Summary
Several alanine mutations in the response regulator Spo0F induce hypersporulation in Bacillus subtilis. L66A, I90A and H101A mutants are purported to be involved in contacts stabilizing the orientation of the α4-helix and hence the β4-α4 kinase recognition loop. Y13A is thought to affect the orientation of the α1-helix and consequently phosphatase action. Using comparative NMR chemical shift analyses for these mutants, we have confirmed these suppositions and isolated residues in Spo0F critical in sensor kinases discrimination. In addition, we discuss how buried residues and intra-protein communication networks contribute to precise molecular recognition by ensuring that the correct surface is presented.
Keywords: Sporulation, Kinases, Spo0F mutants
1. Introduction
The initiation of sporulation in Bacillus subtilis is regulated by a signal transduction pathway referred to as a multi-component phosphorelay [1]. In response to binding an activating ligand, a sporulation specific histidine sensor kinase autophosphorylates (on a conserved histidine) in an ATP-dependent reaction. The phosphorylated kinase transfers the phosphoryl group from the histidine to an aspartic acid binding pocket in the response regulator Spo0F. Spo0F, in turn, transfers the phosphoryl group to a histidine in the phosphotransferase Spo0B that subsequently passes it to the two-domain response regulator transcription factor Spo0A responsible for initiating sporulation [2,3]. In addition to the signal ligand, phosphoryl group flux in the pathway and the ultimate level of phosphorylated Spo0A is controlled by sets of phosphatases acting on Spo0F (RapA/B/E) and Spo0A (YnzD/YisI/Spo0E) [4].
There are several sporulation sensor histidine kinases in Bacillus and it has been suggested that in B. subtilis’ natural, variable environment, Spo0F may target different kinases depending on the conditions at any particular time [5]. There is reason to believe from mutant studies that Spo0F’s ability to choose between multiple kinases may be attributed to some degree to an equilibrium that exists between Spo0F’s various conformations [5]. Spo0F is known to possess conformational flexibility in specific regions and in each of the proteins’ significantly populated conformational sub-states a different kinase interaction may be favored [6]. It is plausible therefore, that specific environments may determine the conformational equilibrium of Spo0F at a particular time - thereby contributing to kinase specificity [5]. In the research presented here, this notion was tested by focusing on Spo0F’s recognition of the sporulation histidine sensor kinases KinA, KinB, KinC and KinD.
Spo0F is a response regulator (124 residues) with a conserved fold [7]. Its structure consists of a central β-sheet core consisting of five parallel β-strands, surrounded by five α-helices. The aspartic acid residues D10, D11 and D54 form an active site with a divalent cation to accept the incoming phosphoryl moiety and form an acyl-phosphate with D54. Surrounding the Asp pocket is a set of β-α loops that connect the strands and helices. Mutational studies of sporulation sensor histidine kinases revealed that wild-type Spo0F was phosphorylated predominantly by KinA and, to a lesser degree, by KinB [5,8,9]. Double knockout mutations of KinA and KinB showed that KinC and KinD phosphorylated wild-type Spo0F and Spo0F mutant Y13A poorly and sporulation was greatly reduced [5,8,9]. However certain Spo0F mutants (L66A, I90A and H101A) suppressed this phenotype and sporulated extremely well in a kinA kinB double knockout mutant strain [5]. Further kinase mutation studies showed that the I90A Spo0F mutant could be phosphorylated by KinD in the absence of KinA, B, and C and that the L66A and H101A Spo0F mutants preferred KinC.
These results suggested that, in the laboratory setting, wild-type Spo0F and Spo0F mutant Y13A preferentially recognizes KinA and KinB, the Spo0F mutants L66A and H101A recognize KinA, KinB, and KinC, while I90A Spo0F has the ability to be recognized by KinA, KinB, KinC and KinD. In principle therefore, comparative structural studies between these mutants may provide the first information identifying which residues in Spo0F drive targeting to specific kinases.
Previous investigations showed that the β4-α4 loop (residues 82–90) in wild-type Spo0F interacts with KinA [9] and it was proposed that the L66A, I90A and H101A mutations resulted in destabilizing contacts for the α4-helix (residues 90–97). This perturbation was suggested to propagate and alter the conformation of critical recognition residues in the β4-α4 loop [9]. In this work, circular dichroism (CD) and tyrosine fluorescence emission studies confirm no major structural perturbations are induced by the mutations and NMR chemical shift perturbations for the Spo0F mutants provide very strong evidence to support this model. Additionally the data identified residues in Spo0F important for specific kinase recognition.
2. Material and Methods
2.1 Spo0F Expression and Purification
The Spo0F mutants were expressed and purified as described previously [9].
2.2 NMR Spectroscopy
All NMR experiments were performed at 298K on a Varian INOVA 600. 1.0 – 2.0 mM protein samples in the following buffer were used: 90%:10% or 1%:99% H2O:D2O, 25 mM Tris (pH6.9), 50 mM KCl, and 0.02% NaN3. Sequential assignments were made from HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO and HN(CA)CO experiments. Side-chains were assigned from H(CCO)NH, (H)C(CO)NH and HCCH-TOCSY experiments [10–14]. The spectra were processed with NMRPIPE and analyzed with NMRVIEW on LINUX workstations running Fedora Core 5. Molecules were visualized and aligned with Pymol [15].
2.3 Surface accessibility area
NACCESS was used to calculate the surface accessibility area on wild-type and mutant Spo0F proteins (1FSP) [7,16]. Percentage of accessible area is reported for all atoms in the residue.
2.4 Circular dichroism
CD spectra were measured using a Jasco J600A spectropolarimeter using a 0.1 cm (far-UV) Hellma cuvette (Hellma Corp.). All measurements were corrected for background signal, and buffer conditions were subtracted under the same experimental conditions. All spectra were recoreded in 25 mM Tris (pH6.9), 50 mM KCl, and 0.02% NaN3. Spectra were recorded in triplicate at 25 ± 1.0 C from 190 to 250 nm.
2.5 Tyrosine Fluorescence Emission
Tyrosine fluorescence emission measurements were performed using a PTI C-61 spectrofluorometer (Photon Technology International) with a 1 mL four-sided Hellma cuvette (Hellma Corp.). Samples were excited at 275 nm (tyrosine fluorescence), and emission was measured from 300 to 400 nm. Spectra were collected in triplicate at 25 mM Tris (pH6.9), 50 mM KCl, and 0.02% NaN3. The instrument was equipped with thermostated cell holders, and the temperature was held constant at 25 ± 1.0 C using a circulating water bath.
3. Results and Discussion
On order to ascertain structural differences in Spo0F mutant proteins, CD, tyrosine emission fluorescence, and the backbone 1HN and 15N chemical shifts of wild type Spo0F were compared to the backbone 1HN and 15N chemical shifts of Spo0F mutants L66A, I90A, H101A and Y13A. CD and tyrosine fluorescence data (Figure 1) confirmed that the mutations do not significantly perturb the structure compared to wild type Spo0F. In all cases, the CD spectra are virtually indistinguishable. Tyrosine fluorescence verifies that the local structural environments surrounding the tyrosine residues of the H101A and I90A Spo0F mutants are similar to that of wild-type Spo0F. The reduction in Y13A’s tyrosine emission can be attributed to the mutation itself. When taken in conjunction with the CD data, the reduction in L66A’s tyrosine emission is most likely due to small, localized structural changes and not to gross conformational changes. However, these experiments were unable to provide specific reasons for the hypersporulation effects of the mutants. To accomplish this, and to complement the CD and fluorescence studies, chemical shift changes between the mutant protein and wild-type were quantified for each residue by calculating the cumulative chemical shift (ΔδNH), where:
Figure 1. Circular dichroism and tyrosine emission fluorescence.
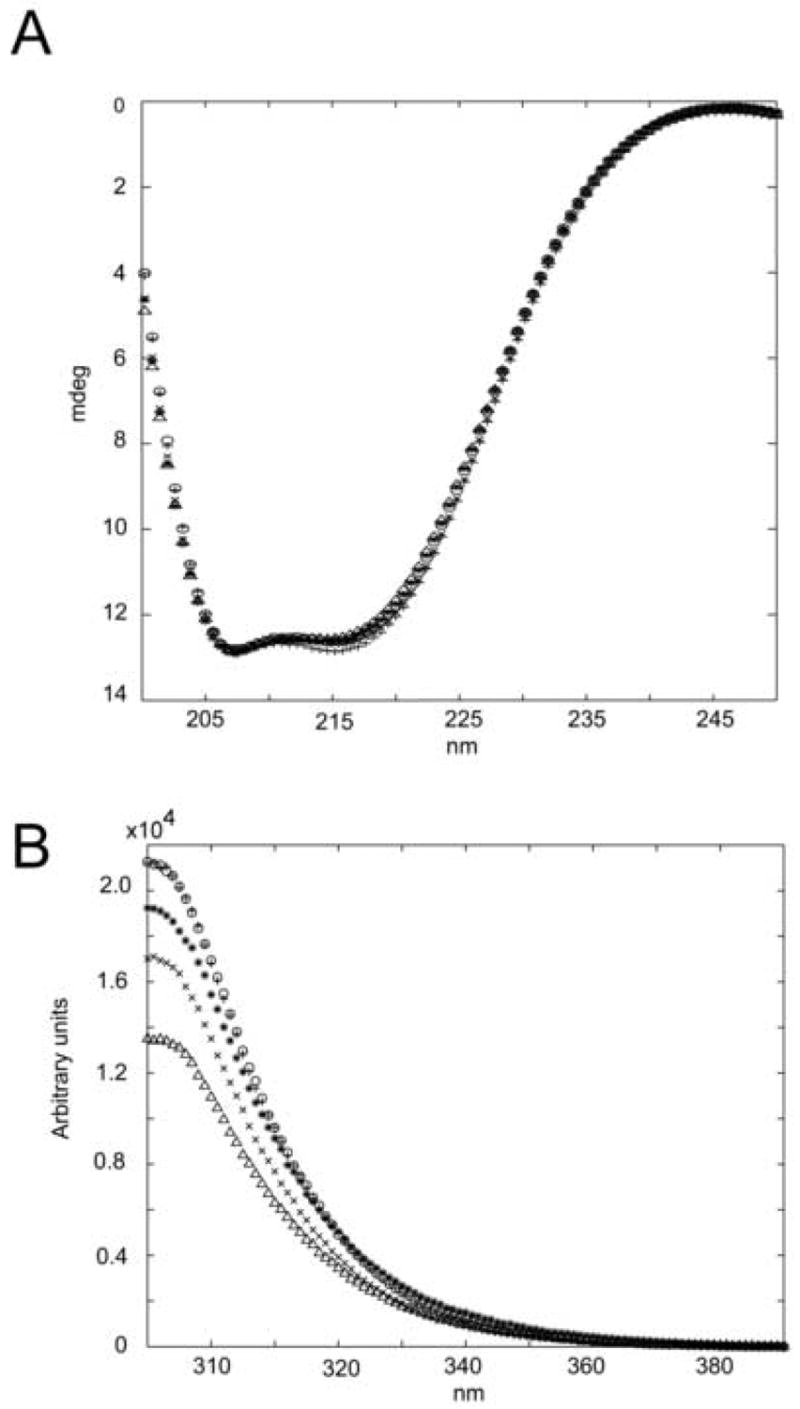
A) Far-UV circular dichroism spectra; B) tyrosine emission fluorescence of Spo0F mutants; “x” Y13A, “^” L66A, “*” I90A, “0” H101A and “+” wild type. Results confirm similar secondary structures between all mutant proteins and wild type, indicating no major conformational changes upon mutation.
| [1] |
Significant chemical shift differences are indicative of structural perturbations and conformational changes at that specific residue. Figure 2 shows ΔδNH plotted for each residue in each mutant protein along with the calculated CSI plot. Simple inspection of the data shows that L66A, H101A and I90A cause perturbations predominantly in the β4-α4 loop/α4-helix, while Y13A causes perturbations predominantly in the β1-α1 loop and N-terminal end of α1-helix.
Figure 2. Chemical shift changes for Spo0F mutants compared to wild-type.
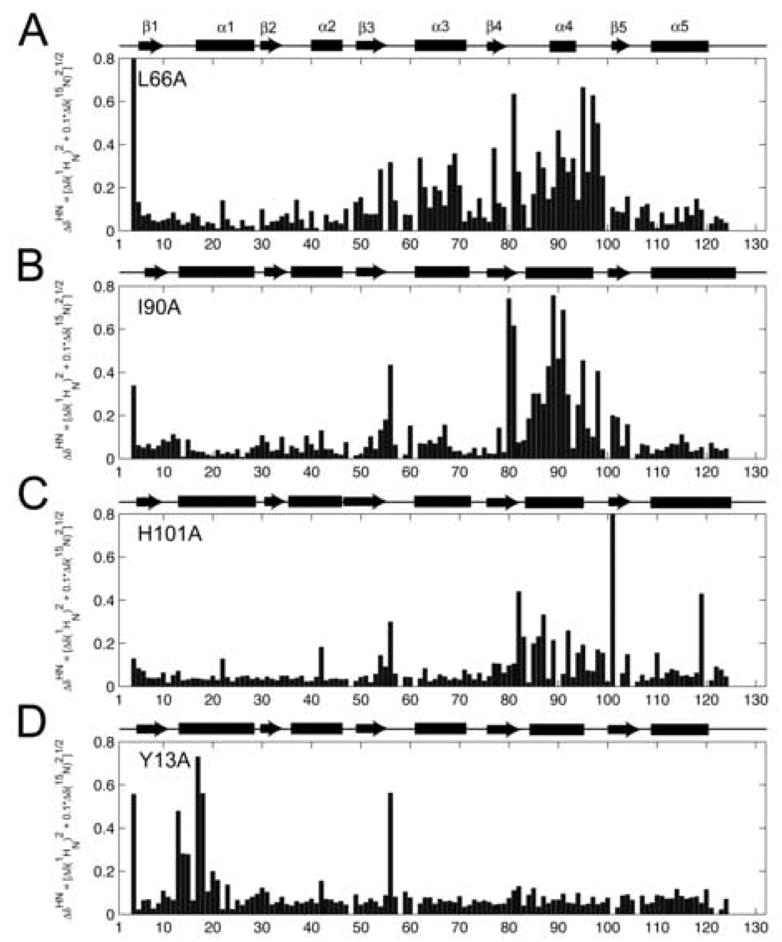
A: L66A; B: I90A; C: H101A; D: Y13A. The secondary structural elements (rectangles for helices, arrows for strands) were determined using the CSI program and 13Cα, 13Cβ, and 13CO chemical shift data. ΔδNH was determined using Eqn. [1].
Concentrating initially on the L66A, I90A and H101A chemical shift data (Figure 2), it is clear that each plot has the same overall form, with the most significant chemical shift changes generally occurring across a range from residues 80 to 103. It is important to note that for I90A, differentially large effects are seen for the region encompassing residues 88–91. These data are shown in Fig. 3, which maps the residues identified as a ΔδNH > 0.4 ppm onto the high-resolution solution structure of Spo0F (1FSP) [7]. Residues shown in blue are common to both L66A and I90A; residues in red are from H101A and residues in orange are solely from L66A. It can be seen that all these residues lie in a well-defined strip on Spo0F, strip “B”. Therefore these residues solely contribute to the KinC recognition, as Spo0F hypersporulation mutants L66A and H101A can be phosphorylated by KinA, KinB and KinC but not KinD. Residues depicted in green are only from I90A. The Spo0F mutant I90A can be phosphorylated by KinD. It is evident that residues 88, 89 and 91 lie outside strip “B” and lie within strips “A” and “C”. Hence, it is likely that residues 88, 89 and 91 play a significant role in KinD recognition, while the other residues in strip “B” are mainly involved in KinA, B and C recognition.
Figure 3. Residues in Spo0F mutants involved in kinase recognition.
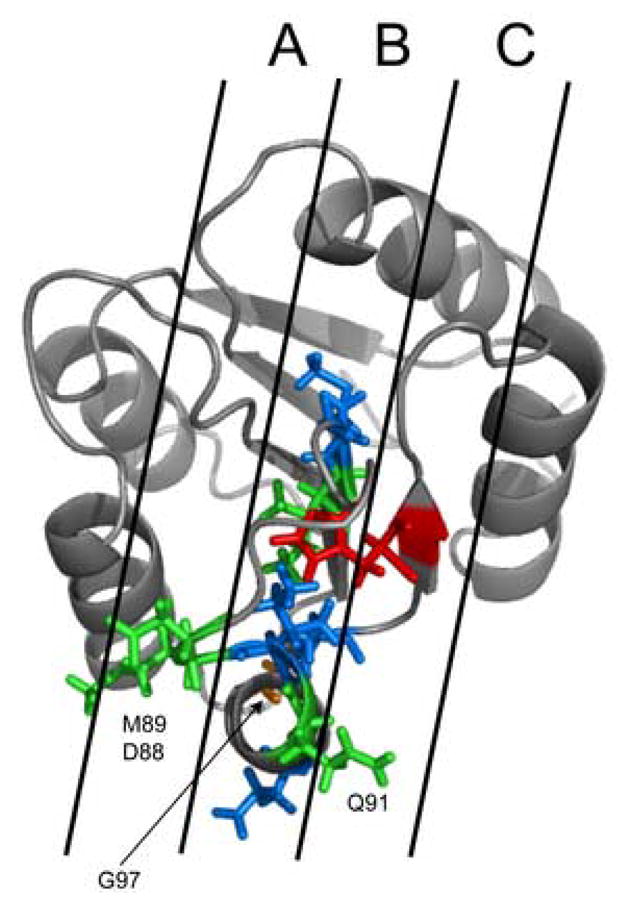
Residues in the β4-α4 kinase recognition loop exhibiting significant chemical shift perturbations (> 0.4ppm) mapped onto the structure of wild-type Spo0F (1FSP) [7]. Residues colored blue are perturbed in both L66A and I90A. Residues colored orange are only perturbed in L66A. Residues colored red are only perturbed in H101A. Residues colored green are only perturbed in I90A.
While the L66A, I90A and H101A mutations are thought to alter the conformation of the β4-α4 kinase recognition loop, not all hyper-sporulating Spo0F mutants are found in this region. The Y13A Spo0F mutant (located at the end of the β1-α1 loop coming into the N-terminal end of the α1-helix) also produces a hyper-sporulation phenotype. Unlike the L66A, I90A and H101A mutants, Y13A has the same kinase recognition properties as the wild-type protein, specifically requiring KinA and KinB to sporulate [5]. KinC and KinD are found to be incapable of phosphorylating Spo0F Y13A. It has been suggested that the increased frequency of Y13A sporulation compared to wild-type Spo0F is most likely due to increased resistance to dephosphorylation by the Rap phosphatases and not due to kinase recognition effects. If this is indeed the case, we would not expect to observe structural perturbations in the β4-α4 kinase recognition loop in the Y13A mutant.
In the Y13A mutation (Fig. 2D) we note the prevalence of perturbations in the β1-α1 loop/α1-helix region. Focusing on this area, significant chemical shift changes are observed for residues Y13(A), G14, I15, I17, L18. These residues reside in the region known to be responsible for interactions with the RapB phosphatase [17,18]. It is likely that the Y13A mutation alters the conformation of this region, prohibiting RapB from making the proper contacts with Spo0F and so hindering its dephosphorylation. It can be seen that there are no notable chemical shift perturbations in the β4-α4 loop or α4-helix. This suggests that the Y13A mutation does not structurally affect this region. Consequently, kinase specificity should be the same as for wild-type Spo0F – which it is [5]. The resultant hypersporulation phenotype for the Y13A mutation is therefore due to the inability of this protein to dephosphorylate via phosphatase interactions and not due to altered kinase specificity.
In all four Spo0F mutants, residue K56 is markedly perturbed, suggesting a notable conformational change in this region. It has been shown that K56 assists in the coordination of a metal ion that is responsible for the binding the incoming, activating phosphoryl group [19]. The metal ion interacts with the backbone carbonyl group of K56 and the side chain oxygen atoms of D54 and D11. Since all of these mutants display increased sporulation frequency compared to wild-type Spo0F [5] in full kinase background, it can be assumed that any conformational changes undergone by K56 only serve to orient its position for more favorable interactions with the metal ion/phosphoryl group.
The Spo0F mutants L66A, I90A and H101A that affect kinase targeting, cause changes in the conformation of the β4-α4 recognition loop. In each mutant, the position of this loop is different, as determined by different chemical shift perturbation patterns compared to wild-type Spo0F. Chemical shift changes are also seen in the α4-helix, suggesting that its re-orientation substantially affects the orientation of the β4-α4 kinase recognition loop.
Interestingly, in wild-type Spo0F, residues L66, I90 and H101 have minimal exposed surface area. The amount of surface area exposed for all atoms of each of these residues is: 3.1% for L66; 7.9% for I90 and 10.4% for H101. Nevertheless, these almost completely buried residues substantially affect kinase specificity. Our previous studies have shown that L66, I90 and H101 in Spo0F move in a concerted fashion on the same timescale and are part of intra-protein communication networks that pass information throughout the core of the protein to the surface [6]. The L66A, I90A and H101A mutants disrupt these communication networks, altering their critical dynamic and packing properties. Consequently, changes normally propagated through the protein are affected and surfaces to be presented to a binding target are altered. This implies that residues that predominantly inhabit the protein core, or at least exist below the surface, are critical in ensuring target specificity.
In this work, we have used NMR chemical shift perturbations of Spo0F mutants to determine individual residues and structural regions important in multiple kinase recognition. Our results show that the conformations of the β4-α4 loop and the α4-helix, dictate Spo0F’s kinase specificity and explain hypersporulation phenotypes previously observed. In turn, these data support the suggestion that, in its natural, variable environment, Spo0F may recognize different kinases depending on the specific circumstance at any given time. We have also shown that hypersporulation driven by the Spo0F Y13A mutant is likely due to Rap phosphatase effects and not kinase recognition. These studies lend substance to the hypothesis that molecular recognition processes are significantly influenced by intra-protein communication networks consisting of residues in the core of the protein dictating the correct surface to be presented to a binding partner. We are currently determining the high resolution NMR solution structures for all the Spo0F mutants discussed in order to determine the extent of conformational change taking place in these regions and the accurate structural reasons for the hypersporulation phenotypes observed. This will include the precise positioning of the α1-helix, β4-α4 loop and α4-helix in each case.
Finally, this work suggests that while the surface of the protein is essential for its ability to recognize a specific kinase, residues in the core of the protein perform critical functions in presenting the required, precise surface. Alterations in the core of the protein, via environmental, mutational or chemical perturbations, can efficiently propagate to the surface via defined communication networks, and alter the surface composition. This is especially important for response regulator proteins as a whole, since: (1) there are numerous two component systems in all bacterial species; (2) response regulators all have similar structures and (3) their targets also have very similar structures (4-helix bundles). Therefore, the chance of a response regulator recognizing the wrong target is significant if recognition is not strictly defined. Having both surface and buried residues contribute to the recognition process allows for a synergistic mechanism to provide greater specificity.
3.1 Data Deposition
The H10A, I90A, L66A and Y13A chemical shifts have been deposited in the BioMagResBank (15008, 15009, 15010 and 15011 respectively).
Acknowledgments
We would like to thank Daniel M. Sullivan for sample preparation of Spo0F mutants and editing of this manuscript. This work was supported in part by NIH grants GM55769 (JC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–52. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 2.Hoch JA. Regulation of the onset of the stationary phase and sporulation in Bacillus subtilis. Adv Microb Physiol. 1993;35:111–33. doi: 10.1016/s0065-2911(08)60098-3. [DOI] [PubMed] [Google Scholar]
- 3.Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–65. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 4.Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol. 2001;42:133–43. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–95. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- 6.Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–93. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- 7.Feher VA, et al. High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition. Biochemistry. 1997;36:10015–25. doi: 10.1021/bi970816l. [DOI] [PubMed] [Google Scholar]
- 8.Trach KA, Hoch JA. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 9.Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–12. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 10.Logan TM, Olejniczak ET, Xu RX, Fesik SW. Side chain and backbone assignments in isotopically labeled proteins from two heteronuclear triple resonance experiments. FEBS Lett. 1992;314:413–8. doi: 10.1016/0014-5793(92)81517-p. [DOI] [PubMed] [Google Scholar]
- 11.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3:225–31. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 12.Montelione GT, Emerson SD, Lyons BA. A general approach for determining scalar coupling constants in polypeptides and proteins. Biopolymers. 1992;32:327–34. doi: 10.1002/bip.360320406. [DOI] [PubMed] [Google Scholar]
- 13.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–67. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 14.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 15.DeLano WL. The PyMol Molecular Graphics. on World Wide Web http://www.pymol.org. 2002
- 16.Hubbard SJTJM. Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. NACCESS. [Google Scholar]
- 17.Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry. 1998;37:16538–45. doi: 10.1021/bi981340o. [DOI] [PubMed] [Google Scholar]
- 18.Perego M, Hoch JA. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:1549–53. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madhusudan Zapf J, Whiteley JM, Hoch JA, Xuong NH, Varughese KI. Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis. Structure. 1996;4:679–90. doi: 10.1016/s0969-2126(96)00074-3. [DOI] [PubMed] [Google Scholar]


