Abstract
CD8+ cells from healthy HIV-1 infected individuals suppress human immunodeficiency virus (HIV) replication in infected cells by a non-cytotoxic mechanism. This activity is associated with the production of a soluble CD8+ cell antiviral factor (CAF) that inhibits viral replication at the level of transcription. Strong CD8+ cell non-cytotoxic anti-HIV responses (CNAR) correlate with an asymptomatic state and long-term survival of HIV-infected individuals. This antiviral activity is lost when the infected individual advances to disease. In attempts to define the gene(s) mediating CNAR we have evaluated differential gene expression between CD8+ cells from infected subjects with high CNAR and CD8+ cells from uninfected controls that lack this activity. The expression analysis, using the Affymetrix® GeneChip® Human Genome U133 set, indicated that 18% of the genes were differentially expressed (DE) of which 9.2% were up-regulated. A total of 568 genes were up-regulated with a >2.0 fold difference in expression levels and a >50% concordance of difference call. Stringent selection criteria narrowed down the list to 52 up-regulated ‘high confidence genes’ (≥75% concordance). These genes function in a wide variety of cellular processes and include 13 associated with immunologic activity.
Keywords: HIV replication, CD8+ cell anti-HIV response, reverse transcriptase, cDNA microarray, differential gene expression, noncytotoxic antiviral activity
Introduction
CD8+ cells from healthy HIV-1 infected individuals can suppress human immunodeficiency virus (HIV) replication in infected CD4+ cells without killing the cell (Levy, Mackewicz, and Barker, 1996; Walker et al., 1986). This CD8+ cell non-cytotoxic antiviral response (CNAR) is not restricted by class I or class II molecules (Levy, Mackewicz, and Barker, 1996; Mackewicz, Garovoy, and Levy, 1998) and is mediated at least in part by production of a soluble CD8+ cell antiviral factor (CAF) that inhibits viral replication at the level of transcription (Copeland, McKay, and Rosenthal, 1995; Mackewicz, Blackbourn, and Levy, 1995; Walker and Levy, 1989). CAF is not retrovirus-specific and lacks identity with other known cytokines, including various chemokines with anti-HIV activity (Brinchmann, Gaudernack, and Vartdal, 1991; Mackewicz, Ortega, and Levy, 1994) (for review, see (Levy, 2003)).
Clinical evidence suggests that CNAR is an innate immune response that helps prevent viral replication as well as control virus during the acute infection phase, even before seroconversion (Levy, 2001; Mackewicz et al., 1994). High CNAR correlates with an asymptomatic state and long-term survival of HIV-infected individuals (Mackewicz, Ortega, and Levy, 1991) (Gomez, Smaill, and Rosenthal, 1994; Landay, Mackewicz, and Levy, 1993). Loss of CNAR over time is usually associated with progression to disease; patients who have developed AIDS do not show this anti-HIV response (Barker et al., 1998; Gomez, Smaill, and Rosenthal, 1994; Landay, Mackewicz, and Levy, 1993; Levy, Mackewicz, and Barker, 1996; Mackewicz, Ortega, and Levy, 1991). These findings suggest that CNAR plays an important role in controlling HIV infection and is one of the major determinants of a beneficial clinical course. Moreover, CNAR has been detected in exposed (high risk) uninfected subjects (within 1 year) suggesting a role in protection from infection (Stranford et al., 1999).
Differential expression data has proven to be useful in the discovery of novel genes or biological functions. In the present study we have chosen this approach in attempts to identify a CNAR-associated gene(s). The gene expression profile of CD8+ cells from HIV-1 infected subjects with high CNAR and CD8+ cells from uninfected controls that lack this antiviral activity has being evaluated using high-density oligonucleotide microarrays (Affymetrix® GeneChip® Human Genome U133 Set). The identification of the gene (s) responsible for this natural anti-HIV response should provide a valuable novel approach for therapy of infected people.
Results
Acute Infection Assay for CD8+ Cell Noncytotoxic Anti-HIV Response (CNAR)
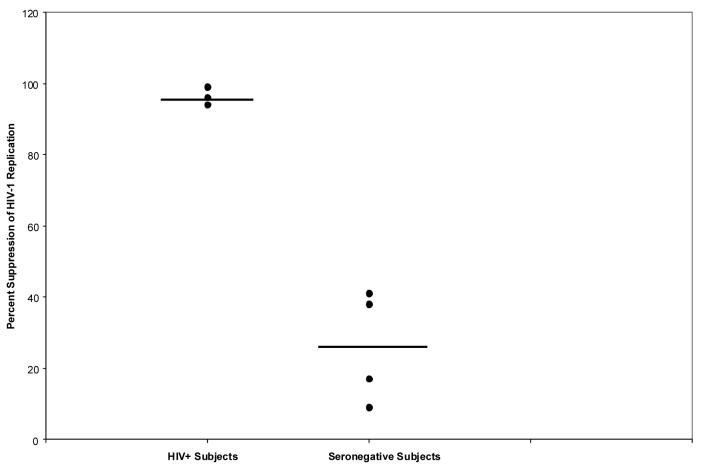
Acute infection assays were performed on CD8+ cells from four healthy HIV-infected subjects and four seronegative subjects. After a five day co-culture of CD8+ cells with CD4+ cells at a 1:1 cell input ratio, the percentage of suppression was calculated by comparing the average value of RT activity in culture fluids from control wells containing replicates (x3) of infected CD4+ cells grown alone with the average RT activity in the co-cultures. The average percent of HIV-1 replication was 96% for HIV-infected individuals (high CNAR) and 26% for seronegative subjects (Figure 1).
Figure 1.
Percent suppression of HIV-1 replication by CD8+ cells from seronegative controls and HIV-1 infected subjects with strong CNAR. Suppression of HIV-1SF33 replication was assessed at day 5 of an acute infection assay at a 1:1 CD8+ cell/CD4+ cell ratio.
Microarray Expression Analysis
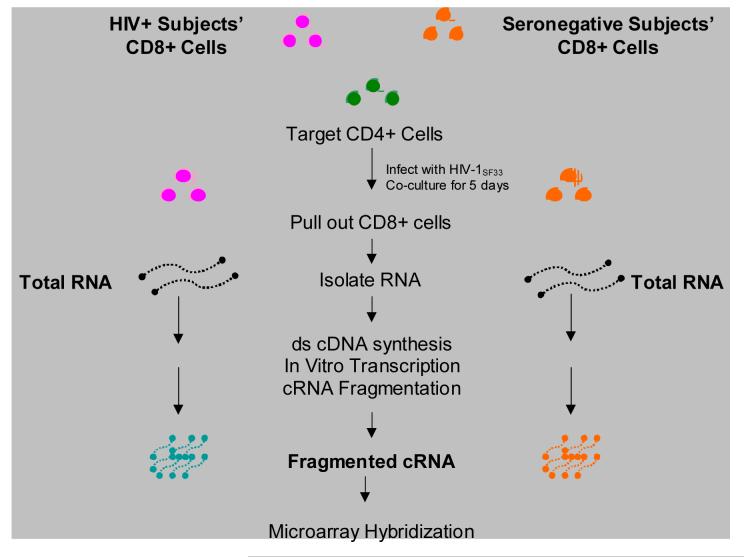
CD8+ cells’ cRNA from four healthy HIV-infected subjects with strong CNAR and four HIV-seronegative individuals that lack this activity were hybridized to the Affymetrix® GeneChip® Human Genome U133 set (representing ∼33,000 human genes, including EST sequences) and data were submitted to gene expression analysis using GeneChip® Analysis Microarray Suite v5, MicroDB™ v3.0 and Data Mining Tool Software v3.0 (Figure 2).
Figure 2.
Preparation of cRNA for microarray hybridization. Total RNA was isolated and purified from CD8+ cells from HIV+ subjects showing high CNAR and seronegative control subjects with none or low CNAR using a combined Trizol / RNeasy method. Complementary DNA (cDNA) was synthesized with the SuperScript™ Double Stranded cDNA Synthesis Kit. The cDNA was then subjected to an in vitro transcription (IVT) reaction and the product fragmented to generate RNA fragments of approximately 35 to 200 bases. Samples were analyzed on 1% agarose gels. See Materials and Methods for details.
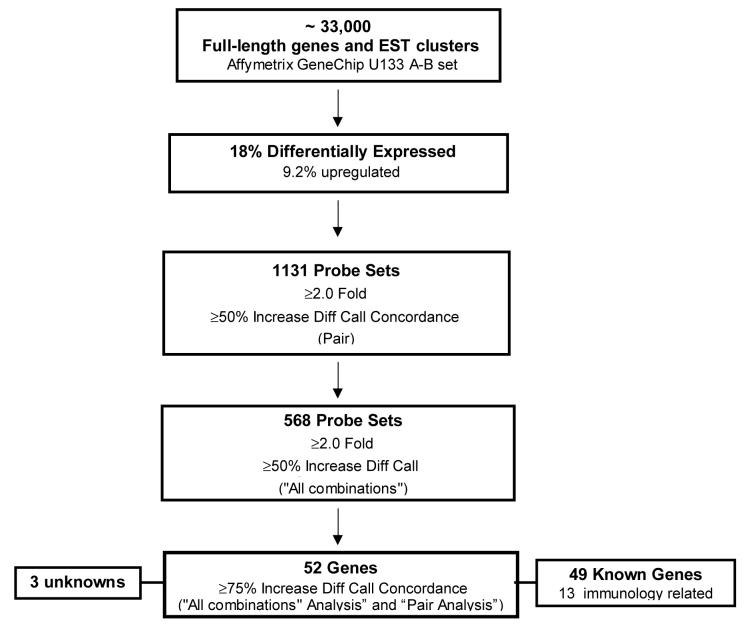
Comparison expression analysis (see Materials and Methods), revealed that 18% of the genes were differentially expressed (DE) of which 9.2% were up-regulated. A total of 568 genes were up-regulated with a >2.0 fold difference in expression levels and a >50% concordance of difference call. A ‘Comparison Ranking Test’ using a cut-off value of ≥75% was used to increase the confidence that the transcripts selected were indeed DE and to generate a more manageable number of genes for further analysis (Figure 3). These stringent selection criteria narrowed down the list to 69 up-regulated ‘high confidence genes’. HLA associated genes (which are not relevant to CNAR) (Levy, Mackewicz, and Barker, 1996; Mackewicz, Garovoy, and Levy, 1998) and 10 sequences with repeats were deleted generating a final list of 52 genes. 13 of these are associated with immune responses and three are unknowns (Figure 3). A summary of the information gathered for each upregulated gene identified that includes the Affymetrix probe identification number, gene title, gene symbol, chromosome location and Entrez reference number is presented in Table 1.
Figure 3.
The differential gene expression between CD8+ cells from HIV+ subjects with high CNAR and CD8+ cells from uninfected controls that lack this activity was evaluated using the Affymetrix® GeneChip® Human Genome U133 Set. Expression analysis indicated that 18% of the genes were differentially expressed (DE); 9.2% up-regulated. Further analysis of DE genes revealed a total of 568 up-regulated genes with a >2.0 fold difference in expression levels and a >50% concordance of difference call. Stringent selection criteria narrowed down the list to 52 up-regulated ‘high confidence genes’ (≥75% concordance). These genes function in a wide variety of cellular processes and include 13 immunologic genes.
Table 1.
Differentially expressed genes, ≥75% Concordance.
| Probe Set ID | Gene Title | Gene Symbol | Chromosomal Location | Entrez Gene |
|---|---|---|---|---|
| 1405_i_at | chemokine (C-C motif) ligand 5 | CCL5 | 17q11.2-q12 | 6352 Entrez gene |
| 200800_s_at | heat shock 70kDa protein 1A | HSPA1A | 6p21.3 | 3303 Entrez gene |
| 200878_at | endothelial PAS domain protein 1 | EPAS1 | 2p21-p16 | 2034 Entrez gene |
| 201889_at | family with sequence similarity 3, member C | FAM3C | 7q22.1-q31.1 | 10447 Entrez gene |
| 202085_at | tight junction protein 2 (zona occludens 2) | TJP2 | 9q13-q21 | 9414 Entrez gene |
| 202206_at | ADP-ribosylation factor-like 7 | ARL7 | 2q37.1 | 10123 Entrez gene |
| 202265_at | polycomb group ring finger 4 | PCGF4 | 10p11.23 | 648 Entrez gene |
| 202551_s_at | cysteine rich transmembrane BMP regulator 1 (chordin-like) | CRIM1 | 2p21 | 51232 Entrez gene |
| 202760_s_at | A kinase (PRKA) anchor protein 2 | AKAP2 | 9q31-q33 | 11217 Entrez gene |
| 203232_s_at | ataxin 1 | ATXN1 | 6p23 | 6310 Entrez gene |
| 203868_s_at | vascular cell adhesion molecule 1 | VCAM1 | 1p32-p31 | 7412 Entrez gene |
| 203946_s_at | arginase, type II | ARG2 | 14q24.1-q24.3 | 384 Entrez gene |
| 203989_x_at | coagulation factor II (thrombin) receptor | F2R | 5q13 | 2149 Entrez gene |
| 204103_at | chemokine (C-C motif) ligand 4 | CCL4 | 17q12 | 6351 Entrez gene |
| 204529_s_at | thymus high mobility group box protein TOX | TOX | 8q12.1 | 9760 Entrez gene |
| 204798_at | v-myb myeloblastosis viral oncogene homolog (avian) | MYB | 6q22-q23 | 4602 Entrez gene |
| 205114_s_at | chemokine (C-C motif) ligand 3 | CCL3 | 17q11-q21 | 414062 Entrez gene |
| 206666_at | granzyme K (granzyme 3; tryptase II) | GZMK | 5q11-q12 | 3003 Entrez gene |
| 207229_at | killer cell lectin-like receptor subfamily A, member 1 | KLRA1 | 12p13-p12 | 10748 Entrez gene |
| 207536_s_at | tumor necrosis factor receptor superfamily, member 9 | TNFRSF9 | 1p36 | 3604 Entrez gene |
| 208690_s_at | PDZ and LIM domain 1 (elfin) | PDLIM1 | 10q22-q26.3 | 9124 Entrez gene |
| 208949_s_at | lectin, galactoside-binding, soluble, 3 (galectin 3) | LGALS3 | 14q21-q22 | 3958 Entrez gene |
| 209348_s_at | v-maf musculoaponeurotic fibrosarcoma oncogene homolog | MAF | 16q22-q23 | 4094 Entrez gene |
| 210116_at | SH2 domain protein 1A, Duncan′s disease | SH2D1A | Xq25-q26 | 4068 Entrez gene |
| 210140_at | cystatin F (leukocystatin) | CST7 | 20p11.21 | 8530 Entrez gene |
| 210321_at | granzyme H (cathepsin G-like 2, protein h-CCPX) | GZMH | 14q11.2 | 2999 Entrez gene |
| 210354_at | interferon, gamma | IFNG | 12q14 | 3458 Entrez gene |
| 210732_s_at | lectin, galactoside-binding, soluble, 8 (galectin 8) | LGALS8 | 1q42-q43 | 3964 Entrez gene |
| 211269_s_at | interleukin 2 receptor, alpha | IL2RA | 10p15-p14 | 3559 Entrez gene |
| 211503_s_at | RAB14, member RAS oncogene family | RAB14 | 9q32-q34.11 | 51552 Entrez gene |
| 212398_at | radixin | RDX | 11q23 | 5962 Entrez gene |
| 213649_at | splicing factor, arginine/serine-rich 7, 35kDa | SFRS7 | 2p22.1 | 6432 Entrez gene |
| 214452_at | branched chain aminotransferase 1, cytosolic | BCAT1 | 12pter-q12 | 586 Entrez gene |
| 214615_at | purinergic receptor P2Y, G-protein coupled, 10 | P2RY10 | Xq21.1 | 27334 Entrez gene |
| 214657_s_at | Trophoblast-derived noncoding RNA | TncRNA | 11q13.1 | 283131 Entrez gene |
| 215001_s_at | glutamate-ammonia ligase (glutamine synthetase) | GLUL | 1q31 | 2752 Entrez gene |
| 216834_at | regulator of G-protein signalling 1 | RGS1 | 1q31 | 5996 Entrez gene |
| 217147_s_at | T cell receptor associated transmembrane adaptor 1 | TRAT1 | 3q13 | 50852 Entrez gene |
| 219371_s_at | Kruppel-like factor 2 (lung) | KLF2 | 19p13.13-p13.11 | 10365 Entrez gene |
| 220990_s_at | transmembrane protein 49 | TMEM49 | 17q23.2 | 406991 Entrez gene |
| 223519_at | sterile alpha motif and leucine zipper containing kinase AZK | ZAK | 2q24.2 | 51776 Entrez gene |
| 224917_at | microRNA 21 | MIRN21 | 406991 Entrez gene | |
| 226683_at | Sorting nexin associated golgi protein 1 | SNAG1 | 5q11.2 | 112574 Entrez gene |
| 228964_at | PR domain containing 1, with ZNF domain | PRDM1 | 6q21-q22.1 | 639 Entrez gene |
| 231776_at | eomesodermin homolog (Xenopus laevis) | EOMES | 3p21.3-p21.2 | 8320 Entrez gene |
| 235003_at | U2AF homology motif (UHM) kinase 1 | UHMK1 | 1q23.3 | 127933 Entrez gene |
| 235310_at | germinal center expressed transcript 2 | GCET2 | 3q13.2 | 257144 Entrez gene |
| 236787_at | CDNA FLJ35091 fis, clone PLACE6005786 | |||
| 239002_at | asp (abnormal spindle)-like, microcephaly associated | ASPM | 1q31 | 259266 Entrez gene |
| 240070_at | hypothetical protein FLJ39873 | FLJ39873 | 3q13.31 | 201633 Entrez gene |
| 243296_at | Pre-B-cell colony enhancing factor 1 | PBEF1 | 7q22.3 | 10135 Entrez gene |
| 244774_at | Phosphatase and actin regulator 2 | PHACTR2 | 6q24.2 | 9749 Entrez gene |
cRNA from the same pair of subjects (HIV+ and seronegative controls) used in this study were also hybridized to the GeneChip® Human Genome U95 Set and analyzed using both GeneChip® Analysis Microarray Suite v4.0 (MSv4) and v5.0 (MSv5). Results obtained using these previous Affymetrix platforms and analysis software were, in general, very similar to the results presented in this paper (data not shown).
Functional Analysis of the upregulated differentially expressed genes
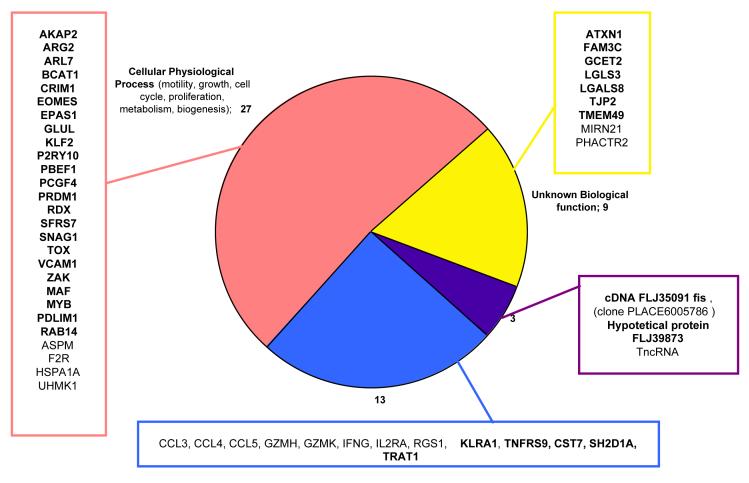
The upregulated gene’s list was submitted through the NetAffx Analysis Center and DAVID (see Materials and Methods) to obtain annotations and functional data that will help in the identification of genes that might be related to CNAR. The genes could be grouped into four categories (Figure 4): 27 genes associated with cellular physiological processes (i.e. cell cycle, motility, apoptosis, biogenesis, proliferation, metabolism), 13 genes linked to immunological processes (both cellular and humoral), 9 genes with unknown biological function (5 of them with defined molecular functions) and three unknown genes. The immunology-associated genes included chemokine ligands 3,4,and 5 (RANTES), granzyme H and K, cystatin F, tumor necrosis factor receptor superfamily member 9, killer cell lectin-like receptor subfamily A, interleukin 2 receptor alpha, and interferon gamma. Some of these immunology genes have been previously reported to be up-regulated in HIV infection (Genin et al., 1999; Ryo et al., 2000).
Figure 4.
Differentially expressed genes classified by biological function. Genes were classified in four main categories: immune response associated genes, genes associated with cellular physiological processes, genes with unknown biological functions and unknown genes. Information is presented as Œgene symbol. Refer to Table 1 for Œgene title, and additional information. Genes marked in bold were further analyzed by kRT-PCR.
The DE list also includes genes involved in a wide range of cellular physiological processes. Among these was the vascular cell adhesion molecule (VCAM) that has been associated with strong CNAR (Diaz et al., 2005). Genes that are not linked to a biological process include a family with sequence similarity 3 with cytokine-like molecular function and microRNA21 (small regulatory RNAs). The unknown genes include a hypothetical protein, FLJ39873, a trophoblast-derived non-coding RNA and a cDNA FLJ35091 fis clone PLACE6005786.
Quantitative PCR analysis of selected differentially expressed genes
In order to confirm the differential expression of genes from the U133 microarray, mRNA expression levels from 36 genes were measured by kRT-PCR. Of these 36 genes, twenty-three are involved in cellular physiological processes, five have been associated with immunological functions, seven are known genes with unknown biological function, one is a hypothetical protein and the other is a cDNA clone (Figure 4).
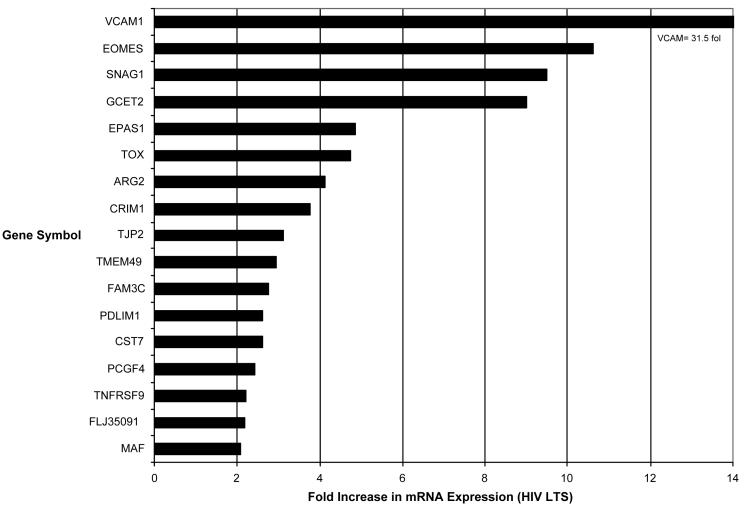
Seventeen (47.2%) genes demonstrated up-regulation of mRNA to a statistically significant level (p ≤ 0.05) in CD8+ cells from seven healthy HIV-infected subjects with strong CNAR compared to CD8+ cells from seven uninfected healthy subjects who lacked CNAR. Ten of these were part of the twenty-three genes that play a role in cellular physiological processes. Of the five genes tested with known immunological functions only CST7 and TNFRS9 were significantly upregulated in the healthy HIV-infected subjects. Four of the seven genes with unknown biological function (FAM3C, TJP2, TMEM49 and GCET2) were significantly up-regulated. The unknown clone screened (FLJ35091 fis, clone PLACE6005786) was also significantly up-regulated in the CD8+ cells from the seven healthy HIV-infected subjects compared to the CD8+ cells from seven healthy uninfected subjects. All of these genes were found to be consistently up-regulated by greater than a 2-fold increase in mRNA expression (Figure 5). Four of the seventeen genes showed greater than a 5-fold increase in expression (EOMES, SNAG1, GCET2, VCAM1), and two of the seventeen had greater than a 10-fold increase in expression (EOMES, VCAM1) (Figure 5).
Figure 5.
Fold Increase in mRNA expression. Seventeen out of thirty six genes (47.2%) tested were found to be consistently upregulated by kRT-PCR. All showed greater than a 2-fold increase in mRNA expression. Four of the seventeen genes showed greater than a 5-fold increase in expression (GCET2, SNAG1, EOMES, VCAM1), and two of the seventeen had greater than a 10-fold increase in expression (EOMES,VCAM1).
Discussion
In an attempt to define CNAR, we evaluated, using Affymetrix® genechips, the differential gene expression between CD8+ cells from infected subjects with high CNAR and CD8+ cells from uninfected controls that lack this activity (Figure 1). We conducted these studies using the Affymetrix GeneChip® U133 with improved annotation, classification and sequence quality (Affymetrix I. Array Design for the GeneChip® Human Genome U133. Santa Clara, CA, 2001) (Figure 2). Comparison ranking analysis narrowed down the up-regulated candidate gene’s list to 52 “high confidence″ genes that overlap when all possible combinations of comparisons analyses for the HIV-infected individuals and the uninfected controls were generated (≥75% concordance) (Figure 3). These genes, involved in a wide range of cellular processes, included thirteen immunology related genes and 3 unknowns (Figure 4). The identification of genes that have been previously reported to be up-regulated during viral diseases and HIV infection (e.g. IFN-γ, CCL 3-5) gave us confidence in our analysis approach. Furthermore, some of the up-regulated genes (e.g. VCAM-1) overlapped with genes identified using the Stanford Lymphochip® in separate studies conducted by our laboratory (Diaz et al., 2005).
The expression profile of the selected identified genes was confirmed using kinetic reverse transcriptase-PCR (kRT-PCR) comparing gene mRNA expression in CNAR+ and CNAR- CD8+ cells. Seventeen out of 36 genes screened (47.2%) were found to be consistently up-regulated by greater than a 2-fold in the HIV-positive subjects showing high CNAR activity (p ≤ 0.05) (Figure 5). In particular, EOMES, SNAG1, VCAM-1 and GCET2 -1, showed greater than a 5-fold increase in expression.
The eomesodermin homolog gene (EOMES) is induced in effector CD8 (+) T cells in vitro and in vivo. EOMES expression is sufficient to bring up functions of effector CD8 (+) T cells, including interferon-gamma perforin and granzyme B (Pearce et al., 2003). Some studies suggested that it is likely to act as a key regulatory gene in the development of cell-mediated immunity (Pearce et al., 2003). SNAG1 is a member of the sorting nexin family and is involved in intracellular trafficking. The specific function of this protein has not been determined (Entrez Gene ID: 112574). The finding of the VCAM gene significantly up-regulated in this study confirms previously reported data on the up-regulation of this gene associated with CNAR (Diaz et al., 2005).
A notable gene is GCET2 also termed HGAL (human germinal center-associated lymphoma). This gene is mainly expressed in germinal center B cells and is stimulated specifically by the interleukin-4 (IL-4). It encodes a cytoplasmic protein of 178 amino acids that contains an immunoreceptor tyrosine-based activation motif (ITAM). The expression of GCET2 was strongly correlated with clinical outcome in diffuse large B-cell lymphoma (DLBCL) patients (Lossos et al., 2003). Studies found that patients with DLBCL expressing high levels of GCET2 mRNA demonstrate significantly longer overall survival than patients with low expression of the gene (Lossos et al., 2003). Therefore, the finding of high expression of GCET2 in peripheral CD8+ T cells from HIV-1 subjects with high CNAR merits further evaluation.
FAM3C could be another candidate for additional investigation in CNAR. It belongs to a novel cytokine-like gene family (Zhu et al., 2002) but its biological function has yet to be determined. An unknown cDNA clone screened (FLJ35091 fis, clone PLACE6005786) was also significantly upregulated in the CD8+ cells showing CNAR. Information found at the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) regarding this cDNA clone suggests that it is expressed in lymph nodes. The protein and function has not yet been determined. Studies to define the association of this unknown protein with CNAR should also be considered.
The immunology response related genes showing greater than 2-fold increase in mRNA expression were CST7 (leukocystatin) and tumor necrosis factor receptor superfamily, member 9 (TNFRS9). CST7 encodes for cystatin F, which is a cysteine protease inhibitor that is found in peripheral blood cells and spleen. TNFRS9 is a member of the tumor necrosis factor (TNF) receptor families which are important regulators of a wide variety of physiologic processes and play a pivotal role in the regulation of immune responses.
The rest of the genes showing greater than 2-fold increase in mRNA expression are involved in transcription regulation, (EPAS1, TOX, PCGF4, MAF), nitric oxide synthesis (ARG-2), response to oxidative stress (PDLIM1, organization of epithelial and endothelial intercellular junctions (TJP2) and neurogenesis (CRIM1). They are likely not directly associated with CNAR but might be indicative of an activated immune system due to chronic infection.
In summary, using the Affymetrix GeneChip® U133 we were able to identify 52 “high confidence” genes that were up-regulated in CD8+ cells from infected subjects with high CNAR when compared to CD8+ cells from uninfected controls that lack this activity. Analysis by kRT-PCR allowed us to identify candidate genes for further analysis to determine their relevance to CNAR. The identification of the gene (s) responsible for this natural anti-HIV response should provide a valuable approach for the treatment of HIV-1 infected people.
Materials and Methods
Study Subjects
Four healthy HIV-infected subjects with strong CNAR and four HIV-seronegative individuals that lack this activity were chosen for this study. The HIV-infected subjects were males, with an average age of 48, and average number of years infected of 15. None had received antiretroviral treatment. Seronegative individuals were also males with an average age of 55 years. All participants came from the San Francisco Bay Area. This study received approval of the UCSF Committee for Human Research and participants signed an informed consent form.
CD8+ Cell Subset Purification
Heparinized wholeblood samples were collected from healthy HIV-1 infected subjects by venipuncture. Blood from uninfected subjects was obtained through the Blood Centers of the Pacific (San Francisco, CA, USA). Peripheral blood mononuclear cells (PBMC) were obtained from the blood by Ficoll/Hypaque (Sigma Chemicals, St. Louis, MO, USA) gradient separation (Castro et al., 1988). The CD8+ cellular fraction was purified from the PBMC by immunomagnetic (IM) bead separations (Miltenyi Biotech, Auburn, CA, USA) (Diaz et al., 2005). The CD8 monoclonal antibody was directed against the α chain. Flow cytometry indicated that the isolated CD8+ cell population was > 97% CD3+. Thus, a very small contamination with other cell types was possible but would not have greatly affected the results. The cells were cultured at a density of 3 × 106 cells/ml in a complete RPMI 1640 medium (Mediatech Cellgro, VA, USA) consisting of 10% (vol/vol) heat-inactivated (56°C, 30 min) fetal bovine serum (FBS) (Gibco-BRL, Gaithersburg, Maryland, USA), 1% antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin), 2 mM glutamine, and 100 units/ml recombinant IL-2 (generously provided by Glaxo Wellcome, Triangle Park, NC, USA).
In Vitro CD4+ Cell Infection
The CD4+ cellular fraction was purified from the PBMC from healthy donors by IM bead separations. The cells were stimulated with phytohemagglutinin-leucoagglutinin (PHA-L) (Sigma Chemicals, St. Louis, MO) for 3 days (3 μg/ml), washed, and treated with polybrene (2 μg/ml) (Sigma Chemicals, St. Louis, MO, USA) for 30 min. The cells (3 × 106 cells/ml) were infected with 10,000 tissue culture 50% infectious dose (TCID50) of the HIV-1SF33 isolate (Mackewicz, Blackbourn, and Levy, 1995). This cytopathic, β-chemokine-insensitive virus has been standardized in our laboratory so that the input used results in substantial virus replication within 7 to 9 days. After 1 hour, these in vitro infected cells were washed to remove free virus and resuspended at a concentration of 2 × 106 cells/ ml in the complete RPMI 1640 medium.
Acute Infection Assay to Measure the CD8+ Cell Noncytotoxic Anti-HIV Response (CNAR)
CNAR activity was evaluated using the acute infection assay (Mackewicz, Ortega, and Levy, 1991). Briefly, HIV-1SF33-acutely infected CD4+ cells, plated in 12 well plates at 106 cells/well, were mixed with either CD8+ cells from HIV-infected subjects or CD8+ cells from the uninfected controls at a 1:1 CD8+ cell: CD4+ cell input ratio. Cultures, in RPMI 1640 complete medium, were incubated for 5 days at 37°C. The extent of viral replication was measured in culture fluids by reduction in particle-associated reverse transcriptase (RT) activity (Hoffman, Banapour, and Levy, 1985) at days three and five. The percentage of suppression was calculated by comparing the average value of RT activity in culture fluids from control wells containing the infected CD4+ cells grown alone with the average RT activity in fluids from wells containing the co-culture of CD8+ and CD4+ cells.
For RNA extraction, the CD8+ cells were removed from the co-culture at day five by IM bead separation, snap frozen in liquid nitrogen and stored at -70°C. CD8+ cell samples from HIV-infected subjects were used if ≥90% of virus replication was reduced in comparison to control HIV-infected CD4+ cells cultured alone. CD8+ cells from uninfected subjects showed ≤25% reduction in virus replication and served as the CNAR negative controls.
cRNA Preparation
Total RNA was isolated and purified from both sets of CD8+ cells using a combined Trizol / RNeasy (Qiagen Inc., Valencia, CA, USA) method. Briefly, total RNA was extracted from 10 ×106 CD8+ cells using 1 ml of Trizol reagent (Invitrogen Corporation, Carlsbad, CA, USA) and 0.2 ml of chloroform. The aqueous phase was then transferred to an RNeasy column (RNeasy Mini Kit) (Qiagen Inc., Valencia, CA, USA) for RNA cleanup per manufacturer’s protocol. Complementary DNA (cDNA) was synthesized with the SuperScript™ Double Stranded cDNA Synthesis Kit (Invitrogen Corporation, Carlsbad, CA, USA) using an oligo (dT) primer containing the T7 RNA polymerase site. The cDNA was then subjected to an in vitro transcription (IVT) reaction using the T7 RNA polymerase to generate a biotin-labeled complementary RNA (cRNA) (Enzo BioArray™ High Yield™ RNA Transcript Labeling kit, Enzo Life Sciences Inc, Farmingdale, NY, USA) according to the manufacturer instructions. The IVT product was then purified (RNeasy Mini Kit, Qiagen Inc., Valencia, CA, USA) and fragmented for 35 minutes at 94°C in a Tris Acetate buffer containing Magnesium acetate (MgOAc) and Potassium Acetate (KOAc) to generate RNA fragments of approximately 35 to 200 bases. Samples were analyzed on 1% agarose gels.
Microarray Hybridization and Scanning
The quality and labeling efficiency of the fragmented cRNA (∼50 μg) from four discordant pairs (HIV-infected / uninfected subjects) was assessed using the GeneChip® Test3 Arrays (Affymetrix Inc., Santa Clara, CA, USA). The target cRNA was then hybridized to the GeneChip® Human Genome U133 Set (Affymetrix Inc., Santa Clara, CA, USA) comprising two microarrays (U133A and U133B) representing ∼33,000 human genes (including EST sequences). Hybridized probe arrays were washed and stained with a phycoerythrin- streptavidin conjugate on the GeneChip® Fluidics Station 400 following the manufacturer’s instructions. GeneChip® probe arrays were then scanned at 488nm in the Agilent GeneArray® Scanner to generate images containing intensity data for each probe cell on the arrays. Hybridization and scanning of the probe arrays was done at the General Clinical Research Center (GCRC) Core Facility, San Francisco General Hospital, San Francisco, CA.
Data Analysis and Statistics
The microarray analysis was performed using the GeneChip® Analysis Microarray Suite v5.0 (MSv5), the MicroDB™ v3.0 software and the Data Mining Tool v3.0 (DMT) software (Affymetrix Inc., Santa Clara, CA, USA). Data was first analyzed following the MSv5 platform for array analysis described in the Affymetrix® Microarray Suite v5.0 User’s Guide. Briefly, microarray images (.dat files) were visually inspected for non-specific hybridization. Background and “noise” were corrected for each array. The cell intensity files (.cel file), generated automatically from the .dat files, were submitted to “absolute and comparison expression” analyses using the default values. The “absolute expression” analysis generated a “signal value” (relative to the abundance of a transcript) and a “detection call” (indicating whether the transcript was detected (P=present), undetected (A=absent), or at the limit of detection (M=marginal) for each transcript represented in the U133 arrays. It also calculated a p-value associated with the “detection call” using a “one-sided Wilcoxon’s signed rank test”. Before running the comparison expression analysis, the arrays (HIV-infected, n=4 / uninfected subjects, n=4) were scaled to the same target intensity (default) to allow a direct comparison between the groups. The “comparison expression” analysis generated a “signal log ratio” for each transcript (relative change in the expression level of a transcript between baseline (uninfected subjects) and experimental (infected subjects) arrays) using the “one-step Tukey’s biweight method. A signal ratio of 1 (log2 = 1) is equivalent to a 2-fold change. A “change call”, with its associated p-value (“Wilcoxon’s signed rank test”) was also calculated indicating if the change in transcript level was increased (“I”), decreased (“D”) or did not change (NC) relative to the baseline value.
To identify differentially expressed genes, expression analysis data were published into MicroDB™ v3.0 and further analyzed with DMT v3.0. Transcripts were selected if they met the following criteria: “detection call” = “P” and “signal log ratio” ≥ 1 (fold change ≥ 2). Genes or transcripts fulfilling the above criteria were subsequently filtered by “change call”, either “I” or “D”, thus generating two separate gene lists. Each list was further analyzed using a “Comparison Ranking Test”. This method, which performs a “count and percentage analysis “ generated all possible combinations of comparison analyses across the data (experiments or replicates) to determined the consistency or “concordance “of “change calls”. The highest the “concordance” (%), the highest the confidence that the transcript was indeed differentially expressed. We selected a cut-off value of 75%. The identified differentially expressed genes were then grouped by gene families and/or functional classes (biological / molecular function) using the resources provided through the NetAffx Analysis Center (www.affymetrix.com) (Liu et al., 2003) and DAVID (http://apps1.niaid.nih.gov/david/upload.asp) (Dennis et al., 2003).
Quantitative RT-PCR
RNA concentration was determined by spectrophotometry and adjusted to a concentration of 200 ng/μl. Five hundred nanograms of total RNA was reverse transcribed using 250U of murine leukemia virus RT (Invitrogen, Carlsbad, CA) in 1× Amplitaq Buffer (Applied Biosystems, Foster city, CA) supplemented with 7.5 mM MgCL2 with 5 μM random hexamers (Invitrogen), 1 mM each of dNTPs (Invitrogen) and 40 U of RNase inhibitor (Promega, Madison, WI). The reaction mixture was incubated at 25°C for 10 min, 48°C for 40 min, and 95°C for 5 min. five microliters of a 1:10 dilution of the RT reaction was subjected to PCR using the GeneAmp 5700 Sequence Detection System, SYBR Green PCR Master Mix (Applied Biosystems), and 4 μM primer mix in a 25 μl reaction. Cycling conditions were as follows: initial denaturation at 95°C for 10 min followed by 50 cycles at 95°C for 15 s, and 60°C for 1 min. Primer pairs from each individual gene were designed using OIigo Primer Analysis Software v6 (Molecular Biology Insights, Cascade, CO). The primer sequences were subject to BLAST analysis to confirm their specificity for a single gene. The generation of a single PCR product by each primer pair was tested using the Universal Human Reference RNA (Stratagene, La Jolla, CA). The reverse transcription and PCR reactions were conducted as described above. Primer sequences for each transcript are available from the authors.
Acknowledgements
We wish to thank Janet Lankard for her encouragement and Affymetrix for providing the chips for the analysis, Chandi Griffin at UCSF for his help with the microarray procedures, and Ann Murai and Kaylynn Peter for their help with manuscript preparation. This work was supported by a grant R01 AI056992 from NIH/NIAID. Beatriz Martinez-Mariño was supported by postdoctoral fellowship from the Hooper Foundation Training Grant T320CAO9043.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker E, Mackewicz CE, Reyes-Teran G, Sato A, Stranford SA, Fujimura SH, Christopherson C, Chang SY, Levy JA. Virological and immunological features of long-term human immunodeficiency virus-infected individuals who have remained asymptomatic compared to those who have progressed to acquired immunodeficiency syndrome. Blood. 1998;92(9):3105–14. [PubMed] [Google Scholar]
- Brinchmann JE, Gaudernack G, Vartdal F. In vitro replication of HIV-1 in naturally infected CD4+ T cells is inhibited by rIFN alpha2 and by a soluble factor secreted by activated CD8+ T cells, but not by rIFN beta, rIFN gamma, or recombinant tumor necrosis factor-alpha. Journal of Acquired Immune Deficiency Syndromes. 1991;4:480–488. [PubMed] [Google Scholar]
- Castro BA, Weiss CD, Wiviott LD, Levy JA. Optimal conditions for recovery of the human immunodeficiency virus from peripheral blood mononuclear cells. Journal of Clinical Microbiology. 1988;26:2371–2376. doi: 10.1128/jcm.26.11.2371-2376.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland KFT, McKay PJ, Rosenthal KL. Suppression of activation of the human immunodeficiency virus long terminal repeat by CD8+ T cells is not lentivirus specific. AIDS Research and Human Retroviruses. 1995;11:1321–1326. doi: 10.1089/aid.1995.11.1321. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Diaz LS, Foster H, Stone MR, Fujimura S, Relman DA, Levy JA. VCAM-1 expression on CD8+ cells correlates with enhanced anti-HIV suppressing activity. J Immunol. 2005;174(3):1574–9. doi: 10.4049/jimmunol.174.3.1574. [DOI] [PubMed] [Google Scholar]
- Genin P, Mamane Y, Kwon H, LePage C, Wainberg MA, Hiscott J. Differential regulation of CC chemokine gene expression in human immunodeficiency virus-infected myeloid cells. Virology. 1999;261(2):205–15. doi: 10.1006/viro.1999.9852. [DOI] [PubMed] [Google Scholar]
- Gomez AM, Smaill FM, Rosenthal KL. Inhibition of HIV replication by CD8+ T cells correlates with CD4 counts and clinical stage of disease. Clinical and Experimental Immunology. 1994;97:68–75. doi: 10.1111/j.1365-2249.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AD, Banapour B, Levy JA. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology. 1985;147:326–335. doi: 10.1016/0042-6822(85)90135-7. [DOI] [PubMed] [Google Scholar]
- Landay AL, Mackewicz C, Levy JA. An activated CD8+ T cell phenotype correlates with anti-HIV activity and asymptomatic clinical status. Clinical Immunology and Immunopathology. 1993;69:106–116. doi: 10.1006/clin.1993.1157. [DOI] [PubMed] [Google Scholar]
- Levy JA. The importance of the innate immune system in controlling HIV infection and disease. Trends in Immunology. 2001;22:312–316. doi: 10.1016/s1471-4906(01)01925-1. [DOI] [PubMed] [Google Scholar]
- Levy JA. The search for the CD8+ cell anti-HIV factor (CAF) Trends in Immunol. 2003;24(12):628–632. doi: 10.1016/j.it.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Levy JA, Mackewicz CE, Barker E. Controlling HIV pathogenesis: the role of noncytotoxic anti-HIV activity of CD8+ cells. Immunology Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31(1):82–6. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood. 2003;101(2):433–40. doi: 10.1182/blood-2002-06-1931. [DOI] [PubMed] [Google Scholar]
- Mackewicz CE, Blackbourn DJ, Levy JA. CD8+ cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackewicz CE, Garovoy MR, Levy JA. HLA compatibility requirements for CD8+ T-cell-mediated suppression of human immunodeficiency virus replication. Journal of Virology. 1998;72(12):10165–10170. doi: 10.1128/jvi.72.12.10165-10170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackewicz CE, Ortega H, Levy JA. Effect of cytokines on HIV replication in CD4+ lymphocytes: lack of identity with the CD8+ cell antiviral factor. Cellular Immunology. 1994;153:329–343. doi: 10.1006/cimm.1994.1032. [DOI] [PubMed] [Google Scholar]
- Mackewicz CE, Ortega HW, Levy JA. CD8+ cell anti-HIV activity correlates with the clinical state of the infected individual. Journal of Clinical Investigation. 1991;87:1462–1466. doi: 10.1172/JCI115153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackewicz CE, Yang LC, Lifson JD, Levy JA. Non-cytolytic CD8 T-cell anti-HIV responses in primary infection. Lancet. 1994;344:1671–1673. doi: 10.1016/s0140-6736(94)90459-6. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041–3. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Ryo A, Suzuki Y, Arai M, Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Sato C, Yamamoto M, Yamamoto N. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res Hum Retroviruses. 2000;16(10):995–1005. doi: 10.1089/08892220050058416. [DOI] [PubMed] [Google Scholar]
- Stranford S, Skurnick J, Louria D, Osmond D, Chang S, Sninsky J, Ferrari G, Weinhold K, Lindquist C, Levy J. Lack of infection in HIV-exposed individuals is associated with a strong CD8+ cell noncytotoxic anti-HIV response. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1030–1035. doi: 10.1073/pnas.96.3.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CM, Levy JA. A diffusible lymphokine produced by CD8+ T lymphocytes suppresses HIV replication. Immunology. 1989;66:628–630. [PMC free article] [PubMed] [Google Scholar]
- Walker CM, Moody DJ, Stites DP, Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R, Zimmerman D, Boyce R, Tierney LA, Hu E, Livi GP, Wolf B, Abdel-Meguid SS, Rose GD, Aurora R, Hensley P, Briggs M, Young PR. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics. 2002;80(2):144–50. doi: 10.1006/geno.2002.6816. [DOI] [PubMed] [Google Scholar]