Abstract
Non-human primates (NHP) provide crucial research models. Their strong similarities to humans make them particularly valuable for understanding complex behavioral traits and brain structure and function. We report here the genetic mapping of an NHP nervous system biologic trait, the cerebrospinal fluid (CSF) concentration of the dopamine metabolite homovanillic acid (HVA), in an extended inbred vervet monkey (Chlorocebus aethiops sabaeus) pedigree. CSF HVA is an index of CNS dopamine activity, which is hypothesized to contribute substantially to behavioral variations in NHP and humans. For quantitative trait locus (QTL) mapping, we carried out a two-stage procedure. We first scanned the genome using a first-generation genetic map of short tandem repeat markers. Subsequently, using >100 SNPs within the most promising region identified by the genome scan, we mapped a QTL for CSF HVA at a genome-wide level of significance (peak logarithm of odds score >4) to a narrow well delineated interval (<10 Mb). The SNP discovery exploited conserved segments between human and rhesus macaque reference genome sequences. Our findings demonstrate the potential of using existing primate reference genome sequences for designing high-resolution genetic analyses applicable across a wide range of NHP species, including the many for which full genome sequences are not yet available. Leveraging genomic information from sequenced to nonsequenced species should enable the utilization of the full range of NHP diversity in behavior and disease susceptibility to determine the genetic basis of specific biological and behavioral traits.
Keywords: comparative genomics, genetic mapping, homovanillic acid, vervet
The genetic investigation of non-human primates (NHPs) has the potential to generate enormous insights into the development, degeneration, and structure of the human brain and the biological basis of human behavior. Like humans, NHPs experience a prolonged period of postnatal development, together with strong family ties and complex social relationships. Furthermore, most features of human behavior have recognizable counterparts in NHPs, enabling the examination of traits such as anxiety and impulsivity, which are central components of human behavioral disorders. The pronounced interindividual variability observed in such traits within pedigreed primate colonies has stimulated interest in identifying genetic variants associated with these variable traits.
There are several advantages in genetically investigating behavior-related traits in NHPs compared with humans. It is feasible to assess systematically a given trait in all members of large multigenerational NHP pedigrees, providing power to detect genetic variants of relatively small effect. It is also possible to obtain measures related to brain structure and function in NHPs that would be infeasible to collect in human subjects; examples include brain sampling for measurement of gene expression patterns or cerebrospinal fluid (CSF) samples, as in the current study. Perhaps most importantly, although studies of humans must account for uncontrolled and unmeasured environmental variation and gene-environment correlations that can bias or obscure genetic effects on complex traits (1), captive NHP populations provide the opportunity to control and manipulate the environment, independent of genotype (2).
The limited DNA sequence divergence between higher primates has permitted the generation of genome-wide genetic linkage maps in widely investigated Old World monkey (OWM) species, including rhesus macaques, baboons, and vervets, based on the adaptation of human short tandem repeat (STR) sequences (3–6). The availability of these maps has in turn spurred genetic mapping studies of complex traits in these OWMs. Such studies are further facilitated by the availability of genome resources, such as BAC libraries, which are now available for at least 18 primate species (http://bacpac.chori.org), and the reference genome sequences of human, chimpanzee, and rhesus (7–9), which permit comparative genomic analyses.
The relative genetic homogeneity of some captive NHP populations, as exemplified by the Vervet Research Colony (VRC) at the University of California, Los Angeles, is another factor spurring interest in using such populations for genetic mapping of complex traits. Vervets, also called African green monkeys, are among the most widely dispersed OWMs, inhabiting much of subSaharan Africa. In the 1600s, a small number of vervets colonized the Caribbean islands of St. Kitts, Nevis, and Barbados, as travelers on ships involved in the African slave trade (10). This emigration marked the first bottleneck of the ancestral VRC population. Vervets adapted well to the environment of the West Indies, and their populations grew rapidly absent any non-human predators. The current VRC population (>500 individuals) descends from 57 wild-caught vervets (29 females and 28 males) trapped on St. Kitts between 1975 and 1987, with 24 of the original matrilines now in their third to eighth generations. The founding of the VRC represented a second bottleneck for this population (11). The VRC vervets can be connected into one large inbred pedigree (see Methods).
Several heritable behavioral traits have been assessed over multiple generations within the VRC pedigree, including measures of impulsivity, aggressiveness, and anxiety that are assessed in a social challenge test (12). In addition to such heritable behaviors, the VRC pedigree has been phenotyped for biochemical measures that correlate with behavioral traits, such as cisternal CSF levels of the dopamine, serotonin, and norepinephrine metabolites homovanillic acid (HVA), 5-hydroxyindolacetic acid (5-HIAA), and 3-hydroxy-4-methoxyphenylglycol (MHPG), respectively (12, 13). These measures are markers for monoamine (MA) turnover in the CNS (14), which is centrally important in behavioral neuroscience and psychiatry, because most commonly used psychotropic drugs alter one or more MA systems (15, 16).
Natural variation in levels of 5-HIAA, in cisternal CSF of vervets (and other OWM) has been associated with impulsive, aggressive, and risk-taking behavior (13, 17–20). Levels of HVA have been related to aggressive behavior, wounding, sexual behavior, and social dominance in vervets and macaques (12, 20– 23). Numerous studies suggest associations between CSF MA metabolite concentrations and various human behavioral disorders (15, 24, 25), but other than relationships of CSF 5-HIAA to suicidal and aggressive behaviors (26), these associations remain mostly equivocal and nonreplicated, in part because of the difficulty in obtaining CSF from sufficient human subjects. In addition, human studies, in contrast to those in NHPs, rely on lumbar CSF, which may less accurately reflect brain MA levels than cisternal CSF (27).
There is much stronger evidence for the heritability of MA metabolite levels in NHPs than in humans, presumably because of the small sample sizes available for human studies. In humans, a relatively small twin study (28) demonstrated high heritability for MHPG (h2 = 0.74) but could not find unequivocal heritability for the other metabolites. Significant genetic contributions to variation in HVA and 5-HIAA have been observed in rhesus macaques, in comparisons of sire families (29, 30). A study of 271 captive baboons in multigenerational pedigrees showed clear heritability of all three of the CSF MA metabolites measured [5-HIAA, h2 = 0.30 (SE 0.17); MHPG, h2 = 0.36 (SE 0.16); and HVA, h2 = 0.50 (SE 0.19) (31)]. Several studies in humans have found only nominal associations between CSF levels of MA metabolites and variants in hypothesis-based candidate genes (32–34). In this study, we demonstrate the heritability of these metabolites within the VRC pedigree and genetically map a quantitative trait locus (QTL) for HVA. We hypothesize that identification of sequence variants associated with HVA level in vervets may permit more precise delineation of the relationship between dopamine systems and a wide range of behavioral variables.
Results
MA Metabolites in CSF in VRC Pedigree Samples.
The concentrations of MA metabolites in the CSF of 347 vervets from the VRC are shown in Table 1. Each of these metabolites is heritable in this pedigree, with HVA displaying the highest estimated narrow sense heritability (Table 1). The mean levels of all three MA metabolites were higher in females than in males. HVA and 5-HIAA levels declined with age, whereas MHPG increased with age. There was a significant interaction between age and sex and mean levels of 5-HIAA and MHPG, but not HVA.
Table 1.
Distribution and estimated heritability of CSF MA metabolite levels of VRC vervets
| Metabolite | n | Mean (±SD), pmol/ml | Range | Kurtosis | Significant covariates | h2 (SE) | P value† |
|---|---|---|---|---|---|---|---|
| HVA | 346 | 568 (±167) | 218–1,026 | −0.38 | Sex, age* | 0.52 (0.11) | P = 1.4 × 10−9 |
| 5-HIAA | 347 | 178 (±50) | 56–364 | −0.06 | Sex, ageb‡, ageb × sex | 0.41 (0.10) | P = 2.3 × 10−6 |
| MHPG | 346 | 74 (±27) | 8–153 | −0.13 | Sex, age, age2 sex × age2 | 0.39 (0.11) | P = 8.3 × 10−6 |
The predicted level of HVA for an individual vervet in the VRC was estimated to be HVApredicted = −0.17 + 0.38F−0.18(A−5.4), where F takes the value 1 if the individual is female and 0 if male, and A is the observed age category for the individual (age category is centered by subtracting the mean age category of 5.4 from the observed category for an individual; see Methods for more details of these categories).
The predicted level of 5-HIAA for an individual vervet in the VRC was estimated to be 5-HIAApredicted = −0.10 + 0.10F−0.34(J−1.8) + 0.69F(J−1.8), where F takes the value 1 if the individual is female and 0 if male, and J takes the value 1 for a juvenile and 2 for an adult (the juvenile/adult age category is centered by subtracting the mean of 1.8 from the observed category for an individual). The interaction between sex and age resulted in higher predicted levels of 5-HIAA for adults than for juveniles and for female but not males.
The predicted level of MHPG for an individual vervet in the VRC was estimated to be MHPGpredicted = −0.44 + 0.59F + 0.08(A−5.4) + 0.08(A−5.4)2 −0.09F(A−5.4)2, where F takes the value 1 if the individual is female and 0 if male, and A is the observed age category for the individual (age category is centered by subtracting the mean age category of 5.4 from the observed category for an individual). The interaction between sex and the square of age category resulted in lower predicted levels of MHPG with increasing age category, for females but not males.
Genome Scan for QTL.
Given the significant heritability estimates for MA metabolites in the VRC, we undertook a genome scan to identify QTL for these metabolites, using 324 markers from the vervet scaffold autosomal genetic map (6). Two-point analysis using the quantitative trait variance component linkage analysis program SOLAR (35) revealed suggestive linkage of HVA levels to two markers that are separated from one another by ≈13 cM (and ordered with odds of >1,000:1) on vervet chromosome 9p (Fig. 1, D10S585, logarithm of odds (lod) score of 2.3, accounted for 26% of heritable trait variance in HVA and D10S1477, lod score of 1.9, accounted for 24% of heritable trait variance [see supporting information (SI) Dataset 1 for more complete marker orders in this region]. An additional marker from this region (D101779) showed much weaker evidence for linkage but is not as well ordered and displays a lower observed heterozygosity compared with the former two markers. No lod scores >2 were observed for 5-HIAA and MHPG; therefore, we focused our mapping efforts on the QTL for HVA. Complete QTL results for all of the MA metabolites are in SI Dataset 1.
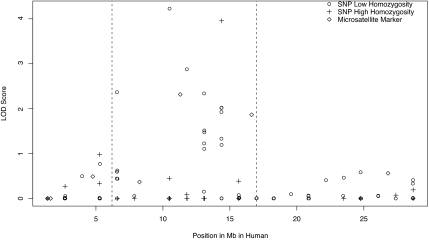
Fig. 1.
Mapping of the HVA QTL. lod scores for SNP and STR markers in the putative QTL region on vervet 9p are shown. On the horizontal axis is the position of the marker, in megabases, taken from the human physical map, and on the vertical axis is the lod score for the test of the null hypothesis that a QTL is at a given position. SNPs with low observed homozygosity (<75% of vervets homozygous) are indicated with open circles, SNPs with high observed homozygosity (>75% of vervets homozygous) are plotted with crosses, and STRs are plotted with solid squares. In the region between ≈7.5 and ≈17.5 Mb (dashed lines), where the highest lod scores are found, low lod scores were found primarily for uninformative SNPs with high homozygosity. Regions with no significant linkage signal contained both informative and noninformative markers, indicating that the lack of a linkage signal in these regions was not likely because of a lack of informative markers.
Generation of SNPs for Fine Mapping the HVA QTL.
To provide support for the linkage signal and to better define the QTL, we performed high-resolution genotyping of the same vervets with additional markers from the QTL region of vervet chromosome 9p. We did not identify any additional human STR markers that map to this chromosomal region, that could be amplified in vervet, and that are polymorphic in the VRC. Therefore, we reasoned that high-resolution mapping could be best achieved by identifying and typing large numbers of vervet SNPs. Because there is no vervet reference genome sequence, we used human and rhesus genome assemblies to drive vervet SNP discovery (Fig. 2). We first aligned the orthologous human and rhesus genomic sequences to identify invariant sequences. We hypothesized that these invariant sequences would also be conserved in vervet and thus could be used for SNP discovery PCR amplicon design, because (i) rhesus and vervet are members of the same subfamily (Cercopithecinae), and both are about equidistantly related to humans; (ii) our genetic map construction had shown a very high degree of conservation of marker orders between vervet and rhesus throughout the genome (6); and (iii) vervet chromosome 9p collinearity was further supported by paired BAC end sequence analysis.
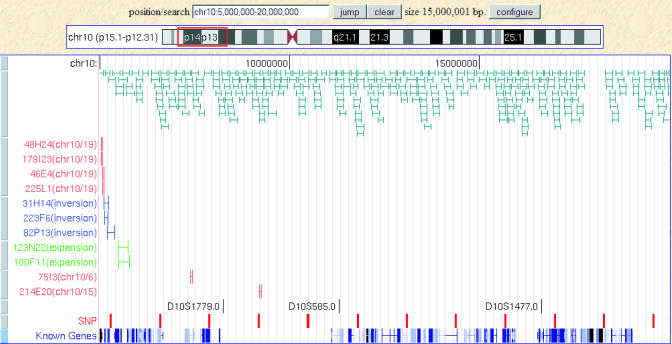
Fig. 2.
Vervet and human genome collinearity. The 15-Mb human region (hg18 chr10:5,000,000–20,000,000) encompassing to the area of highest lod scores (orange box) is shown on the University of California, Santa Cruz, browser (http://genome.ucsc.edu). A series of custom tracks displaying vervet BAC end sequence alignments and inferred BAC clone orthologous relationships are also shown. A total of 315 BAC clones displaying appropriate end sequence orientations and distances are presented in green, collectively covering 93% of the interval. A series of BAC clones with noncolinear end sequence alignment results are also shown: four clones (in red) with one end aligning to human chromosome 10 and the other to human chromosome 19 (in red); three clones with both ends aligning to human chromosome 10 on the same DNA strand (in blue); two clones with inferred sizes more than three standard deviations of the average clone size (in green); and two clones (in red) with one end aligning to human chromosome 10 and the other to a different human chromosome. Vervet-mapping STRs are presented in black, and the positions of SNP clusters are shown in red. Known genes are shown in a dense format, highlighting the scarcity of known genes in the D10S1779–D10S585 interval within the area of significant linkage.
We selected invariant sequences flanking polymorphic regions to enrich for successful primers, which could be used to amplify regions potentially harboring vervet SNPs. The analysis of the proximal 35 Mb of human chromosome 10 and its orthologous region in rhesus yielded 70,076 segments of at least 50 consecutive nucleotides showing 100% identity between human and rhesus. Of these segments, we observed 33,381 cases where the neighboring invariant primer sites were separated by 100–600 nt of polymorphic sequence. The largest distance between these candidate amplicon sites (invariant primer, polymorphic sequence, and invariant primer) was <70 kb, indicating we could design PCR assays for SNP discovery at high resolution across the entire region. We generated amplicons in a hierarchical manner by designing assays in 24 clusters where each cluster was separated by an average physical distance of 1.25 Mb. At each cluster, four independent assays were designed, where each assay was separated by at least 4 kb, but all assays were within a window of <50 kb. Each assay was analyzed for sequence polymorphism on a test set of four to six unrelated vervet monkeys. In total, we identified 105 SNPs, which were then genotyped in all animals for which data on HVA levels were available.
Fine-Mapping Analyses.
As with the original scan, the size and complexity of the VRC pedigree precluded multipoint analyses, and thus only two-point QTL analyses for HVA were conducted on the 105 SNPs (Fig. 2); two SNPs, DHs36-12 and DHs48-08-6, had significant lod scores with HVA (4.23 and 3.96, respectively). These SNPs accounted for 60% and 56% of HVA heritable trait variance, respectively. Within this QTL, seven markers showed at least suggestive evidence for linkage (lod score >2); the linked SNPs indicate a QTL of <10 Mb, encompassing <40 genes in the RefSeq database. Most of the SNPs that did not provide evidence for linkage of HVA to this region were uninformative in the VRC sample.
Discussion
CSF HVA concentration exemplifies a behavior-related trait, amenable to genetic investigation in NHPs but difficult or impossible to study in large human samples because of the invasiveness of the sampling that is required. With a large NHP pedigree, we were able to first identify suggestive linkage, using the vervet STR map, and then, using SNPs, identify a well delineated QTL for this trait on vervet chromosome 9, syntenic with human chromosome 10p. This QTL exceeds the genome-wide threshold for significance in a two-stage linkage study (36).
Linkage has been observed for human disorders in the region on human chromosome 10 syntenic to the vervet HVA QTL region, including traits hypothesized to involve abnormalities of dopamine systems, such as schizophrenia and bipolar disorder (37). There are no obvious candidate genes within the HVA QTL, based on the criterion of a known direct involvement in dopamine metabolism, but several genes in this region could contribute to pathways involved in its regulation. The peak of the QTL (between SNPs DHs36-12 and DHs48-08-6, at locations 10.5 and 14.4 Mb, respectively) includes genes for huntingtin-interacting protein, optineurin, which contributes to metabotropic glutamate receptor desensitization (38), and CUGBP2. The latter gene regulates alternative splicing of several known CNS targets (39), and among its putative targets is frequenin (FREQ or neuronal calcium sensor 1 gene, NCS-1) (40), which inhibits dopamine-induced D2 receptor internalization (41). GATA3, located ≈2.3 Mb telomeric to SNP DHs36–12 participates in transcriptional regulation of serotonergic neuron differentiation in caudal raphe nuclei (42) and transactivation of genes involved in peripheral MA metabolism (43).
Most of the HVA QTL region overlaps an ≈3-Mb gene desert between GATA3 and CUGBP2, which, in addition to one locus for a hypothetical protein, contains three evolutionary conserved regions (minimum of 200 bp with >99% identity between human and rodents; http://ecrbrowser.dcode.org) and a genomic element in which evolution was accelerated in the human lineage (44). These observations suggest that this gene-poor region could be the site of a functional locus, for example, for noncoding RNA or for a cis-acting regulator involved in expression of more distant genes.
The mapping results reported here do not preclude other loci playing a role in HVA. Similarly, although neither the analysis of MHPG and 5-HIAA nor the analysis of ratios of the various MA measures gave significant QTL results, additional genetic-mapping studies of these traits may be warranted either through linkage analysis of an expanded pedigree sample or through association analyses of independent vervets once sufficient mapped SNPs are available.
The two-stage mapping strategy used here succeeded despite the complexity of the pedigree structure and extensive inbreeding (45) in the VRC, which made multipoint linkage analysis unfeasible, and our initial fine-mapping strategy based on clustering of closely spaced SNPs to form multialleic “supermarkers” impractical. However, our success using two-point analyses suggests that the power of very extended pedigrees such as the VRC may be sufficient for QTL analyses more generally, providing that one can identify and genotype a large number of SNPs. As we observed (Fig. 2), with enough SNPs, some of them will be reasonably informative within such a pedigree, even while most of the markers typed display low heterozygosity and thus generate little information regarding linkage.
We hypothesize that the combination of the demographic history and the size and structure of the VRC pedigree enabled us to obtain significant linkage evidence for HVA with a high locus-specific heritability. Linkage analysis in extended NHP pedigrees may be a powerful alternative to genome-wide association analysis for mapping behavior-related QTL. Furthermore, the world-wide availability of samples of independent NHPs, suggests that for many behavior-related traits, it may be possible to conduct association analyses with densely spaced SNPs, to identify the variants contributing to the QTL. For example, such a strategy can be pursued for HVA using CSF samples collected from independent vervets living on St. Kitts.
The mapping of complex behavior-related traits in NHPs has been hampered by a combination of insufficient genetic markers and phenotyping on too few samples. In this report, we describe a method to generate additional genetic markers in a focused manner. Not only can we target specific genomic regions, but we can also develop markers at a desired density and spacing. Our comparative approach used cross-species sequence alignments for three purposes: to identify invariant sequences as a guide for primer design, to identify polymorphic sequences as a guide for SNP discovery, and to use BAC paired end sequences as a guide for evaluating genome collinearity. In mapping HVA, we showed that SNPs could be used successfully to reproduce and refine STR-based mapping results, and that a targeted region in the vervet genome is expected to show strong synteny and similar gene content to the orthologous region of humans.
The use of comparative genomics information can also be applied more broadly to investigate complex traits in other NHP species. Just as we used the rhesus genome assembly as a surrogate for the vervet, it should be equally applicable for other OWM. Outside of the OWM, the reference genomes of a small set of key species could similarly act as references for each entire clade. Alternatively, a species lying at the base of the primate phylogenetic tree could be used as the reference for the entire order. To illustrate this possibility we examined alignments of gray mouse lemur (Microcebus murinus) genomic sequence to human across a 1.6-Mb segment of the Encode ENm001 region (human hg18: chr7:115,597,757–117,475,182). If this region is representative of these two genomes, we calculate it would be feasible to produce amplicons at an average spacing of 50 kb, with 94% of amplicons being separated from each other by <200 kb. This amplicon set could thus support SNP discovery and high-resolution analysis for virtually all NHPs.
Our study also provides evidence of a very high degree of genome collinearity between vervet and human. This observation further supports presumptions of marker order and spacing based on the reference genome, especially because large rearrangements can be subsequently identified after genotyping and genetic mapping. It also accelerates candidate gene analysis, because the reference genome gene set likely represents the vast majority of genes in the region in the species of interest. If genome collinearity tends to hold true for all NHP species at a regional scale (1–10 Mb), then generation of SNP markers can become further simplified to take advantage of massively parallel sequencing strategies and an assumption of conserved marker order. For example, in a currently available scenario, a single experiment to produce 400,000 reads of 250 bp would randomly produce sequence reads at an average of 8 kb across the genome. These sequences could be generated directly from the species or even individuals of choice. Primer design for subsequent SNP discovery assays would be simplified, although the sequences could still be evaluated for evolutionary conservation to enrich for assays flanking polymorphic regions.
Methods
Subjects.
The VRC vervets can be connected into one large pedigree. The pedigree used in the current study contained 673 vervets, including 92 “dummy” parents to take the place of unknown fathers for statistical analysis. Most matings between vervets in this pedigree resulted in only one offspring; however, of 363 matings between two known vervets, 70 produced full siblings. There is extensive inbreeding in the VRC. The average inbreeding coefficient, the probability that a subject receives two alleles at a locus that are identical by descent, for the entire pedigree, is 0.00639. There are 84 individuals with nonzero inbreeding coefficients, and the average inbreeding coefficient among this subset is 0.05122 (range: 0.004–0.25). To put these values in perspective, the average inbreeding coefficient for children born after 1950 from consanguineous marriages in the Old Order Amish is 0.012 (46). There are 107 unique fathers in the pedigree, with an average (range) number of 3.4 (1–11) mates, and an average (range) number of 4.2 (1–17) offspring. There are 182 unique mothers in the pedigree, with an average (range) number of 2 (1–5) mates and an average (range) number of 2.8 (1–11) offspring. There was an average of 4.7 alleles for each STR marker (range, 2–10). The average expected heterozygosity for the markers was 0.64 (range, 0.02–0.84). A partial depiction of the pedigree is included in SI Fig. 3.
MA concentrations were measured in 347 vervet monkeys (128 males and 219 females), 3–23 years of age, who were unambiguously positioned within the VRC pedigree. All subjects were mother-reared and socially housed in large outdoor cages. The phenotyped sample includes the following relative pairs: parent–offspring (n = 240), full siblings (n = 45), and half siblings (n = 920), in addition to other classes of relative pairs.
Colony-management practices at the VRC have been designed to reflect the natural social composition of vervet groups in the wild. All animals are reared by their mothers in large outdoor enclosures. Infants and juveniles remain in the natal group with their mothers and female kin. Males are removed at adolescence and transferred to other groups, and adult males are rotated between groups at 3- to 4-year intervals, to mimic natural processes of emigration and immigration. These policies are intended to promote normal development and provide opportunities for animals to express age- and sex-appropriate species-typical behavior. The maintenance of the population in stable matrilineal groups allows the study of naturally occurring individual differences in behavior and physiology in a developmental and social context that is within the normal range for the species.
CSF Sampling and MA Measurements.
These procedures were carried out under a protocol approved by the Animal Research Committee at University of California, Los Angeles. Animals were captured and anesthetized with 8–10 mg/kg ketamine hydrochloride for CSF sample collection. The hair was shaved on the nape of the neck, the skin was scrubbed with betadine and isopropyl alcohol, and ≈1 cc of CSF was withdrawn from the cisterna magna, within 30 min of anesthesia.
After collection, CSF samples were immediately transferred to amber Eppendorf tubes and placed on wet ice. Samples were centrifuged at 4°C for 15 min at 1,800 × g to remove blood cells, and the supernatant was transferred to new amber cryogenic tubes and frozen at −80°C. All sample collections were performed between mid-December and mid-March. Two-thirds (n = 245) of the animals were sampled in 2 successive years. The remaining third (n = 102) was sampled in 1 of the 2 years.
The samples were assayed for MA metabolites by HPLC with an electrochemical detector (47). A measured aliquot of each sample was mixed with an equal volume of cold mobile phase, the mixture was filtered at centrifugation (6,000 × g for 40 min at 4°C), and part of the filtrate was transferred to a 300-FL microinjection insert. This material was then analyzed by HPLC with electrical detection to allow simultaneous measurement of HVA, 5-HIAA, and MHPG.
To control for annual variation in the results of the MA metabolite assays, the data for each year were standardized by subtracting each subject's value from the mean and dividing by the standard deviation for that year. The mean standardized scores for HVA, 5-HIAA, and MHPG were calculated for the subjects with data from both years.
Heritability Estimates.
The heritability of quantitative CSF phenotypes was estimated by using a variance component approach as modeled in the software package SOLAR (35). Covariates of sex and age at the time the phenotypes were measured were included in all analyses, using stepwise regression. For HVA and MHPG, age was coded as an ordered categorical variable with six levels: age category 7 (vervets born before 1990), age category 6 (vervets born from 1991 to 1994), age categories 5–2 (vervets born between 1998 and 1995, respectively), and age category 1 (vervets too young for CSF sampling). For 5-HIAA, a binary age variable was constructed, indicating whether an individual was juvenile or adult. The analysis of 5-HIAA included an interaction term for sex and age, and the analysis of MHPG included a term for the age–sex interaction, a term for age category squared, and a term for the sex and age category squared interaction. Additive genetic heritability (also called narrow-sense heritability) of the CSF phenotypes was estimated, using the pedigree structure and correlation in trait values between pairs of relatives. No dominance effects were modeled. Examination of plots of the residuals from all models including the covariates indicated above showed no unusual patterns, and the distribution of the residuals did not deviate substantially from a normal distribution.
Marker Development and Genotyping.
We used the human genome (hg17 assembly; National Center for Biotechnology Information build35) and rhesus genome (rheMac2) assemblies and alignments (available at http://genome.ucsc.edu) to identify invariant sequences along human chromosome 10p, syntenic to vervet chromosome 9. Pairs of invariant sequences separated by 100–600 nt of polymorphic sequence were subjected to primer design criteria to control for melting temperature and predicted amplicon length. From 33,381 candidates, 24 clusters of four amplicons were selected at ≈1.25-Mb intervals along human chromosome 10p. For each cluster, a window of 25–50 kb was identified for which four amplicons could be selected, with each amplicon >5 kb distant from its neighbor. The 96 amplicons were assayed on genomic DNAs of four to six unrelated vervet monkeys from the VRC and subjected to DNA sequencing on both strands.
A set of 114 SNPs were genotyped on 347 vervet monkey genomic DNAs by using a Sequenom procedure, giving rise to genotyping data for 105 SNPs from 22 of the 24 clusters. All sequences were compared with a more recent build of the human genome (hg18 assembly; National Center for Biotechnology Information build 36.1) to reconfirm marker order and positions.
QTL Mapping.
Human STR markers genotyped in the vervet pedigree had previously been placed in a scaffold genetic map (6). In these STR data, 455 of the 673 vervets are genotyped for at least one marker, and in the SNP data, 367 of the 673 vervets are genotyped. Maximum-likelihood estimates of marker allele frequencies were calculated by using the SOLAR software, accounting for relationships among individuals. Conditional kinship coefficients, estimating the probability of sharing one or two marker alleles identical by descent, were calculated for each marker, using the observed genotype data and pedigree structure. Two-point linkage analysis compared the likelihood of the null hypothesis that the component of variance because of a QTL at the marker locus was zero against the alternative that it was not zero. The effects of additional, unmeasured QTL on the HVA outcome were absorbed into the residual component of variance. Two-point linkage was performed on 324 autosomal markers. Complete results are in SI Dataset 1.
Supplementary Material
Acknowledgments
We thank Jessica Wasserscheid for physical map bioinformatics and Gary Leveque and Corina Nagy for BAC end sequencing. This work was supported in part by National Institutes of Health Grants MH061852 and RR019963 (to L.A.F.) and RR016300 (to N.B.F.), by Conte Neuroscience Center Grant 5P50 MH62185 (to J.J.M.), and by funds from the University of California, Los Angeles, Semel Institute. K.D. is supported by funding from the National Institutes of Health, Genome Canada, and Genome Québec.
Abbreviations
- NHP
non-human primate
- OWM
Old World monkey
- VRC
Vervet Research Colony
- MA
monoamine
- HVA
homovanillic acid
- 5-HIAA
5-hydroxyindolacetic acid
- MHPG
3-hydroxy-4-methoxyphenylglycol
- CSF
cerebrospinal fluid
- STR
short tandem repeat
- QTL
quantitative trait locus
- lod
logarithm of odds.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707640104/DC1.
References
- 1.Kendler KS, Baker JH. Psychol Med. 2006;19:1–12. [Google Scholar]
- 2.Suomi SJ, Higley JD. NIDA Res Monogr. 1991;114:291–302. [PubMed] [Google Scholar]
- 3.Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- 4.Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A, et al. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Cox LA, Mahaney MC, Vandeberg JL, Rogers J. Genomics. 2006;88:274–281. doi: 10.1016/j.ygeno.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Jasinska AJ, Service SK, Levinson M, Slaten E, Lee O, Sobel E, Fairbanks LA, Bailey JN, Jorgensen MJ, Breidenthal SE, et al. Mamm Genome. 2007;18:347–360. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 9.Rhesus Macaque Genome Sequencing and Analysis Consortium. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 10.McGuire MT. Contributions to Primatology. Vol 1. Basel: Karger; 1974. [Google Scholar]
- 11.Newman TK, Fairbanks LA, Pollack D, Rogers J. Am J Primatol. 2002;56:237–243. doi: 10.1002/ajp.1078. [DOI] [PubMed] [Google Scholar]
- 12.Fairbanks LA, Newman TK, Bailey JN, Jorgensen MJ, Breidenthal SE, Ophoff RA, Comuzzie AG, Martin LJ, Rogers J. Biol Psychiatry. 2004;55:642–647. doi: 10.1016/j.biopsych.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Fairbanks LA, Melega WP, Jorgensen MJ, Kaplan JR, McGuire MT. Neuropsychopharmacology. 2001;24:370–378. doi: 10.1016/S0893-133X(00)00211-6. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan JR, Phillips-Conroy J, Fontenot MB, Jolly CJ, Fairbanks LA, Mann JJ. Neuropsychopharmacology. 1999;20:517–524. doi: 10.1016/S0893-133X(98)00078-5. [DOI] [PubMed] [Google Scholar]
- 15.Castellanos FX, Elia J, Kruesi MJ, Marsh WL, Gulotta CS, Potter WZ, Ritchie GF, Hamburger SD, Rapoport JL. Neuropsychopharmacology. 1996;14:125–137. doi: 10.1016/0893-133X(95)00077-Q. [DOI] [PubMed] [Google Scholar]
- 16.Scheepers FE, Gispen-de Wied CC, Westenberg HG, Kahn RS. Neuropsychopharmacology. 2001;25:468–475. doi: 10.1016/S0893-133X(01)00250-0. [DOI] [PubMed] [Google Scholar]
- 17.Higley JD, Linnoila M. Rec Dev Alcohol. 1997;13:191–219. [PubMed] [Google Scholar]
- 18.Higley JD, Linnoila M. Ann NY Acad Sci. 1997;836:39–56. doi: 10.1111/j.1749-6632.1997.tb52354.x. [DOI] [PubMed] [Google Scholar]
- 19.Fairbanks LA, Fontenot MB, Phillips-Conroy JE, Jolly CJ, Kaplan JR, Mann JJ. Brain Behav Evol. 1999;53:305–312. doi: 10.1159/000006601. [DOI] [PubMed] [Google Scholar]
- 20.Mehlman PT, Higley JD, Faucher I, Lilly AA, Taub DM, Vickers J, Suomi SJ, Linnoila M. Am J Psychiatry. 1994;151:1485–1491. doi: 10.1176/ajp.151.10.1485. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan JR, Manuck SB, Fontenot MB, Mann JJ. Neuropsychopharmacology. 2002;26:431–443. doi: 10.1016/S0893-133X(01)00344-X. [DOI] [PubMed] [Google Scholar]
- 22.Mehlman PT, Higley JD, Fernald BJ, Sallee FR, Suomi SJ, Linnoila M. Psychiatry Res. 1997;72:89–102. doi: 10.1016/s0165-1781(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 23.Howell S, Westergaard G, Hoos B, Chavanne TJ, Shoaf SE, Cleveland A, Snoy PJ, Suomi SJ, Higley JD. Am J Primatol. 2007;69:1–15. doi: 10.1002/ajp.20369. [DOI] [PubMed] [Google Scholar]
- 24.Swann AC, Secunda S, Davis JM, Robins E, Hanin I, Koslow SH, Maas JW. Am J Psychiatry. 1983;140:396–400. doi: 10.1176/ajp.140.4.396. [DOI] [PubMed] [Google Scholar]
- 25.Roy A, Pickar D, Linnoila M, Doran AR, Paul SM. Arch Gen Psychiatry. 1986;43:356–360. doi: 10.1001/archpsyc.1986.01800040066010. [DOI] [PubMed] [Google Scholar]
- 26.Sher L, Oquendo MA, Li S, Huang Y, Grunebaum MF, Burke AK, Malone KM, Mann JJ. Neuropsychopharmacology. 2003;28:1712–1719. doi: 10.1038/sj.npp.1300231. [DOI] [PubMed] [Google Scholar]
- 27.Blennow K, Wallin A, Gottfries CG, Månsson J-E, Svennerholm L. J Neural Trans. 1993;3:1435–1463. doi: 10.1007/BF02260910. [DOI] [PubMed] [Google Scholar]
- 28.Oxenstierna G, Edman G, Iselius L, Oreland L, Ross SB, Sedvall G. J Psychiatr Res. 1986;20:19–29. doi: 10.1016/0022-3956(86)90020-8. [DOI] [PubMed] [Google Scholar]
- 29.Higley JD, Thompson WW, Champoux M, Goldman D, Hasert MF, Kraemer GW, Scanlan JM, Suomi SJ, Linnoila M. Arch Gen Psychiatry. 1993;50:615–623. doi: 10.1001/archpsyc.1993.01820200025003. [DOI] [PubMed] [Google Scholar]
- 30.Clarke AS, Kammerer CM, George KP, Kupfer DJ, McKinney WT, Spence MA, Kraemer GW. Biol Psychiatry. 1995;38:572–577. doi: 10.1016/0006-3223(95)00042-4. [DOI] [PubMed] [Google Scholar]
- 31.Rogers J, Martin LJ, Comuzzie AG, Mann JJ, Manuck SB, Leland M, Kaplan JR. Biol Psychiatry. 2004;55:739–744. doi: 10.1016/j.biopsych.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 32.Jonsson EG, Nothen MM, Gustavsson JP, Neidt H, Bunzel R, Propping P, Sedvall GC. Psychiatry Res. 1998;79:1–9. doi: 10.1016/s0165-1781(98)00027-4. [DOI] [PubMed] [Google Scholar]
- 33.Jonsson EG, Bah J, Melke J, Abou Jamra R, Schumacher J, Westberg L, Ivo R, Cichon S, Propping P, Nothen MM, et al. BMC Psychiatry. 2004;4:4. doi: 10.1186/1471-244X-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, Champoux M, Suomi SJ, Linnoila MV, Higley JD. Mol Psychiatry. 2002;7:118–122. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- 35.Almasy L, Blangero J. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruglyak L, Daly MJ. Am J Hum Genet. 1998;62:994–996. doi: 10.1086/301792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wildenauer DB, Schwab SG, Maier W, Detera-Wadleigh SD. Schizophr Res. 1999;39:107–111. doi: 10.1016/s0920-9964(99)00108-5. [DOI] [PubMed] [Google Scholar]
- 38.Anborgh PH, Godin C, Pampillo M, Dhami GK, Dale LB, Cregan SP, Truant R, Ferguson SS. J Biol Chem. 2005;280:34840–34848. doi: 10.1074/jbc.M504508200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W, Liu H, Han K, Grabowski PJ. RNA. 2002;8:671–685. doi: 10.1017/s1355838202027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faustino NA, Cooper TA. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabbani N, Negyessy L, Lin R, Goldman-Rakic P, Levenson R. J Neurosci. 2002;22:8476–8486. doi: 10.1523/JNEUROSCI.22-19-08476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. J Neurosci. 1999;19:RC12. doi: 10.1523/JNEUROSCI.19-12-j0002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- 44.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wijsman EM, Rothstein JH, Thompson EA. Am J Hum Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khoury MJ, Cohen BH, Diamond EL, Chase GA, McKusick VA. Am J Epidemiol. 1987;125:453–461. doi: 10.1093/oxfordjournals.aje.a114551. [DOI] [PubMed] [Google Scholar]
- 47.Scheinin M, Chang W-H, Kirk KL, Linnoila M. Anal Biochem. 1983;131:246–253. doi: 10.1016/0003-2697(83)90162-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.