Abstract
MHC class II transactivator (CIITA), a co-activator that controls MHC class II (MHC II) transcription, functions as the master regulator of MHC II expression. Persistent activity of the CIITA type III promoter (pIII), one of the four potential promoters of this gene, is responsible for constitutive expression of MHC II by B lymphocytes. In addition, IFN-γ induces expression of CIITA in these cells through the type IV promoter (pIV). Positive regulatory domain 1-binding factor 1 (PRDI-BF1), called B lymphocyte-induced maturation protein 1 (Blimp-1) in mice, represses the expression of CIITA pIII in plasma and multiple myeloma cells. To investigate regulation of CIITA pIV expression by PRDI-BF1 in the B lymphocyte lineage, protein/DNA binding studies, and functional promoter analyses were performed. PRDI-BF1 bound to the IRF-E site in CIITA pIV. Ectopic expression of either PRDI-BF1 or Blimp-1 repressed this promoter in B lymphocytes. In vitro binding and functional analyses of CIITA pIV demonstrated that the IFN regulatory factor-element (IRF-E) is the target of this repression. In vivo genomic footprint analysis demonstrated protein binding at the IRF-E site of CIITA pIV in U266 myeloma cells, which express PRDI-BF1. PRDI-BF1β, a truncated form of PRDI-BF1 that is co-expressed in myeloma cells, also bound to the IRF-E site and repressed CIITA pIV. These findings demonstrate for the first time that, in addition to silencing expression of CIITA pIII in B lymphocytes, PRDI-BF1 is capable of binding and suppressing CIITA pIV.
Keywords: PRDI-BF1, Blimp-1, CIITA, B lymphocytes, multiple myeloma
1. Introduction
The MHC class II transactivator (CIITA) is the primary regulator of both constitutive and inducible expression of MHC class II (MHC II) (Steimle et al., 1993; Steimle et al., 1994; Chang et al., 1994). Interacting with specific DNA-binding and basal transcription factors in the cell nucleus, CIITA acts as a transcriptional co-activator of MHC II genes (Zika and Ting, 2005; Harton and Ting, 2000). To a lesser degree the expression of other genes that function in antigen presentation by MHC II, like invariant chain and DM, are also regulated by CIITA (Chang and Flavell, 1995; Kern et al., 1995; Chin et al., 1997; Westerheide et al., 1997; Nagarajan et al., 2002). As such, CIITA serves as an essential master switch for MHC II antigen presentation and immune responses.
The human CIITA gene, MHC2TA, is regulated primarily at the level of transcription utilizing at least three of four distinct promoters, designated from 5′ to 3′ upstream of the CIITA gene as promoter I–IV (Landmann et al., 2001; Ting and Trowsdale, 2002). Through the use of these multiple promoters, CIITA is regulated in a complex and cell-type specific manner. The CIITA type I promoter (pI) is primarily expressed in myeloid dendritic cells and macrophages cells (Muhlethaler-Mottet et al., 1997; Landmann et al., 2001; Pai et al., 2002). The function of the type II promoter is presently unknown. It is not conserved across species and is only minimally active in human cells (Muhlethaler-Mottet et al., 1997). CIITA type III promoter (pIII) expression is important in B lymphocytes where it is constitutively active, but it can also be induced by IFN-γ stimulation in a number of different cell types (Nikcevich et al., 1997; Piskurich et al., 1999; Soos et al., 2001; Pai et al., 2002; van der Stoep et al., 2002; Nagabhushanam et al., 2003). The CIITA type IV promoter (pIV) is responsive to IFN-γ in many non-professional antigen-presenting cell types and is considered to be the main IFN-γ-inducible promoter of CIITA (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1998; Dong et al., 1999; Piskurich et al., 1999; Soos et al., 2001; Waldburger et al., 2001). Importantly, we have recently shown that CIITA pIV is also active and regulated by IFN-γ in B lymphocytes (Piskurich et al., 2005).
CIITA pIV has multiple, highly conserved upstream regulatory elements that control its activity. These include an IFN-γ activation factor DNA-binding sequence (GAS) element, E-Box, and IFN regulatory factor-element (IRF-E) (Muhlethaler-Mottet et al., 1997; Muhlethaler-Mottet et al., 1998). Several mechanisms have been described for CIITA pIV activation by IFN-γ, including the binding of STAT1 to the GAS site, the binding of the IFN regulatory factors (IRFs) IRF-1 and IRF-2 to the IRF-E, and the binding of USF-1 to the E box (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1999; Xi et al., 1999; Morris et al., 2002; Xi and Blanck, 2003).
Positive regulatory domain I-binding factor 1(PRDI-BF1), called B lymphocyte-induced maturation protein 1 (Blimp-1) in mice, acts as a transcriptional repressor in a variety of cell types (Kakkis and Calame, 1987; Kakkis et al., 1989; Lin et al., 1997). This repressor plays an important role in B lymphocyte development since it triggers the terminal differentiation of B cells into plasma cells (Turner, Jr. et al., 1994; Shaffer et al., 2002; Shapiro-Shelef et al., 2003). A recent report indicates that during plasmacytic differentiation, this repressor might regulate more than 250 genes either directly or indirectly (Shaffer et al., 2002). However, only a few direct targets for PRDI-BF1/Blimp-1 have been identified so far including IFN-β, c-Myc, spi-B, Id3 and Pax-5 (Kelly et al., 1991; Lin et al., 1997; Lin et al., 2002; Shaffer et al., 2002). In addition, we have shown that PRDI-BF1/Blimp-1 suppresses constitutive expression of MHC II by repressing CIITA pIII in B lymphocytes and multiple myeloma cells (Piskurich et al., 2000; Ghosh et al., 2001). Although only a handful of direct targets of repression have been verified, a recent report indicates that the DNA binding site for Blimp-1 resembles sites recognized by IRF-1 and IRF-2 (Kuo and Calame, 2004). Since the CIITA pIV promoter contains an IRF-E sequence that binds both IRF-1 and IRF-2, it represents a good candidate for regulation by PRDI-BF1/Blimp-1 (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1999; Xi et al., 1999; Morris et al., 2002; Xi and Blanck, 2003). We have recently shown that this promoter is active and regulated by IFN-γ in B lymphocytes (Piskurich et al., 2006).
The purpose of this study is to investigate the ability of PRDI-BF1 to regulate the CIITA type IV promoter in cells of the B lymphocyte lineage. This is important since CIITA expression controls the ability of B lymphocytes to present antigen. We demonstrate for the first time that besides silencing constitutive CIITA expression by repressing the CIITA type III promoter, PRDI-BF1/Blimp-1 also suppresses the basal and inducible activity of the CIITA type IV promoter in these cells. These studies provide new evidence that PRDI-BF1 is an attractive target for the development of therapies to increase the ability of the immune system to recognize B cell neoplasms, especially multiple myeloma.
2. Materials and methods
2.1. Cell culture
The human B lymphocyte cell lines, Raji and CA46, and the human myeloma cell lines, U266 and NCI-H929, were grown according to American Type Culture Collection specifications in RPMI 1640 with 10% fetal calf serum, 2mM L-glutamine, and 100 U/ml penicillin and streptomycin.
2.2. Constructs
The human CIITA gene (MHC2TA) pIV luciferase reporter constructs have been described previously (Piskurich et al., 1999). The CIITA pIV and mIV luciferase reporter constructs were formerly called pIVCIITA.Luc and pmIRF.IVCIITA.Luc (contains the IRF-E site mutations, GAActTagAAG, with mutations designated by lowercase type). The pcDNA-PRDI-BF1 and PRDI-BF1β plasmids, which express the full-length and PRDI-BF1β isoforms respectively, have been described previously (Ghosh et al., 2001, Gyory et al., 2003). The pBJ-neo-Blimp-1 plasmids, kindly provided by Dr. K. Calame, have also been described previously (Lin et al., 1997).
2.3. In vitro transcription and translation
The PRDI-BF1 and PRD-BF1β proteins were generated using the TnT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer’s specifications. Briefly, 1 μg of plasmid DNA, 40 μl of TnT Quick Mix and 2 μl of methionine were mixed, and the total volume was adjusted to 50 μl with nuclease-free water. The mixture was incubated at 30 C for 90 min. The pcDNA3.1 vector plasmid (Invitrogen) was used as a negative control.
2.4 Transfections and luciferase assays
Raji cells were transiently transfected by electroporation and luciferase assays were performed as previously described (Piskurich et al., 1998) with the following modification: 6 h after electroporation, cells were treated with human recombinant IFN-γ (500 U/ml) for 24 h and then harvested. For all transfection experiments, co-transfections were also performed with the pGL2-Basic plasmid (Promega) as a luciferase reporter negative control. No induction or repression of this plasmid was observed. Luciferase activity was measured using a Tropix TR717 luminometer (PE Applied Biosystems); as an internal control, each transfection included 300 ng of the pRL-TK Vector (Promega), which directs the expression of Renilla luciferase. Renilla luciferase activity was measured using the Renilla Luciferase Assay System (Promega) according to the manufacturer’s protocol and used to normalize the luciferase activity for transfection efficiency.
2.5. Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared from 2 × 106 cells according to the manufacturer’s directions using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pierce). The WT CIITA pIV oligonucleotide probe was as follows: 5′-CTGCAGAAAGAAAGTGAAAGGGAAAAAGAACT-3′. EMSAs were performed as previously described with the following modifications: EMSAs using antibodies were performed in a reaction mixture that contained 50 mM KCl, 5 mM NaCl, 5% glycerol, 10 mM Tris (pH 7.9), 1.5 mM MgCl2, 1 mM dithiothreitol, and 3 μg of poly (dI)(dC) (Piskurich et al., 1999); EMSAs using cold competitors were performed in a reaction mixture contained 80 mM NaCl, 10% glycerol, 20 mM Tris (pH 7.5) and 0.5 μg of sheared salmon sperm DNA (Invitrogen) (Piskurich et al., 2000). Antibodies were incubated with nuclear extract or in vitro translated proteins in the reaction mixture for 30 min before addition of probe. The antibody to Blimp-1, which recognizes both the full-length and truncated forms of PRDI-BF1, and isotope control antibody were purchased from Novus. Antibodies to IRF-1 and IRF-2 were purchased from Santa Cruz Biotechnology. Oligonucleotide probes used in 200-fold molar excess as cold competitors were the Blimp-1-binding site of the c-Myc promoter, MYC, 5′-CGCGTACAGAAAGGGAAAGGACTAG-3′; CIITA pIV containing a mutation in the IRF-E site that disrupts both GAAAG sequence motifs, MUT CIITA pIV, 5′-CTGCAGAAAGAActTagAAGGGAAAAAGAACT-3′ (mutations shown in lowercase type); CIITA pIV containing a mutation in the 5′ GAAAG sequence motif, 5′MUT CIITA pIV, 5′-CTGCAGAAAGAActTGAAAGGGAAAAAGAACT-3′; CIITA pIV containing a mutation in the 3′ GAAAG sequence motif, 3′MUT CIITA pIV, 5′-CTGCAGAAAGAAAGTagAAGGGAAAAAGAACT-3′.
2.6. In vivo genomic footprinting
The dimethyl sulfate (DMS) treatment, genomic DNA preparation, amplification of human CIITA (MHC2TA) pIV using ligation-mediated PCR, electrophoresis and visualization were performed as previously described (Piskurich et al., 1999). Three MHC2TA locus-specific primers were used to amplify cleaved fragments from the upper (coding) strand of CIITA pIV. Ligation-mediated PCR was also performed on the lower (noncoding) strand, but no significant protections or enhancements were observed.
3. Results
3.1 PRDI-BF1 binds to the CIITA type IV promoter
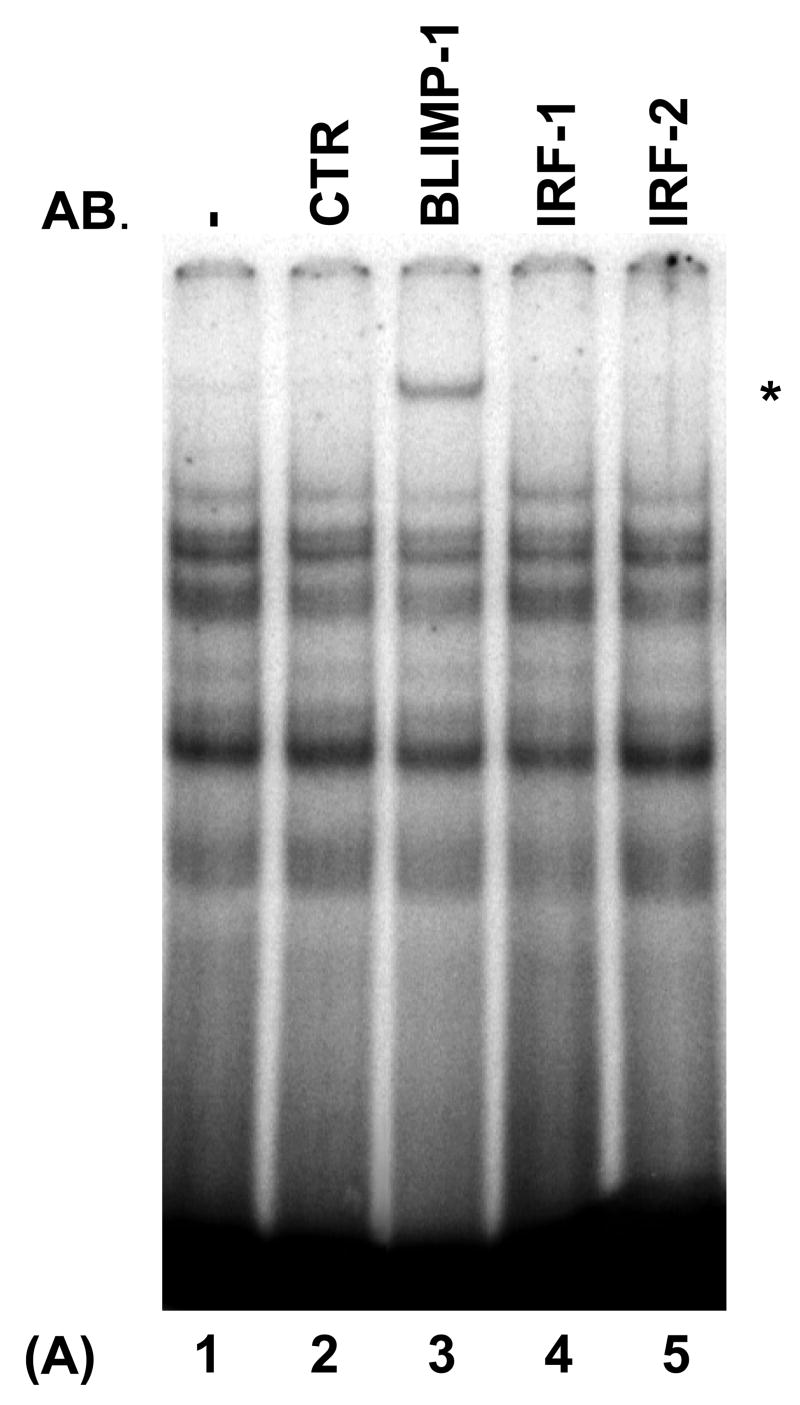
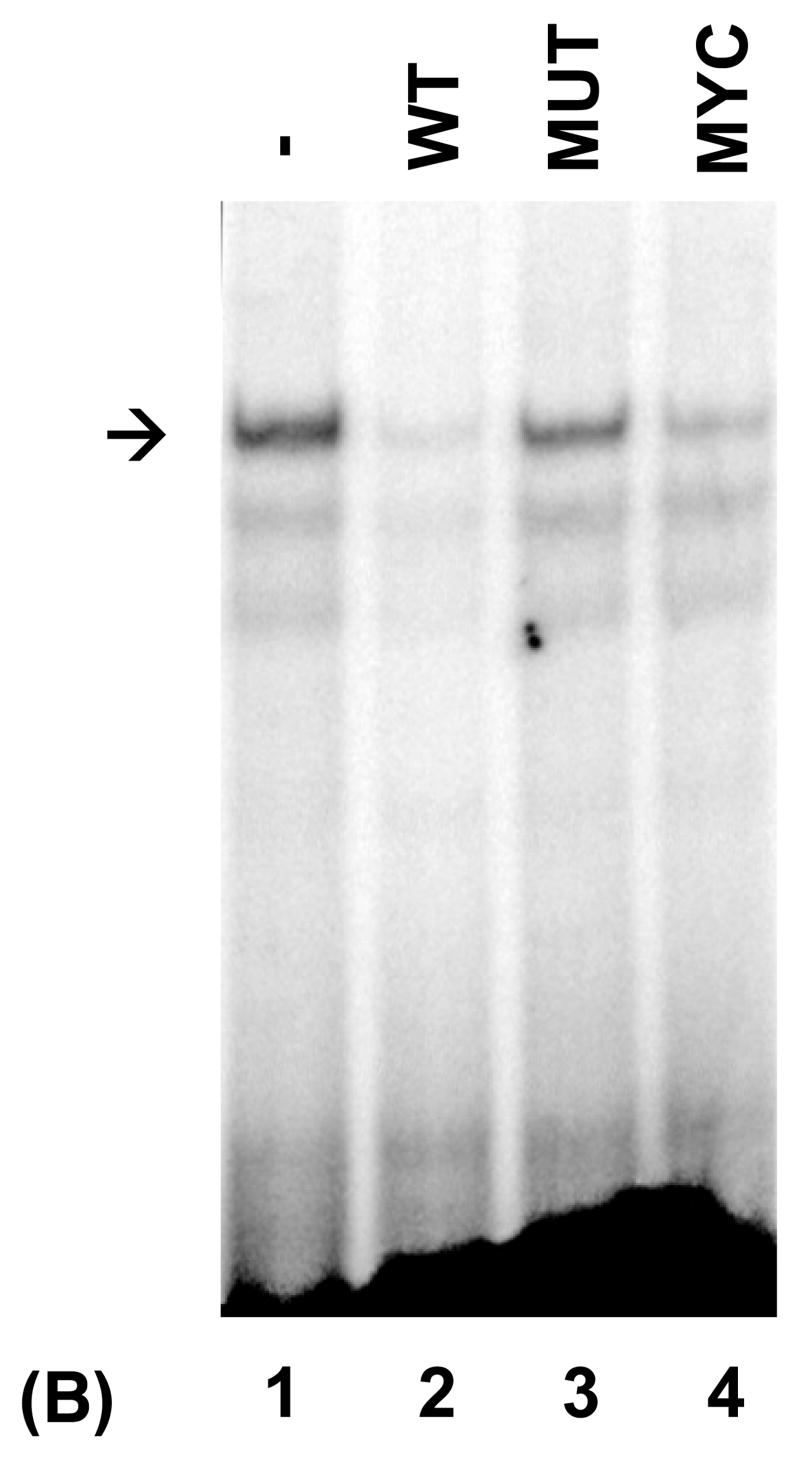
Recent evidence indicates that the PRDI-BF1/Blimp-1 DNA recognition sequence resembles the IRF-E sequences found in genes regulated by IRF-1 and IRF-2 (Kuo et al., 2004). To determine if the IRF-E site of the type IV promoter of the human CIITA gene (MHC2TA) is bound by PRDI-BF1, electrophoretic mobility shift analyses (EMSAs) were performed using nuclear extract from U266 myeloma cells that express PRDI-BF1 and a probe that spans sequences of the IRF-E site (from −77 to −30) of CIITA pIV. Multiple protein/DNA complexes were observed (Fig. 1A). To determine if these complexes contain PRDI-BF1, nuclear extracts were pre-incubated with a monoclonal antibody directed against Blimp-1 that is cross reactive with PRDI-BF1. Pre-incubation with this antibody clearly detected PRDI-BF1 within the complexes (Fig. 1A) (lane 3, asterisk). Consistent with the absence of IFN-γ stimulation, antibodies directed against IRF-1 or IRF-2 did not detect the presence of these factors within the complexes. To determine if PRDI-BF1 can directly bind to CIITA pIV, EMSAs were performed using recombinant PRDI-BF1 protein. As shown in Figure 1B, a single strong complex was formed. Specific cold competitors consisting of the wild-type CIITA pIV (WT) or of the known Blimp-1 binding site of the c-Myc promoter (MYC) diminish the formation of this complex (Fig. 1B, compare lane 1 with lanes 2 and 4). In contrast, it is not competed by a CIITA pIV competitor oligonucleotide with a site-specific mutation of the IRF-E site that disrupts both GAAAG sequence motifs (MUT) (compare lanes 1 and 3), which indicates that this complex is specific for the IRF-E. Lower faint bands are produced by truncated products in the in vitro translated PRDI-BF1 preparation. Similar results were seen in EMSAs using nuclear extracts from human U266 or NCI-H929 human myeloma cells (data not shown). These experiments demonstrate that PRDI-BF1 directly recognizes and binds to the IRF-E site of CIITA pIV.
Fig. 1.
PRDI-BF1 binds to the IRF-E site of the human CIITA (MHCIITA) type IV promoter. The probe spans the CIITA pIV IRF-E site. (A) EMSA analysis detects PRDI-BF1. Nuclear extract is from U266 human myeloma cells. Preincubation with a monoclonal antibody against Blimp-1, that detects both Blimp-1 and PRDI-BF1, induces a supershifted complex (lane 3, asterisk). Preincubation with an IgG1 isotype control (CTR), or with anti-IRF-1 or anti-IRF-2 antibodies does not result in a supershifted complex. Antibodies (AB.) are indicated at the top. (B) EMSA analysis using in vitro translated PRDI-BF1 protein. One specific complex is severely reduced by incubation with cold wild-type CIITA pIV oligonucleotide (WT) (lane 2, arrow) or oligonucleotide bearing the Blimp-1-binding site of the c-Myc promoter (MYC) (lane 4). This reduction is not observed using a CIITA pIV cold competitor oligonucleotide with a mutated IRF-E site (MUT) (lane 3).
3.2 PRDI-BF1/Blimp-1 represses the CIITA type IV promoter in B lymphocytes
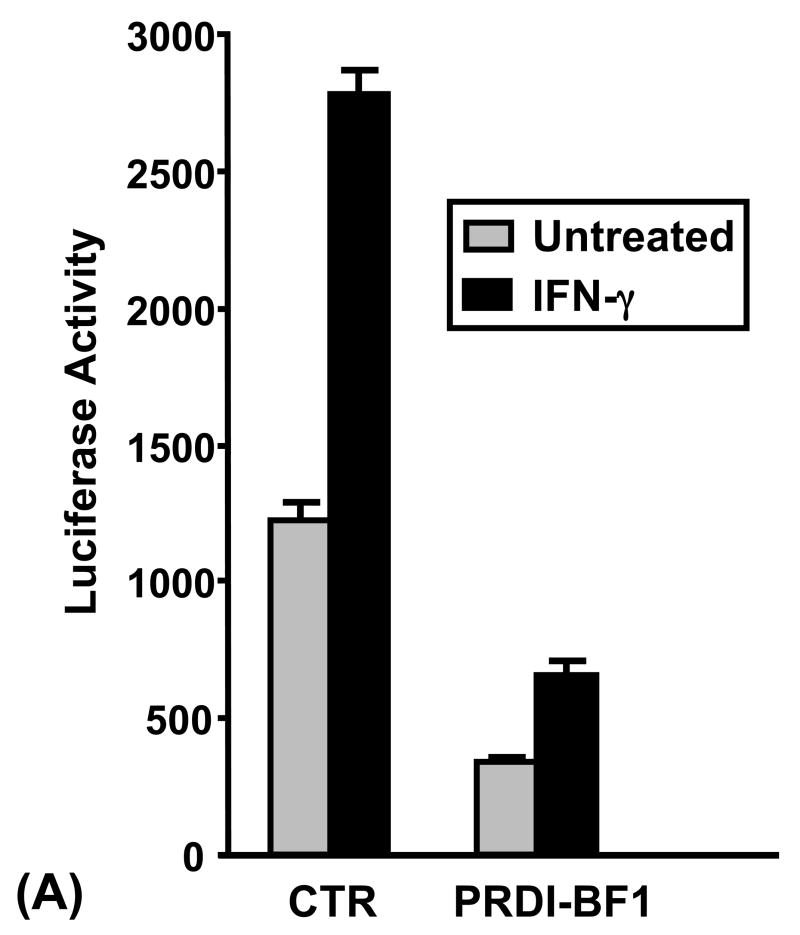
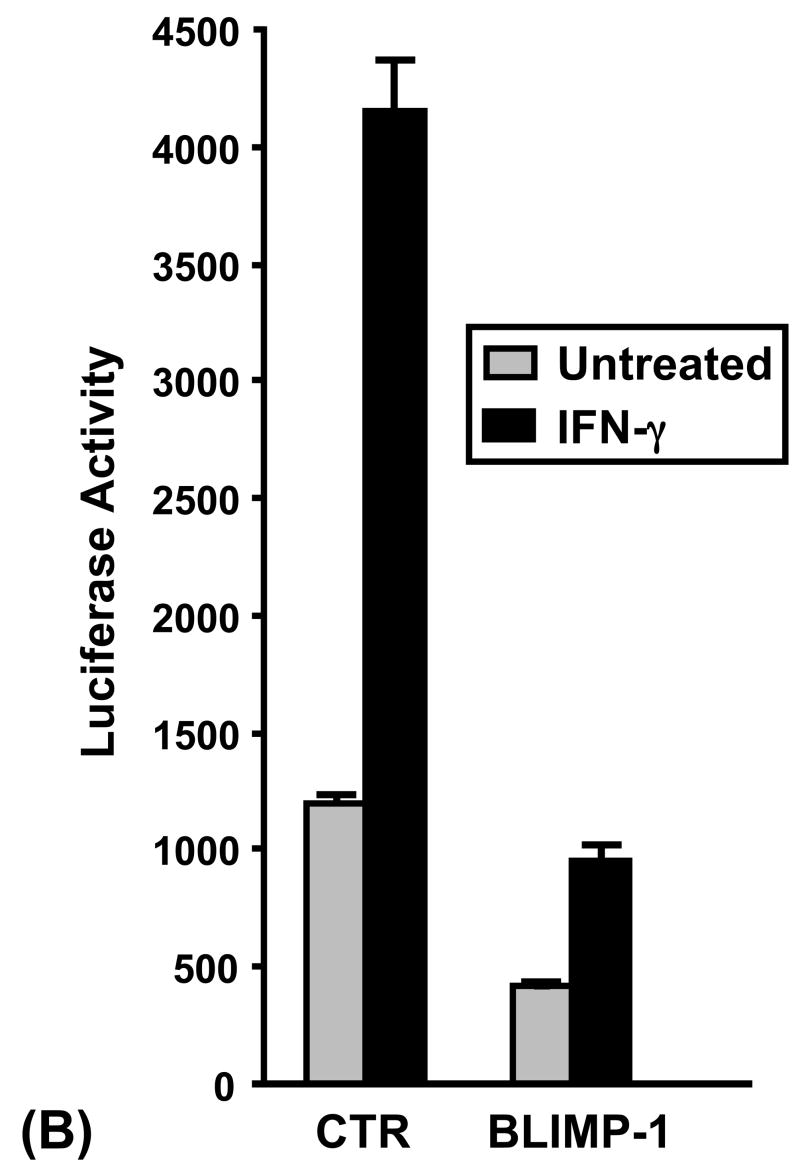
To functionally test whether CIITA pIV is repressed by PRDI-BF1, Raji human B cells were co-transfected by electroporation with a reporter plasmid where luciferase expression was under the control of CIITA pIV and a PRDI-BF1 expression plasmid. Ectopic expression of PRDI-BF1 repressed both the basal and IFN-γ-induced activity of CIITA pIV in B lymphocytes (Fig. 2A). Ectopic expression of PRDI-BF1 resulted in a 3–8 fold suppression (3.6-fold and 4.2-fold, respectively, for the representative experiment that is shown) of both basal and IFN-γ-inducible CIITA pIV activity. IFN-γ treatment resulted in over 2-fold induction of CIITA pIV activity for co-transfections with either the vector control plasmid or the PRDI-BF1 expression plasmid.
Fig. 2.
Expression of PRDI-BF1/Blimp-1 represses the CIITA type IV promoter. (A) Transient co-transfections of Raji B cells were performed by electroporation using 10 μg of the CIITA pIV luciferase reporter plasmid and 10 μg of the PRDI-BF1 expression plasmid or the empty vector control. After a 6 h recovery period, half of the cultures were treated with 500 U/ml of recombinant human IFN-γ (R&D Systems) for 24 h before before the cultures were harvested. Ectopic expression of PRDI-BF1 resulted in a 4-fold reduction in activity of both the treated and untreated samples. Luciferase activity was measured and normalized to Renilla luciferase activity. Bars show the S.E.M., n = 3. At least three independent experiments have been performed with similar results. (B) Co-transfections of Raji B cells were also performed as described above using 10 μg of the CIITA pIV luciferase reporter plasmid and 10 μg of the Blimp-1 expression or control plasmid. Ectopic expression of Blimp-1 resulted in a 3–4-fold reduction of activity in both the treated and untreated samples.
Previous studies for the human CIITA type III promoter indicate that it is functionally repressed across species by both the human, PRDI-BF1, and the murine, Blimp-1, forms of the repressor (Piskurich et al., 2000). To determine if this is the case for the type IV promoter, co-transfections of Raji cells were performed using the CIITA pIV luciferase reporter plasmid and an expression plasmid for Blimp-1. Consistent with the finding for PRDI-BF1, a similar fold induction of CIITA pIV activity by IFN-γ was observed for both the control and Blimp-1 expression plasmids (Fig. 2B). In addition, Blimp-1 over expression resulted in suppression of basal and inducible CIITA pIV activity, comparable to the suppression seen with PRDI-BF1. These results demonstrate that, like CIITA pIII, CIITA pIV is repressed by both the human and murine forms of this repressor in B lymphocytes.
3.3 Repression and binding of the CIITA type IV promoter by PRDI-BF1 requires sequences within the IRF-E site
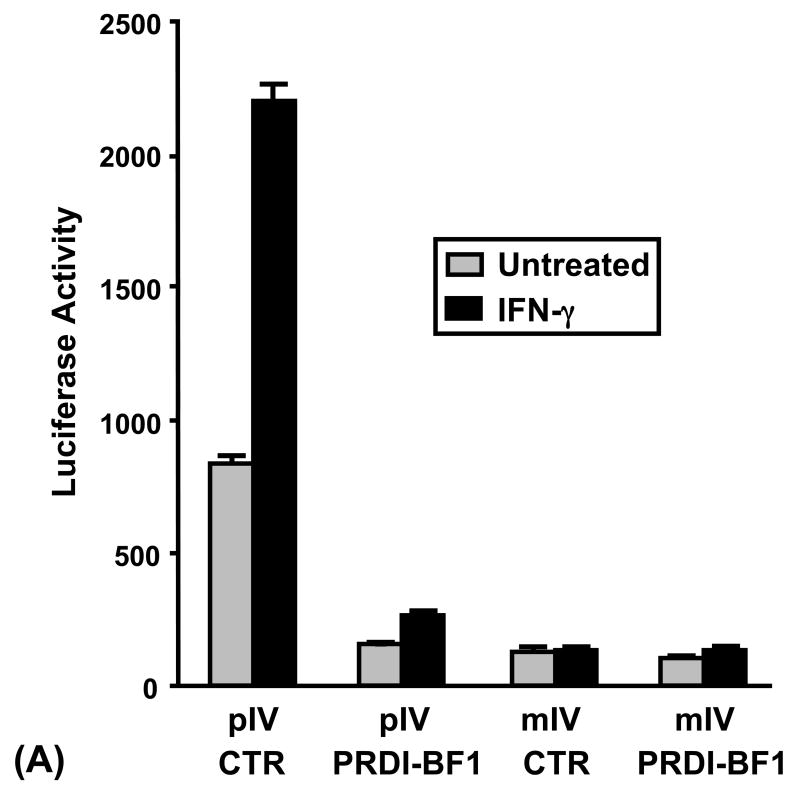
EMSA binding competition experiments performed with a CIITA pIV oligonucleotide containing a mutated IRF-E site did not diminish the specific complex that was formed using a wild-type CIITA pIV probe (Fig. 1B). To functionally determine if this IRF-E site is required for the repression of CIITA pIV by PRDI-BF1, Raji B cells were co-transfected with a PRDI-BF1 expression plasmid and a CIITA pIV luciferase reporter plasmid with an identical mutation of the IRF-E site. Ectopic expression of PRDI-BF1 did not result in a change in the basal or inducible activity of this mutated CIITA promoter (Fig. 3A). The activity of the wild-type CIITA pIV reporter plasmid is shown for comparison. We have previously shown in both B cells and 2fTGH fibrosarcoma cells that IFN-γ does not induce the activity of this mutated promoter and that the basal level of activity is lower compared to the wild-type promoter (Piskurich et al., 2005, and data not shown). These experiments provide additional support for the idea that PRDI-BF1 represses the activity of CIITA pIV by binding to sequences contained in the IRF-E site of this promoter.
Fig. 3.
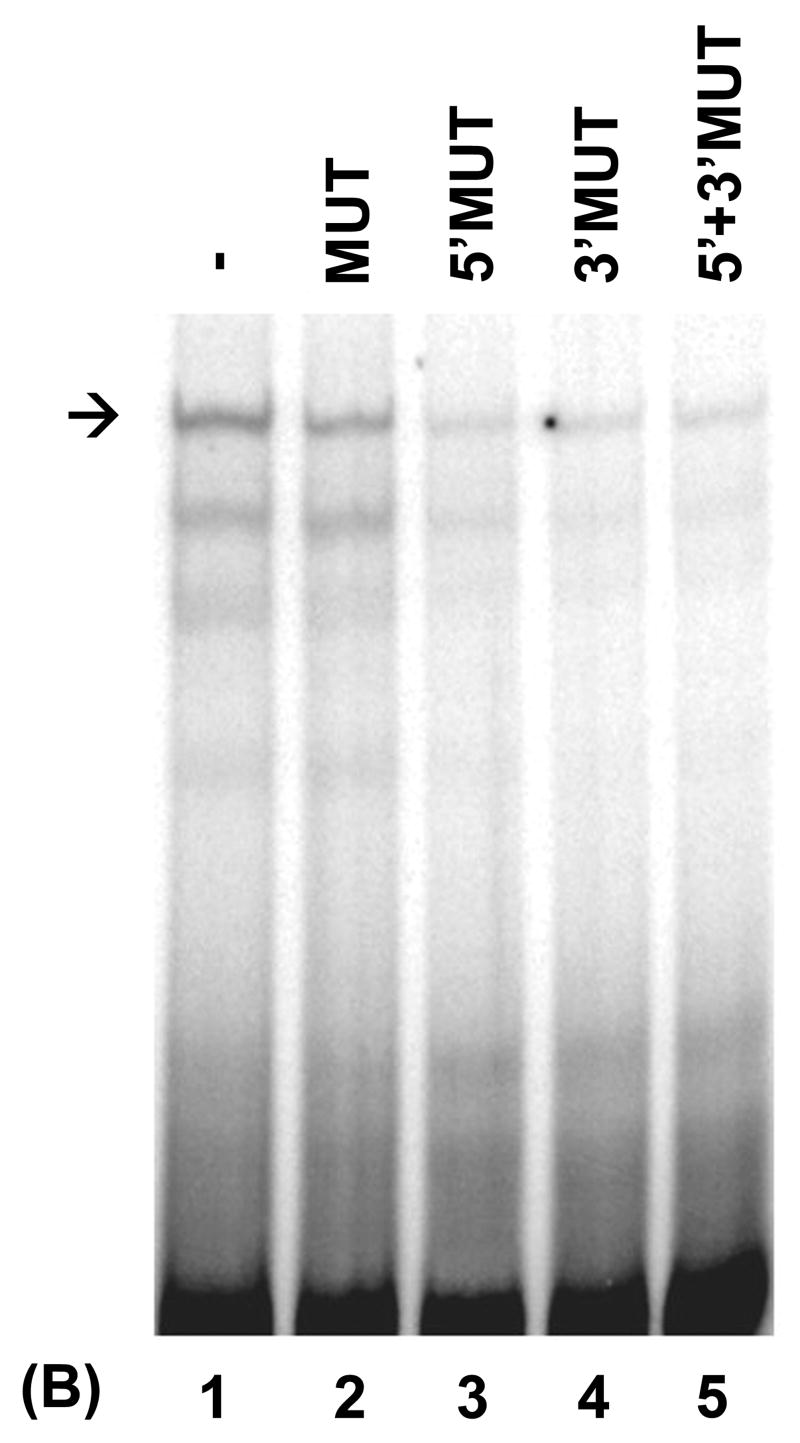
Repression of the CIITA type IV promoter by PRDI-BF1 requires sequences within the IRF-E site. (A) Raji B cells were transiently co-transfected by electroporation using 10 μg of the wild-type or mutated CIITA pIV luciferase reporter plasmids and 10 μg of the PRDI-BF1 expression plasmid or the empty vector control. After a 6 h recovery period, half of the cultures were treated with 500 U/ml of recombinant human IFN-γ for 24 h before the cultures were harvested. Luciferase activity was measured and normalized to Renilla luciferase activity. Bars show the S.E.M., n = 3. At least three independent experiments have been performed with similar results. (B) EMSA analysis performed using wild-type CIITA pIV probe, in vitro translated PRDI-BF1 protein and specific cold competitors. The cold competitors include a CIITA pIV oligonucleotide with a mutation that disrupts both GAAAG motifs in the IRF-E site (MUT), and oligonucleotides with mutations that disrupt the first (5′MUT) or second (3′MUT) GAAAG motif used alone or in combination. Incubation with either of the cold competitor oligonucleotides in which a single unmutated GAAAG motif is present is capable of diminishing the formation of the specific complex (arrow), compare lanes 3 and 4 with lanes 1 and 2. No additional reduction is seen when both mutated oligonucleotides were used in combination (compare lane 5 with lanes 3 and 4).
Only a handful of gene targets for repression by PRDI-BF1/Blimp-1 have been verified (Kelly et al., 1991; Lin et al., 1997; Lin et al., 2002; Shaffer et al., 2002). Analysis of the promoter sequences of these genes and extensive binding analyses aimed at determining the consensus DNA-binding site for Blimp-1 indicate that this site resembles the IRF-E and contains a GAAAG sequence motif, except in the unique case of CIITA pIII (Kuo and Calame, 2004). The CIITA pIV IRF-E sequence, GAAAGTGAAAG, contains two GAAAG motifs. The IRF-E mutation used in the EMSAs and transfections described in this report so far disrupted both of these motifs. To further study the mechanism by which PRDI-BF1 regulates CIITA pIV, EMSA binding competition analyses were performed to determine which of the GAAAG motifs function in the binding of CIITA pIV by PRDI-BF1. These experiments used wild-type CIITA pIV as probe and in vitro translated PRDI-BF1 protein (Fig. 3B). The effects of specific cold competitors consisting of the CIITA pIV IRF-E site with mutations in both GAAAG motifs (MUT), or the CIITA pIV IRF-E site with a mutation in either the first (5′MUT) or second (3′MUT) motif were compared. Each of the oligonucleotides with a single mutated GAAAG motif is capable of diminishing the formation of a specific complex (Fig. 3B, compare lanes 3 and 4 with lanes 1 and 2). No additional reduction was seen when both mutated oligonucleotides were used in combination as competitors (compare lane 5 with lanes 3 and 4). These results provide additional support that the IRF-E site is required for binding and repression of CIITA pIV by PRDI-BF1 and demonstrate that both GAAAG motifs can participate in the binding of this repressor.
3.4 Genomic footprint analysis detects protein-DNA interactions at the IRF-E site of the CIITA type IV promoter in myeloma cells
In vivo genomic footprinting is the method of choice to identify physiologically relevant sequences within a small region of DNA such as the region surrounding the CIITA pIV IRF-E. DMS-treated genomic DNA was obtained from U266 myeloma cells and CA46 B cells. Significant protections indicating protein-DNA contacts were observed in U266 myeloma cells at both GAAAG sequence motifs in the IRF-E site (Fig. 4). These protections were not observed in cells that do not express PRDI-BF1 including Raji and CA 46 B cells (Piskurich et al., 2005, and data not shown). These findings are consistent with PRDI-BF1 binding to the CIITA pIV IRF-E in myeloma cells and lend support to the notion that both GAAAG motifs can participate in the binding of this repressor.
Fig. 4.
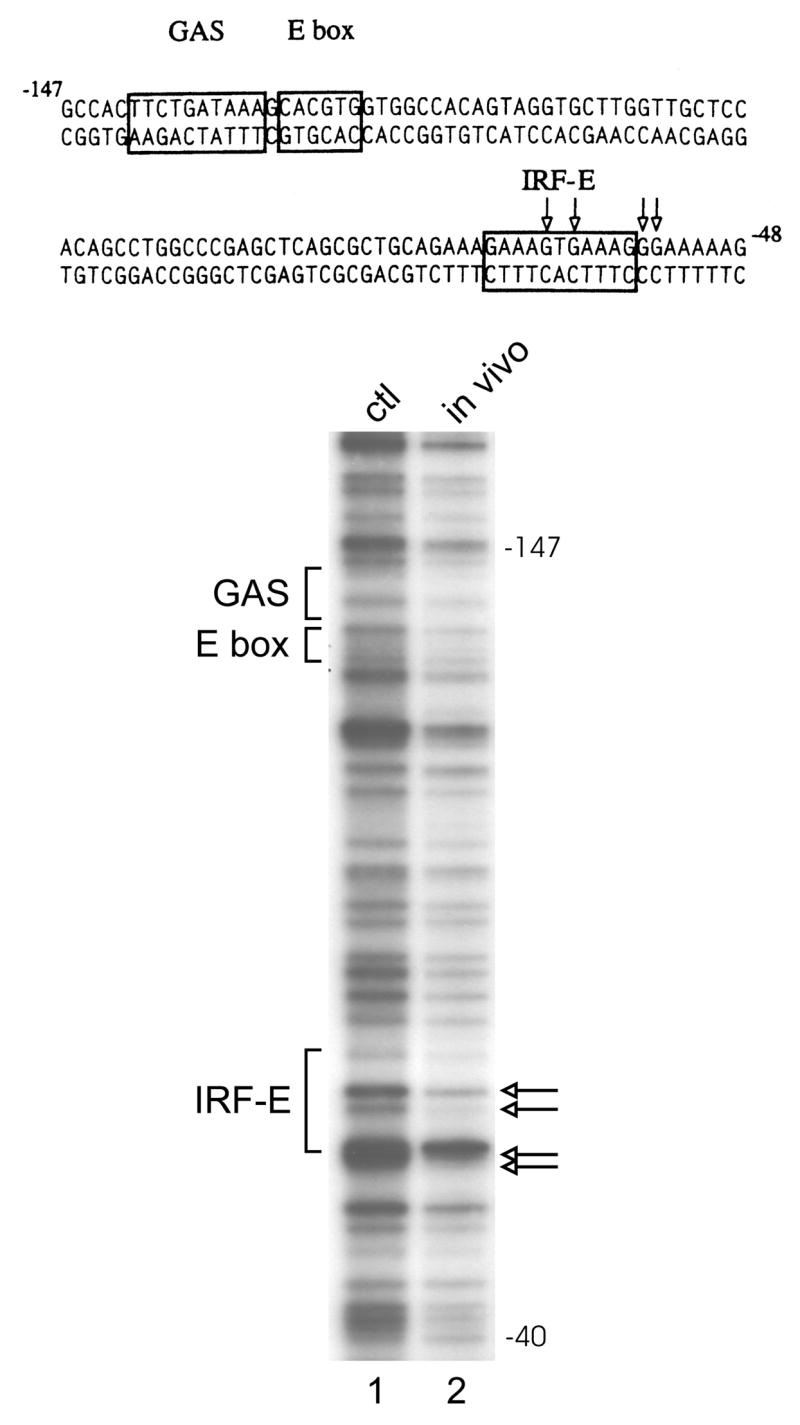
Genomic footprint analysis detects protein-DNA interactions at the IRF-E site of the CIITA type IV promoter in myeloma cells. The sequence of the promoter region is shown with boxes indicating the cis-regulatory elements that have been identified for this promoter, including the IRF-E. A genomic footprint of the upper strand is shown in the lower panel. Lane 1 shows genomic DNA that was methylated in vitro to show the complete guanine ladder. Lane 2 shows the results of U266 myeloma cells treated with DMS in culture. Open arrows indicate bases protected from modification. Four bases are protected in U266 cells compared to DNA methylated in vitro. These bases are not protected in DNA that was treated in vivo and isolated from CA46 or Raji B cells (see text). The protected bases lie within or adjacent to the IRF-E site (see arrows). No differences in protections were noted for the GAS or E box sites.
3.5 PRDI-BF1β, an alternative isoform of PRDI-BF1 expressed in myeloma cells, binds and represses the CIITA type IV promoter
An alternative protein product was recently described for the PRDI-BF1 gene, PRDM1 (Gyory et al., 2003). In addition to the full-length form of PRDI-BF1, myeloma cells co-express a protein isoform called PRDI-BF1β that arises from initiation of the transcription of this gene at an alternative site. Compared to the originally described full-length form of PRDI-BF1, the PRDI-BF1β isoform lacks 101 amino acids at the amino-terminus but retains the zinc finger DNA binding motif of this protein. To examine the ability of this isoform to bind CIITA pIV, EMSAs were performed using CIITA pIV as probe and in vitro translated PRDI-BF1β protein. One predominant complex was shifted by pre-incubation with anti-Blimp-1, which recognizes an epitope that is conserved in PRDI-BF1β (Fig. 5A) (lane 3, asterisk). The incomplete shifting of this band reflects the activity of the antibody to Blimp-1 and also occurs with the full-length form of PRDI-BF1. The wild-type CIITA pIV oligonucleotide (WT, lane 2) and an oligonucleotide bearing the Blimp-1 binding site of the c-Myc gene promoter (MYC, lane 4) greatly reduced this complex, while a CIITA pIV oligonucleotide bearing a mutated IRF-E did not (MUT, lane 3) (Fig. 5B). Oligonucleotides with mutations that disrupt either the first or second GAAAG motif also reduced this complex (Fig. 5C). These results demonstrate that the PRDI-BF1β isoform has a similar binding interaction with CIITA pIV compared to full-length PRDI-BF1.
Fig. 5.
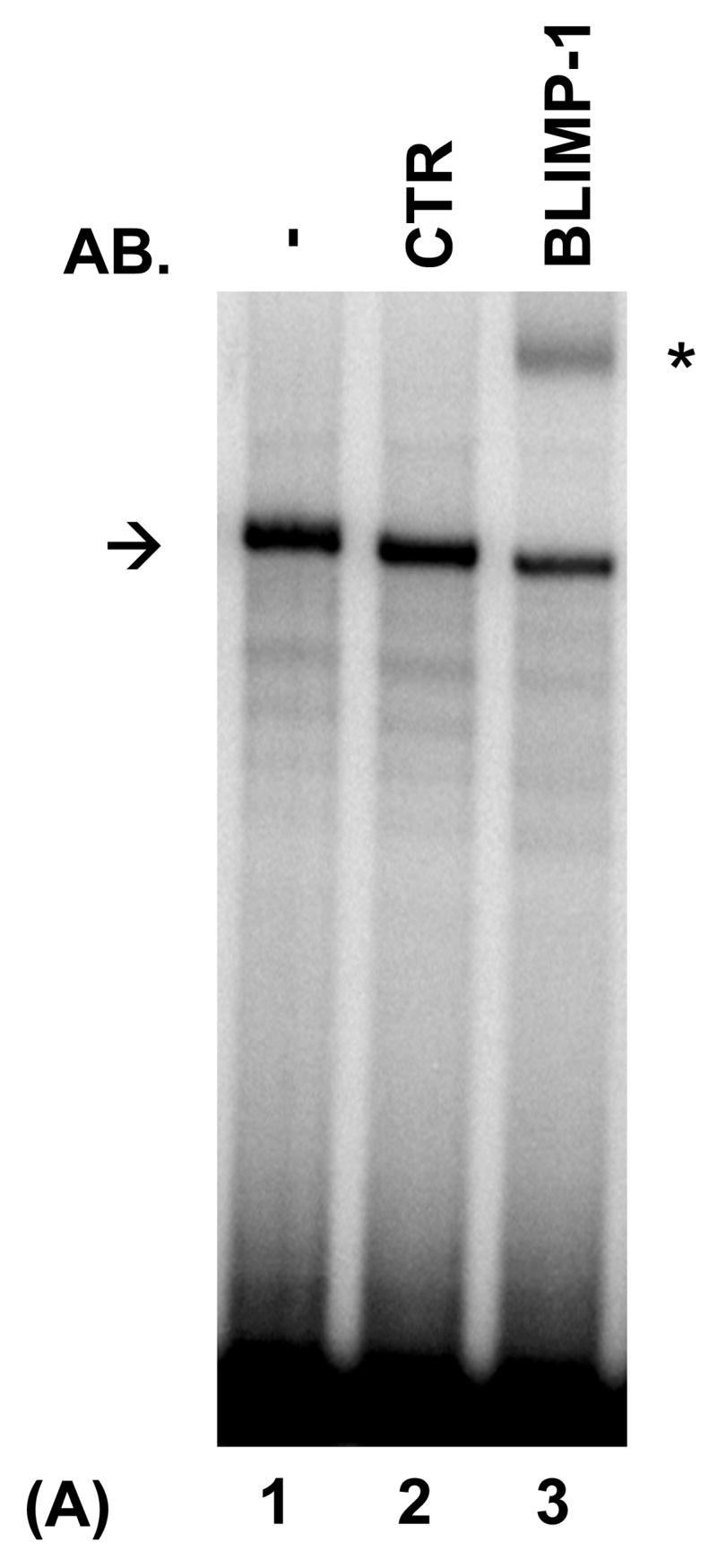
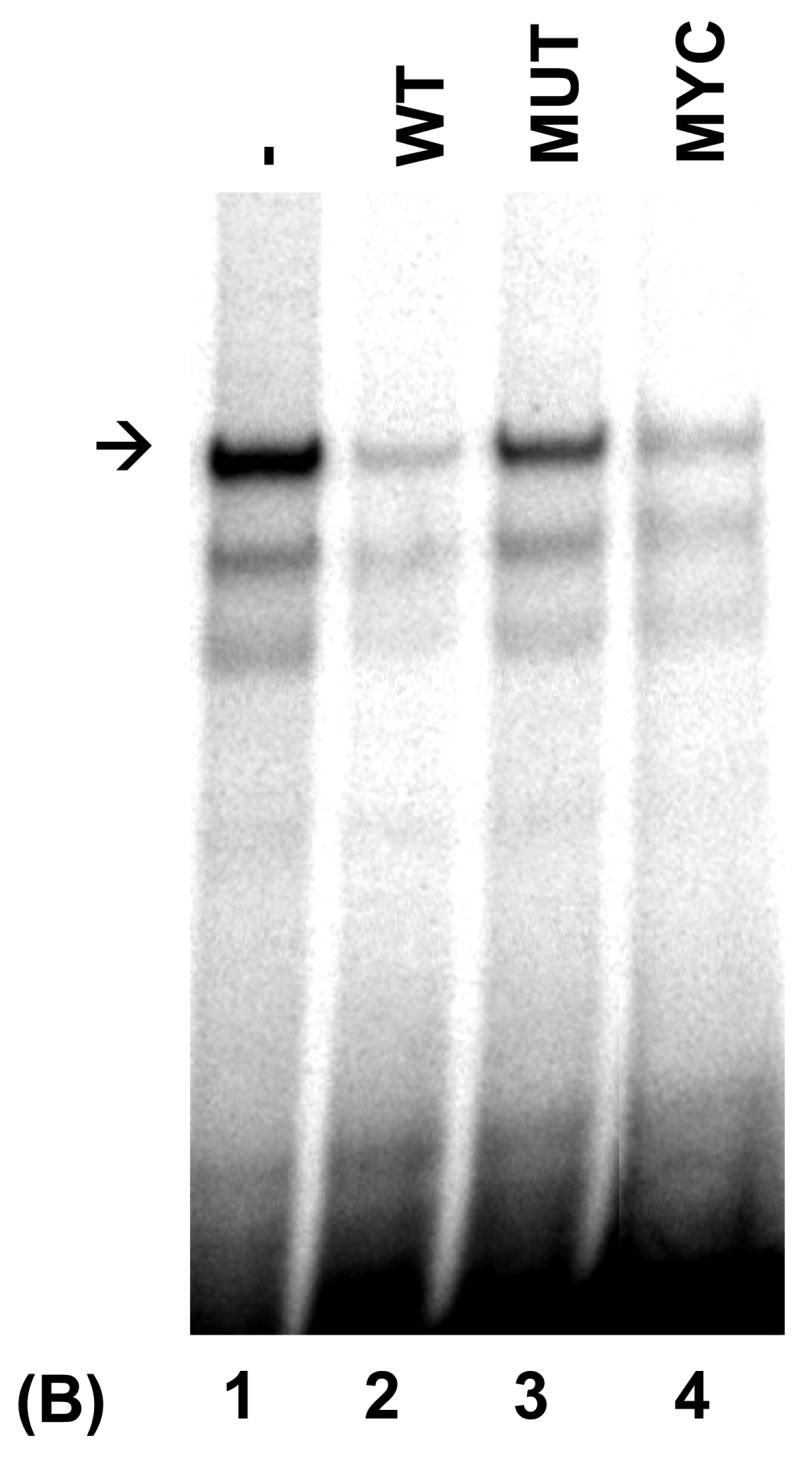
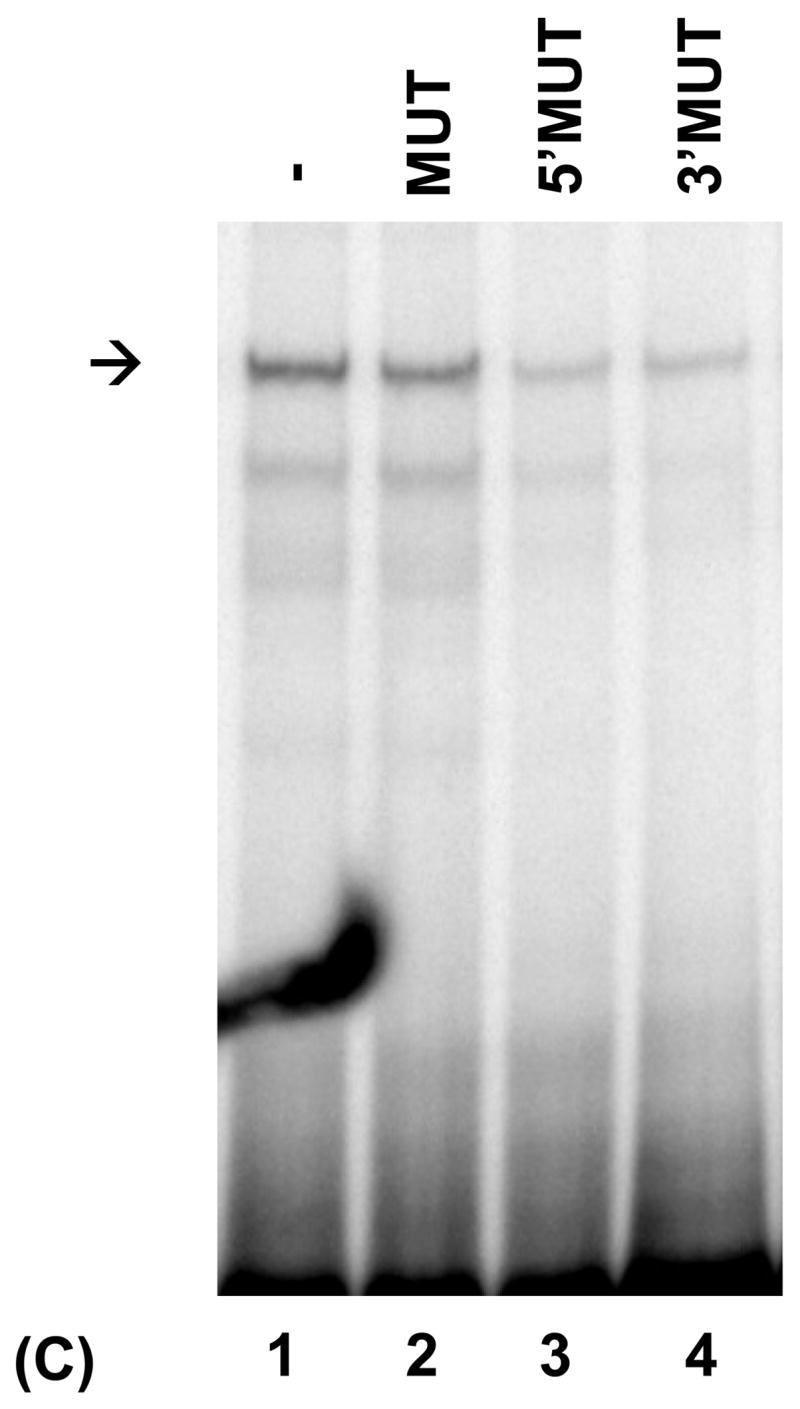
PRDI-BF1β, an alternative isoform of PRDI-BF1 expressed in myeloma cells, binds the CIITA type IV promoter. (A) EMSA analysis performed using the wild-type CIITA pIV probe and in vitro translated PRDI-BF1β protein demonstrates one predominant protein complex (lane 1, arrow). Preincubation with an antibody (AB.) specific for Blimp-1, which recognizes an epitope that is conserved in PRDI-BF1β, results in a shift in the mobility of this complex (lane 3, asterisk). Preincubation with an IgG1 isotype control (CTR) does not result in a supershifted complex. (B) EMSA analysis of in vitro translated PRDI-BF1β protein, using the wild-type CIITA pIV probe and cold competitors. The specific complex (arrow) is diminished by incubation with wild-type CIITA pIV oligonucleotide (WT, lane 2) or an oligonucleotide bearing the Blimp-1 binding site of the c-Myc promoter (MYC, lane 4), but is not diminished by a CIITA pIV oligonucleotide with a mutated IRF-E site (MUT, lane 3). (C) EMSA analysis using in vitro translated PRDI-BF1β protein, the wild-type CIITA pIV probe and mutated cold competitors. The cold competitors include a CIITA pIV oligonucleotide with a mutation that disrupts both GAAAG motifs in the IRF-E site (MUT), and oligonucleotides with mutations that disrupt the first (5′MUT) or second (3′MUT) GAAAG motif. Incubation with either of the cold competitor oligonucleotides in which a single unmutated GAAAG motif is present is capable of diminishing the formation of the specific complex (arrow), compare lanes 3 and 4 with lanes 1 and 2.
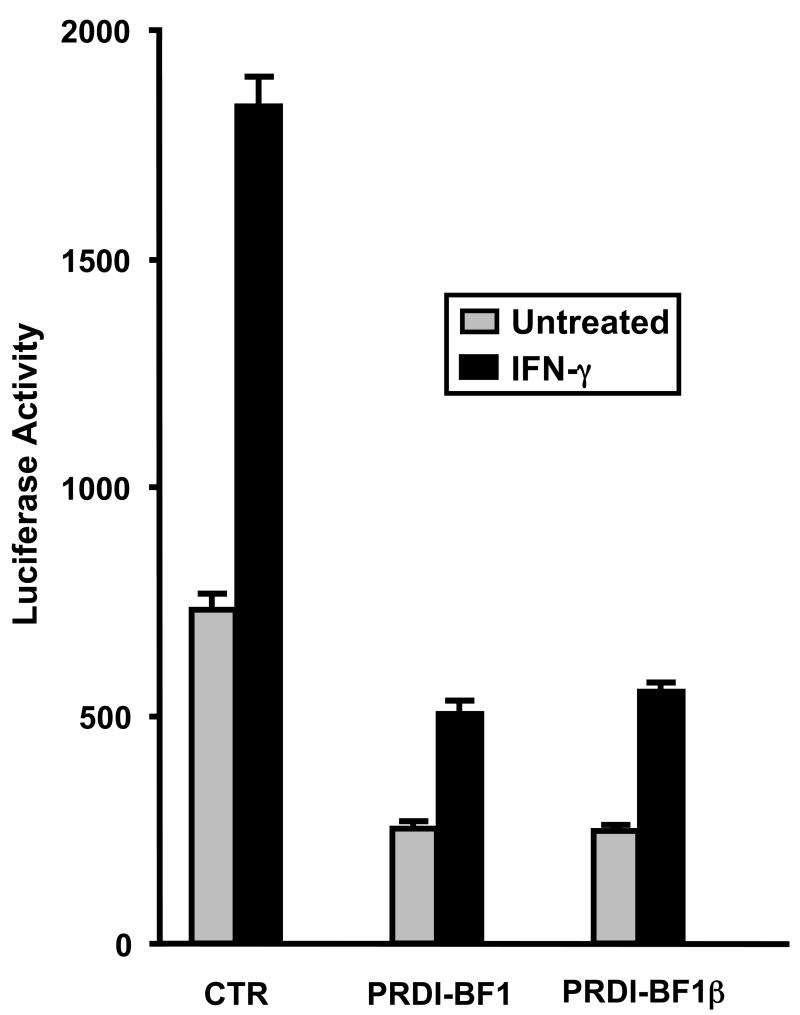
PRDI-BF1β does not act as a classical transdominant repressor of PRDI-BF1 and has actually been shown to retain about 50% of the ability to repress CIITA pIII, and 72% of the ability to repress the c-Myc promoter compared to the full-length protein (Gyory et al., 2003). To determine the ability of PRD-BF1β to repress CIITA pIV, co-transfections were performed in Raji B cells using plasmids expressing either the PRDI-BF1 full-length or β-form, and the CIITA pIV luciferase reporter plasmid. PRD-BF1β exhibits a similar ability to repress CIITA pIV compared to the full-length protein (Fig. 6). Taken together, the EMSA and promoter analyses demonstrate that the PRD-BF1β form of PRDI-BF1 binds and potentially represses CIITA pIV activity in myeloma cells.
Fig. 6.
PRDI-BF1β represses the CIITA type IV promoter. Transient co-transfections of Raji B cells were performed by electroporation using 10 μg of the wild-type CIITA pIV luciferase reporter plasmid and 10 μg of the full-length PRDI-BF1expression plasmid, the PRDI-BF1β expression plasmid, or the empty vector control. After a 6 h recovery period, half of the cultures were treated with 500 U/ml of recombinant human IFN-γ for 24 h before cultures were harvested. Ectopic expression of either PRDI-BF1 or PRFI-BF1β resulted in a 3-fold reduction of activity in both the treated and untreated samples. Luciferase activity was measured and normalized to Renilla luciferase activity. Bars show the S.E.M., n = 3. At least three independent experiments have been performed with similar results.
4. Discussion
MHC class II genes encode proteins involved in the presentation of antigenic peptides to CD4+ T lymphocytes and therefore play a central role in immune responses. CIITA is of crucial importance for the regulation of the expression of MHC class II genes at the level of transcription. As such, its expression is a major factor in the control of antigen presentation (van den Elsen et al., 2004). The expression pattern of the CIITA gene reflects this important role. Constitutive expression is confined to professional antigen presenting cells like B lymphocytes and dendritic cells (Muhlethaler-Mottet et al., 1997). For instance, the CIITA type III promoter is constitutively active in B lymphocytes. CIITA expression is also inducible by IFN-γ in other cell types. Although CIITA pIII activity can be induced by IFN-γ, much of the inducible expression of CIITA occurs through induction of the CIITA type IV promoter and we have recently shown this to be the case for B cells (Piskurich et al., 2006).
CIITA expression is also under developmental control. For example, its expression is silenced when B cells differentiate into plasma cells (Latron et al., 1988; Dellabona et al., 1989; Sartoris et al., 1996). PRDI-BF1, a transcriptional repressor that triggers the terminal differentiation of B cells into plasma cells, is expressed in multiple myeloma cells (Turner, Jr. et al., 1994; Shaffer et al., 2002). We have previously shown that PRDI-BF1 suppresses CIITA expression by repressing the constitutive activity of the CIITA type III promoter in B lymphocytes and myeloma cells (Piskurich et al., 2000; Ghosh et al., 2001). Since they generally express low levels of MHC class II proteins, it is possible that multiple myeloma cells and plasmacytoma cells escape the immune system by suppressing the CIITA expression (Liu et al., 1999; Walter et al., 2000; Prod’homme et al., 2004). For this reason, we have been interested in studying the ways in which CIITA expression is controlled in the B lymphocyte lineage. In this study, we demonstrate for the first time that PRDI-BF1 suppresses the activity of the CIITA type IV promoter. This finding lends new support to the idea that PRDI-BF1 is a key factor in the control of CIITA expression in B lymphocytes and myeloma cells.
A recent report identified a consensus site for the binding of Blimp-1, the murine homolog of PRDI-BF1, using random oligonucleotide selection (Kuo and Calame, 2004). It concluded that the binding site for this repressor resembles an IRF-E site, but that Blimp-1 does not bind all IRF-Es. It also suggested that the GAAAG sequence motif was important for repressor binding. The DNA sequence of the IRF-E of the CIITA type IV promoter, GAAAGTGAAAG, contains two GAAAG sequence motifs. When both of these motifs are mutated, activation of this promoter by IFN-γ and its ability to be bound and suppressed by PRDI-BF1 is severely diminished. We further demonstrate that the GAAAG motifs within this IRF-E can participate in PRDI-BF1 binding. These data provide new support for the idea that PRDI-BF1 binds IRF-E sites that contain the GAAAG motif. These findings provide knowledge that may assist in the identification of novel gene targets for repression by PRDI-BF1/Blimp-1. While PRDI-BF1 suppresses both the basal and IFN-γ-induced activity of the CIITA type IV promoter, some induction of this promoter by IFN-γ is retained. Importantly, this finding demonstrates that it may still be possible to therapeutically up regulate the activity of the CIITA type IV promoter and MHC II expression in myeloma cells using IFN-γ.
PRDI-BF1 belongs the PRDM family of proteins. Unifying features of this family are that they all function as transcriptional repressors and contain a PR domain, although the exact function of this domain is unknown (Keller and Maniatis, 1991; Alvarez-Venegas and Avramova, 2002). Elevated expression of truncated isoforms of PRDM proteins, lacking the PR domain and amino terminus of the protein, have been demonstrated in some types of neoplastic cells and may play a role in disease (Schneider et al., 2002). A truncated isoform of PRDI-BF1 was recently described (Gyory et al., 2003). This isoform, called PRDI-BF1β, is 101 amino acids shorter, lacks the amino-terminal acidic domain and contains an incomplete PR domain compared to full-length PRDI-BF1. Myeloma cells co-express both the full-length PRDI-BF1 and PRDI-BF1β forms. PRDI-BF1β represses the CIITA (MHCIITA) type III and c-Myc promoters, but this suppression is 25–50% less than full-length PRDI-BF1. We now demonstrate that the PRDI-BF1β isoform binds to the CIITA type IV promoter and its ability to repress CIITA pIV is not impaired. These results suggest that the amino-terminal acidic and PR domains do not play an important role in the suppression of the CIITA type IV promoter and are consistent with the previous finding that PRDI-BF1β does not act as a classical transdominant negative of PRDI-BF1.
In summary, we have investigated the ability of the transcriptional repressor, PRDI-BF1/Blimp-1, to suppress the CIITA type IV promoter in B lymphocytes. Our results indicate that this promoter is bound and suppressed by PRDI-BF1. Binding and suppression are dependent on the IRF-E site of CIITA pIV. These findings provide new evidence that PRDI-BF1 is a key factor in the control of CIITA expression in the B lymphocyte lineage. CIITA controls the expression of MHC class II genes that are required for the recognition of antigens by the immune system. Knowledge of the ways in which it is controlled in myeloma cells may prove useful in the design of therapies aimed at increasing the ability of the immune system to recognize B cell neoplasms, especially multiple myeloma.
Acknowledgments
The authors wish to thank Kathryn Calame for providing the pBJ-neo-Blimp-1 and control plasmids. This investigation was supported in part by research grants from the National Cancer Institute (RO1CA102203 and RO1CA114504) and the MEDCEN Community Health Foundation of Central Georgia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Venegas R, Avramova Z. SET-domain proteins of the Su(var)3–9, E(z) and trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- Chang CH, Fontes JD, Peterlin M, Flavell RA. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765–767. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC, Li G, Ting JP. Activation and transdominant suppression of MHC class II and HLA-DMB promoters by a series of C-terminal class II transactivator deletion mutants. J Immunol. 1997;159:2789–2794. [PubMed] [Google Scholar]
- Dellabona P, Latron F, Maffei A, Scarpellino L, Accolla RS. Transcriptional control of MHC class II gene expression during differentiation from B cells to plasma cells. J Immunol. 1989;142:2902–2910. [PubMed] [Google Scholar]
- Dong Y, Rohn WM, Benveniste EN. IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J Immunol. 1999;162:4731–9. [PubMed] [Google Scholar]
- Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J Immunol. 2003;170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E, Calame K. A plasmacytoma-specific factor binds the c-myc promoter region. Proc Natl Acad Sci USA. 1987;84:7031–7035. doi: 10.1073/pnas.84.20.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E, Riggs KJ, Gillespie W, Calame K. A transcriptional repressor of c-myc. Nature. 1989;339:718–721. doi: 10.1038/339718a0. [DOI] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta- interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Kelly AP, Monaco JJ, Cho SG, Trowsdale J. A new human HLA class II-related locus, DM. Nature. 1991;353:571–573. doi: 10.1038/353571a0. [DOI] [PubMed] [Google Scholar]
- Kern I, Steimle V, Siegrist CA, Mach B. The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int Immunol. 1995;7:1295–1299. doi: 10.1093/intimm/7.8.1295. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Landmann S, Muhlethaler-Mottet A, Bernasconi L, Suter T, Waldburger JM, Masternak K, Arrighi JF, Hauser C, Fontana A, Reith W. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J Exp Med. 2001;194:379–391. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latron F, Jotterand-Bellomo M, Maffei A, Scarpellino L, Bernard M, Strominger JL, Accolla RS. Active suppression of major histocompatability complex class II gene expression during differentiation from B-cells to plasma cells. Proc Natl Acad Sci USA. 1988;85:2229–2233. doi: 10.1073/pnas.85.7.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Takahashi M, Toba K, Zheng Z, Hashimoto S, Nikkuni K, Furukawa T, Koike T, Aizawa Y. Regulation of the expression of MHC class I and II by class II transactivator (CIITA) in hematopoietic cells. Hematol Oncol. 1999;17:149–160. doi: 10.1002/(sici)1099-1069(199912)17:4<149::aid-hon645>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a Gamma Interferon Response: Expression and Assembly of CIITA Promoter IV and Inhibition by Methylation. Mol Cell Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- Nagabhushanam V, Solache A, Ting L-M, Escaron CJ, Zhang JY, Ernst JD. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J Immunol. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- Nagarajan UM, Peijnenburg A, Gobin SJ, Boss JM, van den Elsen PJ. Novel mutations within the RFX-B gene and partial rescue of MHC and related genes through exogenous class II transactivator in RFX-B-deficient cells. J Immunol 2000. 2002;164:3666–3674. doi: 10.4049/jimmunol.164.7.3666. [DOI] [PubMed] [Google Scholar]
- Nikcevich KM, Gordon KB, Tan L, Hurst SD, Kroepfl JF, Gardinier M, Barrett TA, Miller SD. IFN-gamma-activated primary murine astrocytes express B7 costimulatory molecules and prime naive antigen-specific T cells. J Immunol. 1997;158:614–621. [PubMed] [Google Scholar]
- Pai RK, Askew D, Boom WH, Harding CV. Regulation of Class II MHC Expression in APCs: Roles of Types I, III, and IV Class II Transactivator. J Immunol. 2002;169:1326–33. doi: 10.4049/jimmunol.169.3.1326. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Wang Y, Linhoff MW, White LC, Ting JP. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- Piskurich JF, Linhoff MW, Wang Y, Ting JPY. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor beta. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Gilbert CA, Ashley BD, Zhao M, Chen H, Wu J, Bolick SC, Wright KL. Expression of the MHC class II transactivator (CIITA) type IV promoter in B lymphocytes and regulation by IFN-gamma. Mol Immunol. 2005;43:519–528. doi: 10.1016/j.molimm.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod’homme T, Drenou B, De RC, Barbieri G, Wiszniewski W, Bastard C, Charron D, Alcaide-Loridan C. Defective class II transactivator expression in a B lymphoma cell line. Leukemia. 2004;18:832–840. doi: 10.1038/sj.leu.2403315. [DOI] [PubMed] [Google Scholar]
- Sartoris S, Tosi G, De Lerma Barbaro A, Cestari T, Accolla RS. Active suppression of the class II transactivator-encoding AIR-1 locus is responsible for the lack of major histocompatibility complex class II gene expression observed during differentiation from B cells to plasma cells. Eur J Immunol. 1996;26:2456–2460. doi: 10.1002/eji.1830261028. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, Heyzer-Williams LJ, Liao J, Heyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Soos JM, Krieger JI, Stuve O, King CL, Patarroyo JC, Aldape K, Wosik K, Slavin AJ, Nelson PA, Antel JP, Zamvil SS. Malignant glioma cells use MHC class II transactivator (CIITA) promoters III and IV to direct IFN-gamma-inducible CIITA expression and can function as nonprofessional antigen presenting cells in endocytic processing and CD4(+) T-cell activation. Glia. 2001;36:391–405. doi: 10.1002/glia.1125. [DOI] [PubMed] [Google Scholar]
- Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N. Transcriptional regulation of antigen presentation. Curr Opin Immunol. 2004;16:67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- van der Stoep N, Biesta P, Quinten E, van den Elsen PJ. Lack of IFN-gamma-mediated induction of the class II transactivator (CIITA) through promoter methylation is predominantly found in developmental tumor cell lines. Int J Cancer. 2002;97:501–507. doi: 10.1002/ijc.1623. [DOI] [PubMed] [Google Scholar]
- Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W. Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene. J Exp Med. 2001;194:393–406. doi: 10.1084/jem.194.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W, Lingnau K, Schmitt E, Loos M, Maeurer MJ. MHC class II antigen presentation pathway in murine tumours: tumour evasion from immunosurveillance? Br J Cancer. 2000;83:1192–1201. doi: 10.1054/bjoc.2000.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Louis-Plence P, Ping D, He XF, Boss JM. HLA-DMA and HLA-DMB gene expression functions through the conserved S-X-Y region. J Immunol. 1997;158:4812–4821. [PubMed] [Google Scholar]
- Xi H, Eason DD, Ghosh D, Dovhey S, Wright KL, Blanck G. Co-occupancy of the interferon regulatory element of the class II transactivator (CIITA) type IV promoter by interferon regulatory factors 1 and 2. Oncogene. 1999;18:5889–5903. doi: 10.1038/sj.onc.1202969. [DOI] [PubMed] [Google Scholar]
- Xi H, Blanck G. The IRF-2 DNA binding domain facilitates the activation of the class II transactivator (CIITA) type IV promoter by IRF-1. Mol Immunol. 2003;39:677–684. doi: 10.1016/s0161-5890(02)00214-6. [DOI] [PubMed] [Google Scholar]
- Zika E, Ting JP. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr Opin Immunol. 2005;17:58–64. doi: 10.1016/j.coi.2004.11.008. [DOI] [PubMed] [Google Scholar]