Abstract
Posttranslational acetylation of core histone amino termini has long been associated with transcriptionally active chromatin. Recent reports have demonstrated histone acetyltransferase activity in a small group of conserved transcriptional regulators directly linked to gene activation. In addition, the presence of a putative acetyltransferase domain has been discovered in a group of proteins known as the MYST family (for its founding members MOZ, YBF2/SAS3, SAS2, and Tip60). Members of this family are implicated in acute myeloid leukemia (MOZ), transcriptional silencing in yeast (SAS2 and YBF2/SAS3), HIV Tat interaction in humans (Tip60), and dosage compensation in Drosophila (MOF). In this report, we express a yeast ORF with homology to MYST family members and show it possesses histone acetyltransferase activity. Unlike the other MYST family members in Saccharomyces cerevisiae this gene is essential for growth.
It has long been recognized that a correlation exists between acetylation of lysines in the core histones and states of transcription (1–4). More recently it has become apparent that the Nɛ-acetylation of highly conserved lysines on the amino-terminal tails of histones is associated with a variety of cellular processes, including the deposition of newly synthesized histones onto DNA, the activation of transcription, and the establishment or maintenance of heterochromatin (for review, see refs. 2 and 3). However, there is little understanding of the molecular processes that relate histone acetylation to these functions.
The first two histone acetyltransferases (HATs) to be cloned and characterized were the deposition-related HAT1 (5) from yeast and the transcription-related HAT from Tetrahymena, p55, that was highly similar to yeast Gcn5p (6). Although Hat1p and Gcn5p have different substrate specificities (3), they share a series of motifs with the superfamily of N-acetyltransferases (referred to as GNAT for GCN5-related N-acetyltransferases), a group that includes several amino-terminal acetyltransferases (7). Members of a recently discovered group of proteins, known as the MYST family (7, 8) for its founding members MOZ (8), YBF2/SAS3 (9), SAS2 (9, 10), Tip60 (11), have a subset of the motifs that characterize the GNAT superfamily (7, 12). One of these, Tip60, was recently shown to acetylate histone substrates (13). Two of these proteins, Sas2p (9, 10) and Sas3p (9), are apparently involved in the control of transcriptional silencing in yeast. Due to the relationships between transcriptional silencing and the acetylation of histones (14–16), it is possible that SAS2 and SAS3 could be HATs, i.e., Nɛ-lysine acetyltransferases. However, mutations in the amino-terminal acetyltransferases NAT1 and ARD1 also affect silencing of the mating type loci (17). Therefore, without any biochemical evidence, it is not clear whether particular MYST family members are Nα- or Nɛ-acetyltransferases.
In addition to the effects on silencing of sas2 and sas3 mutants, other MYST family members are implicated in transcriptional control. MOZ was discovered due to its involvement in a chromosomal translocation that fuses this gene product in-frame with the transcriptional coactivator CBP, resulting in a form of acute myeloid leukemia (8). Tip60 was cloned due to its ability to interact with the viral transactivator HIV Tat in the two-hybrid system (11). MOF was discovered in a genetic screen for gene products necessary for the hypertranscription and the acetylation of H4 at Lys-16 that is associated with Drosophila dosage compensation (18). Finally, a yeast ORF (ESA1 for essential SAS2-related acetyltransferase) was identified on the basis of its strong homology to MOF and Tip60. Herein, we report that this essential yeast gene ESA1 encodes a histone acetyltransferase that acetylates nonrandomly internal lysines of specific histones in a pattern that distinguishes it from other known HATs.
MATERIALS AND METHODS
Recombinant Proteins.
The sequence of the yeast ORF ScYOR244w was obtained from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). PCR primers with restriction site extensions were used to amplify ScYOR244w from wild-type strain EJ66 (19), and this product was cloned into the NheI–EcoRI sites of pRSET (Invitrogen). Esa1p/pRSET was transformed into DH5αF′ (BRL) and induced with isopropyl β-d-thiogalactoside and T7 phage infection according to Invitrogen’s protocols. Bacterial pellets were sonicated in 300 mM NaCl/1 mM DTT/1 mM phenylmethylsulfonyl fluoride/50 mM Tris⋅HCl, pH 8.0, and purified on Ni2+ agarose (Qiagen, Chatsworth, CA). Gcn5p and Hat1p were made as described (6, 20). The point mutant G315E was made according to standard procedures (21) and confirmed by sequencing.
HAT Assays.
Chicken erythrocyte core histones (10 μg) were incubated with a mixture of enzyme, 0.1 μCi of [3H]acetyl CoA (4.3 Ci/mmol; 1 Ci = 37 GBq; Amersham), 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA, 10% glycerol, and 50 mM Tris⋅HCl (pH 8.0) in 50 μl. Reactions were carried out at 30°C for 10 min, and aliquots were spotted to Whatman P81 filters and processed as described (22). Equivalent aliquots were separated by SDS/PAGE, stained with Coomassie blue, and fluorographed with Entensify (NEN) to identify which histones were labeled. Chicken erythrocyte and yeast histones were gifts from Craig Mizzen and Min-Hao Kuo, respectively (University of Rochester, Rochester, NY). Deacetylated histones from Tetrahymena were made by treating macronuclei with a mixture of 10 mM EDTA, 10 mM Tris⋅HCl (pH 8.0), and 1 mM phenylmethylsulfonyl fluoride for 2 hr at 4°C followed by extraction with 0.2 M H2SO4 and trichloroacetic acid precipitation. Drosophila histones were acid-extracted from 0- to 20-hr embryonic nuclei. Individual histones were purified from a mixture of core histones by reverse-phase HPLC.
Protein Sequencing.
Each histone (5 μg) was incubated with 5 μCi of [3H]acetyl CoA and about 150 ng of Esa1p in 400 μl for 20 min at 30°C. The pH was then raised to 10 with NaOH to destroy acetyl CoA and neutralized after 10 min with HCl, and the H4 was repurified by reverse-phase HPLC and processed for sequencing as described (23). Drosophila H4 and chicken H2A were deblocked with trifluoroacetic acid before sequencing as described (23).
Yeast Knockout Mutagenesis.
KanMX was amplified from pFA6-kanMX2 by using the following primers: (5′-CTACCATTCTCGGAAATACTGCAAGAAATCATCGATGTCCCCAGCTGAAGCTTCGTACGC-3′ and 5′-AGTTGTTTGAATGTAAGTTTAGGAAAGCACTACATAGCGCATAGGCCACTAGTGGATCTG-3′) with sequences flanking ESA1 in genomic DNA (24). AE100 (FY23 × FY73) (25) was transformed with 2 μg of this PCR product, and disruptions were selected on G418 and confirmed by PCR. Sporulation was performed on sporulation plates at 30°C for 5–6 days, and tetrads were dissected on YPD.
RESULTS AND DISCUSSION
Substrate Usage by Esa1p.
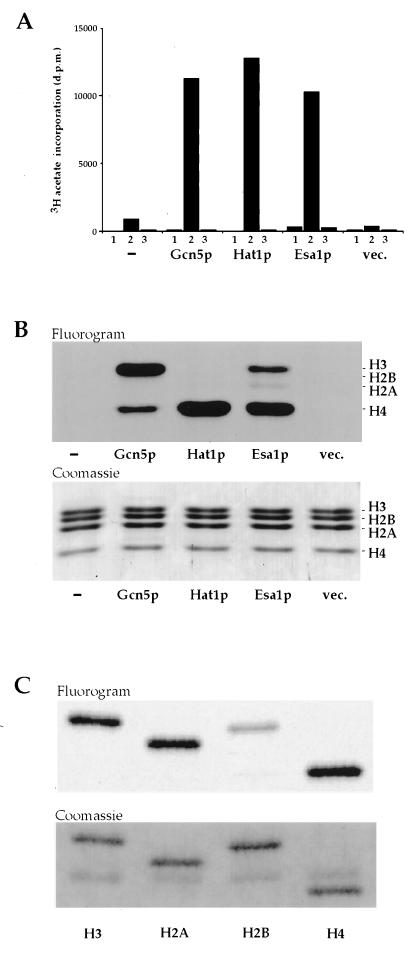
We expressed a Tip60/MOF-related yeast ORF ScYOR244w (18) (hereafter referred to as ESA1 for essential SAS2-related acetyltransferase) in bacteria and assayed it for HAT activity relative to two well-studied yeast HATs, Gcn5p (6, 19) and Hat1p (5, 20) (Fig. 1A). Our results demonstrate clearly that Esa1p has significant HAT activity when assayed with a mixture of chicken histones as substrate. Acetate incorporation was not observed in the absence of histone or when BSA was used as a nonhistone substrate. The histones acetylated by these enzymes were identified by fluorography (Fig. 1B). As expected from published studies, Gcn5p (6, 19) and Hat1p (5, 20) prefer histone H3 and H4, respectively. Under these assay conditions, Esa1p shows a clear preference for histone H4 and to a lesser extent H3 followed by weak acetylation of H2A. Thus, when tested with a mixture of histones, the substrate specificity of Esa1p is similar to that described for the human enzyme Tip60 (13). Consistent with data obtained with yeast Gcn5p (19), differences are observed when purified histones are reacted with Esa1p. H2A, H3, and H4 are all acetylated relatively well when incubated individually with Esa1p (Fig. 1C). Although H2B is not acetylated in a mixture of histones from a variety of species, chicken H2B is acetylated at a modest level when presented to the enzyme in a purified form (Fig. 1C). However, Esa1p does not acetylate pure yeast or Drosophila H2B (data not shown). This difference likely reflects the poor conservation of H2B amino termini in different species. The strong acetylation of H4 is notable given the reported enrichment of H4Ac16 acetylation on the male X chromosome of Drosophila, which is in part mediated by the Esa1p/Tip60-related MOF (18, 26, 27).
Figure 1.
Esa1p is a histone acetyltransferase. (A) No enzyme (bars −) and bacterial-expressed Gcn5p, Hat1p, Esa1p, or vector only (bars vec.) were incubated without substrate (bars 1), with chicken erythrocyte core histones (bars 2), or with BSA (bars 3), and [3H]acetate incorporation was measured by a filter binding assay. (B) Specificity for core histone acetylation was assessed by separating reaction products on a 15% SDS/PAGE gel, staining with Coomassie blue, and fluorography. (Upper) Fluorogram. (Lower) The corresponding Coomassie-stained gel. Although the level of H2A acetylation by Esa1p is relatively low, it is reproducible. (C) Individual chicken histones were purified by reverse-phase HPLC, and equivalent amounts of each histone were incubated with Esa1p and processed for fluorography as above. H4, H2A, and H3 are well acetylated by Esa1p when presented individually.
Nonrandom Site Usage by Esa1p Differs from That of Gcn5p.
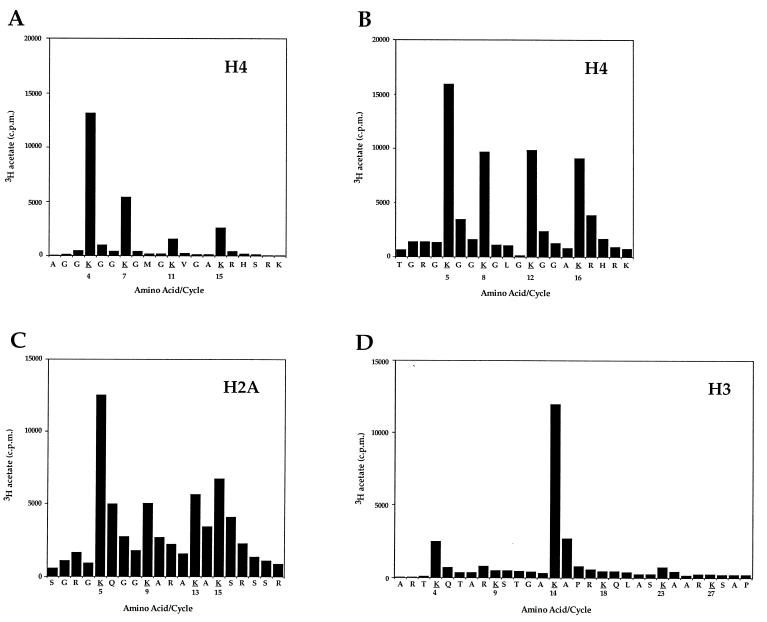
To identify rigorously the lysine(s) acetylated by Esa1p, individual histones H4, H3, or H2A were acetylated in vitro and subjected to microsequence analyses followed by direct determination of the radioactivity at each cycle/position of the sequence. In these analyses, the 3H label is in the acetyl moiety proper, precisely identifying the site of acetylation. Esa1p acetylates H4, H3, and H2A when presented with histones from chicken erythrocytes, Drosophila, Tetrahymena, and yeast (data not shown). However, Tetrahymena histones are easily prepared in the deacetylated state and are, therefore, convenient substrates for these analyses to avoid potential complications arising from preexisting acetylation at specific sites (for a discussion, see ref. 19). Moreover, the Tetrahymena H4 is unique among H4s in being “nonblocked” at its amino terminus lending itself well to microsequence analyses (23).
When Tetrahymena H4 is incubated with Esa1p and acetyl CoA, most of the acetate incorporation is found at Lys-4 (Fig. 2A). Tetrahymena H4 contains a single deletion at amino acid 3, so each acetylation site is numbered one less than that observed in most other H4s. The next highest site of acetate incorporation is at Lys-7, followed by lower levels at Lys-15 and Lys-11. To determine whether Esa1p acetylates Lys-5 on an H4 closer in primary sequence to yeast H4, we used H4 purified from Drosophila embryos [yeast H4 has proven to be a poor substrate for this type of study because it is largely preacetylated at Lys-16 (see ref. 19)]. Drosophila H4 was acetylated in vitro and subjected to microsequencing as described above, with the exception that the amino terminus was deblocked by incubation with trifluoroacetic acid before sequencing (Fig. 2B). The acetylation pattern on Drosophila H4 is similar to the pattern obtained with Tetrahymena H4, such that Lys-4/5 is still a highly preferred site of acetylation, but with both H4s, all four sites are used especially when repetitive yields are calculated (data not shown). Although Lys-4 and Lys-7 were identified as the most highly used transcription associated sites in Tetrahymena macronuclei (28), Lys-4/5 is also a deposition site in numerous species (28, 29). Therefore, it is unclear from this site specificity whether Esa1p’s role is in transcription or other processes. Importantly, the H4 acetylation site utilization pattern of Esa1p is clearly distinct from that of Gcn5p, a HAT with a strong preference for Lys-7/8 and Lys-15/16, under similar experimental conditions (19). These data suggest that the functional consequence of Esa1p-mediated HAT activity in yeast is distinct from that of Gcn5p.
Figure 2.
Esa1p acetylates specific histones nonrandomly in H4, H3, and H2A. (A) Deacetylated H4 from Tetrahymena was acetylated in vitro by recombinant Esa1p and subjected to protein sequencing. Products from each sequencing cycle were used for scintillation counting and HPLC identification of the residue. The cpm for each cycle are plotted against the amino acid detected for that cycle. Potential acetylation sites are in boldface type and underlined. Because Tetrahymena H4 contains a single deletion at amino acid 3, each acetylation site is numbered one less than in most other H4 proteins. (B) Drosophila H4 was acetylated by Esa1p and processed as above, except that the amino terminus was deblocked before microsequencing. For both substrates we observe the most [3H]acetate on Lys-4/5. However, when repetitive yield is taken into account, we observe with Drosophila H4 that Lys-16 has the most dpm per pmol, but with Tetrahymena H4, Lys-4 has the most dpm per pmol. (C) Chicken erythrocyte H2A was acetylated by Esa1p and processed as above, with deblocking of the amino terminus before microsequencing. (D) Tetrahymena H3 was acetylated by Esa1p and processed as above, without deblocking of the amino terminus.
Given our inability to discriminate whether Esa1p HAT activity is more transcription- or deposition-related, based on H4 site data (Fig. 2 A and B), we examined the site utilization pattern when Tetrahymena H3 was acetylated by Esa1p (Fig. 2D). Clearly most of the acetate incorporation is found at Lys-14, a site also strongly preferred by the transcriptional coactivators Gcn5p and TAFII250 (19, 30). Known sites of H3 acetylation in mammals are Lys-9, -14, -18, and -23 (31), with Lys-9 of H3 being identified as a site of deposition-related acetylation in yeast (17).
There is, at present, less information available on the acetylation states of H2A and H2B than there is for H3 and H4. To our knowledge, newly synthesized H2A and H2B are not deposited into nuclei in an acetylated form (3), and Lys-5 of H2A is the only identified site of acetylation in vivo (32). Because H2A is also a good substrate for Esa1p when reacted as a pure histone (Fig. 1C), we sought to determine its pattern of site utilization. Under these conditions, H2A is acetylated by Esa1p at Lys-5, -9, -13, and -15 (Fig. 2C). From the collective site utilization data presented in Fig. 2, we conclude that Esa1p is a HAT in yeast that is clearly distinct from Gcn5p. On the basis of the relatively widespread use of acetylation sites in H2A and H4 (Fig. 2 B and C) and the specific Gcn5p/TAF250-like acetylation of H3 at Lys-14 (Fig. 2D), we favor that Esa1p is functioning more as a transcription-related HAT. However, functions of the protein in other processes cannot be ruled out.
Comparison of Chromatin and Free Histone Substrates.
Esa1p does not acetylate chromatin substrates in vitro, a property shared with Gcn5p and Hat1p (ref. 19 and data not shown). Gcn5p, however, is the catalytic component of large protein complexes that acetylate histones H3 and H2B in chromatin (33). Recently, Grant et al. (33) reported the existence of several Gcn5p-independent HAT complexes in yeast by using nucleosomal substrates. Interestingly, one of these complexes, a 1.4-MDa complex (referred to as complex 2 in ref. 33), shows strong specificity for nucleosomal H2A and H4. Similarly, Tetrahymena macronuclei also contain a second HAT activity distinct from p55 (p55 is the Tetrahymena homolog of Gcn5p, see ref. 6) that acetylates H2A and H4 well in the context of nucleosomes (R. Ohba and C.D.A., unpublished results). Côté and colleagues (J. Côté, A. Clarke, L. Pillus, and J. Workman, personal communication) have evidence suggesting Esa1p is the catalytic subunit of a distinct protein complex that acetylates histones H2A and H4 in the context of chromatin. Efforts are in progress to determine the precise relationships between these activities in yeast and Tetrahymena. It will be informative to determine to what extent the site utilization patterns obtained with recombinant Esa1p and free histones (Fig. 2) reflect patterns obtained when the above mentioned HAT complexes are used to acetylate nucleosomal substrates.
Functional Conservation of an Acetyltransferase Motif.
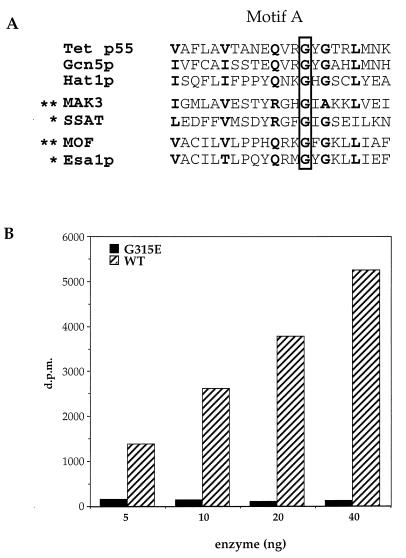
Comparison of the sequence of the mutant allele of mof with the normal wild-type gene reveals that a single amino acid change, Gly → Glu (G691E), is responsible for the mutant phenotype that led to the discovery of the MOF gene (18). This mutation maps within the putative acetyltransferase domain identified in MYST family members (8–10), a finding whose significance is underscored by the demonstration that replacement of the homologous Gly with Asp in human spermidine/spermine acetyltransferase interferes with acetyl CoA binding (34) (Fig. 3A). Although it is not yet known whether MOF has HAT activity, this seems likely given that the G691E mutation leads to a loss of H4Ac16 staining of the male X chromosome (18).
Figure 3.
Point mutant abolishes HAT activity of Esa1p. (A) Alignment of a motif found in a wide variety of acetyltransferases (referred to as domain A in the GNAT superfamily; ref. 7). Note that a specific conserved Gly residue (G315 in Esa1p) has been shown to be necessary for enzymatic function in vitro (marked by ∗) or necessary for gene function in vivo (marked by ∗∗; for a more extensive review, see ref. 7). (B) Various levels of recombinant wild type (WT) and mutant (G315E) Esa1p were assayed for HAT activity with chicken histones. [3H]Acetate incorporation is plotted versus the concentration of enzyme.
To test whether the HAT activity of Esa1p would be abolished by a similar point mutation, we engineered a single G315E mutation into ESA1, expressed the mutant form of the enzyme in bacteria, and assayed for HAT activity as described above. When directly compared with wild-type enzyme by using chicken histones as substrate, it is clear that the G315E mutation severely affects in vitro HAT activity (Fig. 3B). This supports the prediction that this motif is functionally related to the similar region in MAK3, SSAT, and GCN5 acetyltransferases (8–10), despite significant differences between MYST family and GNAT superfamily members (7, 12).
ESA1 Is an Essential Gene.
Finally, we wished to determine whether Esa1p, unlike Gcn5p, is a vital gene product. To this end, ESA1 was disrupted in diploid cells by using the kanMX cassette system. After sporulation of the esa1Δ∷kanMX strain, each dissected tetrad yields two wild-type spores and two spores that have severe growth and morphological phenotypes. These esa1Δ∷kanMX spores germinate, divide four or five times, once approximately every 2 days (30–40 times slower than their sister wild-type spores) before dying, and display a terminal phenotype of giant and dumbbell-shaped cells [reminiscent of the cell cycle phenotypes conferred by some cdc mutants and by a mutation in a putative acetyltransferase PAT1 (35)]. Pioneering studies in yeast have demonstrated the importance of the conserved Lys residues for the control of transcription but have left unresolved whether histone acetylation is essential for growth (15, 16). Although HAT1 and GCN5 are both nonessential, the finding that the histone acetyltransferase Esa1p is required for growth suggests that histone acetylation is required for growth in yeast. However, when the wild-type ESA1 gene is replaced with the G315E mutant, there is no detectable phenotype (data not shown). This raises the possibility that the catalytic activity is not required for viability and that the essential phenotype is due to structural or other functions that do not depend on the catalytic activity of Esa1p. A more likely possibility is that the G315E mutant could be stabilized sufficiently in vivo to maintain HAT activity, whereas our in vitro conditions are destabilizing to the mutant form of the enzyme (12).
In conclusion, we have demonstrated in vitro HAT activity in a member of the MYST family of acetyltransferases. The presence of MYST homologs in organisms as diverse as yeast and humans suggests that these proteins function in one or more important cellular processes. It is becoming increasingly clear that many of the newly identified HAT catalytic subunits function as part of larger multisubunit complexes that allow acetylation of chromatin substrates (33, 36) and this may also be true of MYST family members. In this respect, the function of the zinc finger (8, 18) and chromo domains (18) in this group of proteins is not known. The presence in yeast of a homolog of another Drosophila dosage compensation factor, MSL-3, provides further support for the notion that these proteins are part of an evolutionarily conserved system of targeted acetylation for gene activation.
Acknowledgments
We thank Jacques Côté, Lorraine Pillus, and Jerry Workman for sharing unpublished data; Sue Jinks-Robertson for counsel and assistance with the yeast tetrad analysis; Gray Crouse for providing the pFA6-kanMX2 plasmid; Craig Mizzen for providing chicken histone and nucleosomal substrates; and Min-Hao Kuo for helpful comments on the manuscript. This research was supported by grants from the National Institutes of Health to C.D.A. (GM 53512) and to J.C.L. (GM 15691).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HAT, histone acetyltransferase; MYST, named for its founding members MOZ, YBF2/SAS3, SAS2, and Tip60.
References
- 1.Allfrey V G, Faulkner R, Mirsky A E. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 3.Roth S Y, Allis C D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 4.Wade P A, Wolffe A P. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 5.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 6.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Neuwald A F, Landsman D. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 8.Borrow J, Stanton V P, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, et al. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 9.Reifsnyder C, Lowell J, Clarke A, Pillus L. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenhofer-Murray A E, Rivier D H, Rine J. Genetics. 1997;145:923–934. doi: 10.1093/genetics/145.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 12.Kuo, M.-H., Zhou, J., Jambeck, P., Churchill, M. E. & Allis, C. D. (1998) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 13.Yamamoto T, Horikoshi M. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 14.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Megee P C, Morgan B A, Mittman B A, Smith M M. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 16.Durrin L K, Mann R K, Kayne P S, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 17.Mullen J R, Kayne P S, Moerschell R P, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi J C. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 20.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Brownell J E, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobel R S, Cook R G, Allis C D. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 24.Wach A, Brachat A, Pohlmann R, Phillipsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 25.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 26.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 27.Bone J R, Lavender J S, Richman R, Palmer M J, Turner B M, Kuroda M I. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 28.Chicoine L G, Schulman I G, Richman R, Cook R G, Allis C D. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- 29.Sobel R S, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizzen C A, Yang X-J, Kokubo T, Brownell J B, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 31.Thorne A W, Kmiciek D, Mitchelson K, Sautiere P, Crane-Robinson C. Eur J Biochem. 1990;193:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- 32.Panagiotis P, Bonner W M. J Biol Chem. 1981;256:4669–4675. [PubMed] [Google Scholar]
- 33.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 34.Lu L, Berkey K A, Casero R A. J Biol Chem. 1996;271:18920–18924. doi: 10.1074/jbc.271.31.18920. [DOI] [PubMed] [Google Scholar]
- 35.Lin R, Allis C D, Elledge S J. Genes Cells. 1996;1:923–942. doi: 10.1046/j.1365-2443.1996.d01-215.x. [DOI] [PubMed] [Google Scholar]
- 36.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]