Ora3–Ora4 clade member found in Petromyzon marinus (L.R. Saraiva and S.I. Korsching, unpubl.). The most parsimonious explanation for the absence, presence, and location of the different intron/exon borders in ora3 vs. ora4 is an independent gain of introns posterior to the genesis of the ora3–ora4 gene pair, but prior to the teleost speciation taken into account here.
Several publications recently have demonstrated a reduction in the percentage of intron-containing genes in higher vertebrate GPCRs. This is supported by the evidence presented here. In teleost fish about one half of all ora genes contain introns, whereas in Xenopus no introns are present (data not shown), and in mammalian V1Rs no incidence of introns has been reported. However, the explanations given for this reduction appear only partially applicable to the ora/V1R group of genes. A loss of introns has been suggested by Bryson-Richardson et al. (2004), whereas the genesis of new, intronless genes in higher vertebrates has been emphasized by Fridmanis et al. (2007). The large expansion of the intronless Ora1–Ora2 clade in Xenopus is consistent with the interpretation by Fridmanis et al. (2007). We do not observe any intron loss in the Ora/V1R family; on the contrary, our data support the gain of four introns early in the teleost lineage and of further four introns at different later times, the latest gain restricted to within the pufferfish family. These intron gains in the slowly evolving teleost Ora family support the observation by Carmel et al. (2007), who found that such gains preferentially occur in evolutionarily conserved genes.
The other genomic feature novel for olfactory receptor genes is the occurrence of tightly linked symmetrical ora gene pairs, which are conserved throughout teleost evolution. The emergence of these gene pairs is not completely resolved, but the hypothesized whole genome duplications in the vertebrate lineage cannot be responsible, as the gene pairs are a local structure. In other cases of such gene pairs regulatory elements of one gene have been shown to lie in the other gene, enforcing linked evolution. Quite possibly some of these elements might even be shared among both genes of the pair (cf. Sumiyama et al. 2002), which would explain the maintenance of the symmetrical arrangement of the gene pairs (head-to-head and tail-to-tail). It is conceivable that initially ora5 and ora6 also occurred as such a gene pair, but that their association was degraded long before teleost speciation. Mutual dependency of expression for the gene pair would result in a slowed down evolution of the Ora1–Ora2 and Ora3–Ora4 clades compared to the Ora5–Ora6 clade, whose ortholog identities indeed tend to be distinctly lower than that of the other four genes. Conversely, the rapid gene expansion in the Xenopus Ora1–Ora2 clade predicts a loss of the gene pair arrangement in this species. Indeed, no pairwise arrangement is observed in this species (cf. Supplemental Table 1 and Supplemental Fig. 4). Even though 13 of the 15 Xenopus ora genes, including the ora1 ortholog, are clustered together in a 272-kb small genomic region, the ora genes nearest to ora1 are 35 and 45 kb apart, and all ora genes except the most distant one share the same orientation. Interestingly, for many genes in this cluster their genomic location correlates with their phylogenetic relationship (Supplemental Table 1; Fig. 1), consistent with a recurrent duplication of the cluster element most distant from ora1.
Taken together we have identified a novel family of six olfactory receptor genes in teleost fish. These genes are highly conserved between five evolutionarily distant fish species, in stark contrast to the frequent gene gains and gene losses seen in the related mammalian V1R family and other olfactory receptor families, both teleost and tetrapod.
Methods
Data mining
All annotated V1R sequences were extracted either from the National Center for Biotechnology Information (NCBI) database resources or from the articles in which they were first published (for cow, dog, frog, fish, and opossum genes) and used as query sequences in subsequent analysis. Two combined different strategies were used to search the databases for new V1R-like candidate genes in five fish species and in frog. First, the algorithm TBLASTN was applied to compare amino acid query sequences to the DNA databases (http://www.ensembl.org/index.html) with a nonstringent expectation cutoff value of 10−10; second and last, the automatically ortholog predicted genes in the fish species were retrieved from each of the query sequences.
To be considered as validated ora/V1R-like genes, a triage of the candidates was performed using different inclusion criteria. The confirmed ora/V1R-like genes were then included in subsequent analysis as new query sequences, until no new candidates were found.
The inclusion criteria used were (1) position within the Ora/V1R-like clade in the phylogenetic analysis; (2) application of the BLASTP algorithm in the NCBI nonredundant database should result in annotated V1Rs or some other Ora/V1R-like candidates as first hits (expectation cutoff value of 10−10); (3) presence of typical V1R family motifs; (4) CDS length between 850 and 1250 amino acids; (5) presence of seven trans-membrane domains (based on the consensus of the prediction results obtained by using the TMHMM: http://www.cbs.dtu.dk/services/TMHMM/ and TMpred: http://www.ch.embnet.org/software/TMPRED_form.html servers).
Duplicates genes were removed and the resulting genes were subjected to the analyses described below. The Xenopus tropicalis automatically annotated orthologs of the fish ora genes and mammalian V1Rs were also retrieved and included in the subsequent phylogenetic analysis. For GenBank accession numbers see Supplemental Table 1.
Phylogenetic analysis
MAFFT, version 5.8 (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/), was employed for multiple protein alignments using the E-INS-i strategy with the default parameters.
Phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei 1987) and the reliability of each tree node was assessed by the bootstrap method with 1000 replications. Alignment of the T2R and OR genes used as outgroup was manually verified for proper alignment of conserved residues common to these groups and the ora genes (cf. Fig. 2). Subclades within the ora gene family were determined from the tree as the largest clades that fulfilled two criteria: The clade had >50% bootstrap support and all members within the clade had at least 40% protein identity to each other. Six such subclades were identified, each containing all orthologs of a particular ora gene.
Identity and Similarity matrix
Pairwise alignments of the 28 fish Ora amino acid sequences were performed using the EMBOSS Pairwise Alignment Algorithm (http://www.ebi.ac.uk/emboss/align/) and both the Identity and Similarity values from all the possible comparisons were retrieved and used to make the matrix.
Sequence logos
Sequence logos were generated using a Web-based program, Weblogo, version 2.8.2., developed by http://weblogo.berkeley.edu/logo.cgi. A logo was generated with the 28 fish-specific Ora amino acid sequences representing full-length ORFs. Sequence alignments were manually edited using MEGA 3.1 (Kumar et al. 2004) and highly divergent pieces between the start codon and the beginning of TM1 and seven amino acids downstream from the conserved proline (P) in the TM7 were trimmed to avoid N- and C-terminal length heterogeneity. This did not affect significantly conserved residues. Gap positions present in >85% of the sequences were deleted completely.
dN/dS analysis
The global dN/dS ratios for the full-length ORF of the 28 fish ora receptor coding sequences were determined using the DnaSP 4.10 software package (Rozas et al. 2003), which implements previously published methods (Nei and Gojobori 1986). The nucleotide alignment was manually edited to match the amino acid alignment used in the phylogenetic trees and sequence logo.
To make inferences about selective pressure (positive and negative selection) on individual codons (sites) within the coding sequence of the 28 fish ora genes, the Single Likelihood Ancestor Counting (SLAC) package (http://www.datamonkey.org), which implements the Suzuki-Gojobori method (Suzuki and Gojobori 1999), was used.
The algorithm is briefly outlined. First, a best-fitting nucleotide substitution model was automatically selected by fitting several such substitution models to both the data and a neighbor-joining tree generated from the alignment described above. Taking the obtained substitution rates and branch lengths as constant, a codon model was employed to fit to the data and a global dN/dS ratio was calculated. Then a codon-by-codon reconstruction of the ancestral sequences was performed using maximum likelihood. Afterward, the expected normalized (ES) and observed numbers (EN) of synonymous (NS) and nonsynonymous (NN) substitutions were calculated for each nonconstant site. dN = NN/EN and dS = NS/ES were then computed, and if dN < dS (negative selection) or dN > dS (positive selection), a P-value derived from a two-tailed extended binomial distribution was used to assess significance. Tests on simulated data (S.L.K. Pond and S.D.W. Frost, methods available at http://www.datamonkey.org) show that P-values ≤0.1 identify nearly all true positives, with a false-positive rate generally below the nominal P-value; for actual data, the number of true positives at a given false-positive rate is lower.
In the present study, two thresholds for significance (0.1 and 0.2) were taken into account in order to identify residues potentially involved in odorant-binding activities.
RT-PCR
Ten zebrafish (mix of male and female Danio rerio, strain Ab/Tü) were dissected and the following tissues were pooled for each RNA extraction: barbels + lips (BL), olfactory epithelium (OE), olfactory bulb (OB), brain (B), gills (G), heart (H), and liver (L).
cDNA was generated by using standard protocols with an anchored oligo18(dT) reverse primer. PCR amplifications were performed by using the following primers: Dr_ actin (forward, CCCCATTGAGCACGGTATT; reverse, TCATGGAAGTCCACATG GCAGAAG), Dr ora1 (forward, ATGGACCTGTGTGTCACCAT CAAAGGCGT; reverse, TCATGGAAGTCCACATGGCAGAAG), Dr ora2 (forward, ATGATTGCGGAGGCTGTG; reverse, TCCACGTT GATGGCGTTC), Dr ora3 (forward, ATGGCGCCTCAAAAGA AACCC; reverse, AGATGAAGGCAGGGATGGAGT), Dr ora4 (forward, ATGTCTGAGGTCCTGACGGTG; reverse, GTGGTGCAGC TAATCACCATC), Dr ora5 (forward, ATGCAGCTCCAAGAC TGGGTT; reverse, GGAGTTGGGAATTTTTCCTCA), Dr ora6 (forward, ATGGTGATGGAGCAGATACAGGTGAATC; reverse, AG CACACTCGTCACCGTGA).
Regions chosen for PCR primers did not exhibit any appreciable sequence identity to each other, thereby excluding cross-amplification. Whenever possible, intron-spanning primer pairs were chosen. The following conditions were used: 3 min at 96°C, followed by 35 cycles of 30 sec at 96°C, 30 sec at 60°C, and 60 sec at 72°C, and a final extension of 10 min at 72°C. All the ora PCR fragments were cloned into pDRIVE (Qiagen) and confirmed by sequencing (Supplemental Fig. 3).
In situ hybridizations
The templates for the probes were amplified from cDNA cloned fragments obtained by RT-PCR with the T3 promoter site (TAT TAACCCTCACTAAAGGGAA) attached to the 5′ of the previously described primers. Digoxigenin (DIG) probes were synthesized according to the DIG RNA labeling kit supplier protocol (Roche Molecular Biochemicals). Sections were fixed in 4% paraformaldehyde for 10 min at room temperature. Hybridizations were performed overnight at 60°C using standard protocols. Anti-DIG primary antibody coupled to alkaline phosphatase (Roche Molecular Biochemicals) and NBT-BCIP (Roche Molecular Biochemicals) were used for signal detection.
Acknowledgments
We thank Dr. Franco Weth for advice and help with the in situ hybridizations and Mehmet Saltürk for taking good care of the fish. L.R.S. is supported by the International Graduate School for Genetics and Functional Genomics (IGSGFG). Means for the project came from the University of Cologne, the IGSGFG, and the Deutsche Forschungsgemeinschaft (grant to S.I.K.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6553207
References
- Alioto T.S., Ngai J., Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: Candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschat C., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Luscher C., Broillet M.C., Rodriguez I., Broillet M.C., Rodriguez I., Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson R.J., Logan D.W., Currie P.D., Jackson I.J., Logan D.W., Currie P.D., Jackson I.J., Currie P.D., Jackson I.J., Jackson I.J. Large-scale analysis of gene structure in rhodopsin-like GPCRs: Evidence for widespread loss of an ancient intron. Gene. 2004;338:15–23. doi: 10.1016/j.gene.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Carmel L., Rogozin I.B., Wolf Y.I., Koonin E.V., Rogozin I.B., Wolf Y.I., Koonin E.V., Wolf Y.I., Koonin E.V., Koonin E.V. Evolutionarily conserved genes preferentially accumulate introns. Genome Res. 2007;17:1045–1050. doi: 10.1101/gr.5978207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Punta K., Rothman A., Rodriguez I., Mombaerts P., Rothman A., Rodriguez I., Mombaerts P., Rodriguez I., Mombaerts P., Mombaerts P. Sequence diversity and genomic organization of vomeronasal receptor genes in the mouse. Genome Res. 2000;10:1958–1967. doi: 10.1101/gr.10.12.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C., Axel R., Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Fridmanis D., Fredriksson R., Kapa I., Schioth H.B., Klovins J., Fredriksson R., Kapa I., Schioth H.B., Klovins J., Kapa I., Schioth H.B., Klovins J., Schioth H.B., Klovins J., Klovins J. Formation of new genes explains lower intron density in Rhodopsin G protein-coupled receptors. Mol. Phylogenet. Evol. 2007;43:864–880. doi: 10.1016/j.ympev.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Friedrich R.W., Korsching S.I., Korsching S.I. Chemotopic, combinatorial, and noncombinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J. Neurosci. 1998;18:9977–9988. doi: 10.1523/JNEUROSCI.18-23-09977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T. Codon substitution in evolution and the “saturation” of synonymous changes. Genetics. 1983;105:1011–1027. doi: 10.1093/genetics/105.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus W.E., Shi P., Zhang Y.P., Zhang J., Shi P., Zhang Y.P., Zhang J., Zhang Y.P., Zhang J., Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl. Acad. Sci. 2005;102:5767–5772. doi: 10.1073/pnas.0501589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y., Nishida M., Nishida M. Evolution of vomeronasal-type odorant receptor genes in the zebrafish genome. Gene. 2005;362:19–28. doi: 10.1016/j.gene.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Herrada G., Dulac C., Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Hoegg S., Brinkmann H., Taylor J.S., Meyer A., Brinkmann H., Taylor J.S., Meyer A., Taylor J.S., Meyer A., Meyer A. Phylogenetic timing of the fish-specific genome duplication with the diversification of teleost fish. J. Mol. Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Okada S., Naito H., Nagai T., Yasuoka A., Matsumoto I., Abe K., Okada S., Naito H., Nagai T., Yasuoka A., Matsumoto I., Abe K., Naito H., Nagai T., Yasuoka A., Matsumoto I., Abe K., Nagai T., Yasuoka A., Matsumoto I., Abe K., Yasuoka A., Matsumoto I., Abe K., Matsumoto I., Abe K., Abe K. Two families of candidate taste receptors in fishes. Mech. Dev. 2005;122:1310–1321. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M., Tamura K., Nei M., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lane R.P., Young J., Newman T., Trask B.J., Young J., Newman T., Trask B.J., Newman T., Trask B.J., Trask B.J. Species specificity in rodent pheromone receptor repertoires. Genome Res. 2004;14:603–608. doi: 10.1101/gr.2117004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J.E., Axelsson E., Ellegren H., Axelsson E., Ellegren H., Ellegren H. Fast-X on the Z: Rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H., Buck L.B., Buck L.B. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Meyer A., Schartl M., Schartl M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Niimura Y., Nei M., Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister P., Rodriguez I., Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc. Natl. Acad. Sci. 2005;102:5489–5494. doi: 10.1073/pnas.0402581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I., Feinstein P., Mombaerts P., Feinstein P., Mombaerts P., Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez I., Del Punta K., Rothman A., Ishii T., Mombaerts P., Del Punta K., Rothman A., Ishii T., Mombaerts P., Rothman A., Ishii T., Mombaerts P., Ishii T., Mombaerts P., Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat. Neurosci. 2002;5:134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J.C., Messeguer X., Rozas R., Sanchez-DelBarrio J.C., Messeguer X., Rozas R., Messeguer X., Rozas R., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Ryba N.J., Tirindelli R., Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saito H., Mimmack M.L., Keverne E.B., Kishimoto J., Emson P.C., Mimmack M.L., Keverne E.B., Kishimoto J., Emson P.C., Keverne E.B., Kishimoto J., Emson P.C., Kishimoto J., Emson P.C., Emson P.C. Isolation of mouse vomeronasal receptor genes and their co-localization with specific G-protein messenger RNAs. Brain Res. Mol. Brain Res. 1998;60:215–227. doi: 10.1016/s0169-328x(98)00183-1. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schiöth H.B., Fredriksson R., Fredriksson R. The GRAFS classifcation system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005;142:94–101. doi: 10.1016/j.ygcen.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Shi P., Zhang J., Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey N., Chojnacki A., Narayanan A., Cole T., Murphy C., Chojnacki A., Narayanan A., Cole T., Murphy C., Narayanan A., Cole T., Murphy C., Cole T., Murphy C., Murphy C. Hormonally derived sex pheromones in fish: Exogenous cues and signals from gonad to brain. Can. J. Physiol. Pharmacol. 2003;81:329–341. doi: 10.1139/y03-024. [DOI] [PubMed] [Google Scholar]
- Sumiyama K., Irvine S.Q., Stock D.W., Weiss K.M., Kawasaki K., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Irvine S.Q., Stock D.W., Weiss K.M., Kawasaki K., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Stock D.W., Weiss K.M., Kawasaki K., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Weiss K.M., Kawasaki K., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Kawasaki K., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Shimizu N., Shashikant C.S., Miller W., Ruddle F.H., Shashikant C.S., Miller W., Ruddle F.H., Miller W., Ruddle F.H., Ruddle F.H. Genomic structure and functional control of the Dlx3-7 bigene cluster. Proc. Natl. Acad. Sci. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Gojobori T., Gojobori T. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen P., Schurmans S., Vassart G., Parmentier M., Schurmans S., Vassart G., Parmentier M., Vassart G., Parmentier M., Parmentier M. Olfactory receptors are displayed on dog mature sperm cells. J. Cell Biol. 1993;123:1441–1452. doi: 10.1083/jcb.123.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weth F., Nadler W., Korsching S., Nadler W., Korsching S., Korsching S. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc. Natl. Acad. Sci. 1996;93:13321–13326. doi: 10.1073/pnas.93.23.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Sharp P.M., Sharp P.M. Mammalian gene evolution: Nucleotide sequence divergence rat. J. Mol. Evol. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- Zhang X., Rodriguez I., Mombaerts P., Firestein S., Rodriguez I., Mombaerts P., Firestein S., Mombaerts P., Firestein S., Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]