Abstract
The study of RNA structure using x-ray crystallography or NMR has yielded a wealth of detailed structural information; however, such approaches do not generally yield quantitative information regarding long-range flexibility in solution. To address this issue, we describe a solution-based method that is capable of characterizing the global flexibilities of nonhelix elements in RNA, provided that such elements are flanked by helix (e.g., bulges, internal loops, or branches). The “phased τ ratio” method is based on the principle that, for RNA molecules possessing two variably phased bends, the relative birefringence decay times depend on the flexibility of each bend, not simply the mean bend angles. The method is used to examine the overall flexibility of the yeast tRNAPhe core (as unmodified transcript). In the presence of magnesium ions, the tRNA core is not significantly more flexible than an equivalent length of RNA helix. In the absence of divalent ions, the tRNA core gains flexibility under conditions where its secondary structure is likely to be largely preserved. The phased τ ratio approach should be broadly applicable to nonhelix elements in both RNA and DNA and to protein–nucleic acid interactions.
Yeast tRNAPhe is a paradigm for the study of tertiary structure in RNA: its crystal structure is well-established (1–3), and under comparable ionic conditions, the crystal and solution structures possess nearly identical anticodon–acceptor (mean) interstem angles (1, 4, 5). However, relatively little is known regarding the global flexibility of the tRNA core. Harvey and coworkers (6) have considered the issue of hinge-type flexibility from the computational perspective, noting that large-amplitude bending motions between the acceptor and anticodon domains are not energetically prohibitive. However, a more recent normal-mode analysis of the bending motions for yeast tRNAPhe suggests that the actual range of hinge-type distortions is likely to be small in the absence of external forces (7), a result that is consistent with the small temperature factors for the crystal structure (1, 2). Therefore, in the absence of direct experimental information in solution, the issue of the intrinsic global flexibility of the tRNA core remains unresolved.
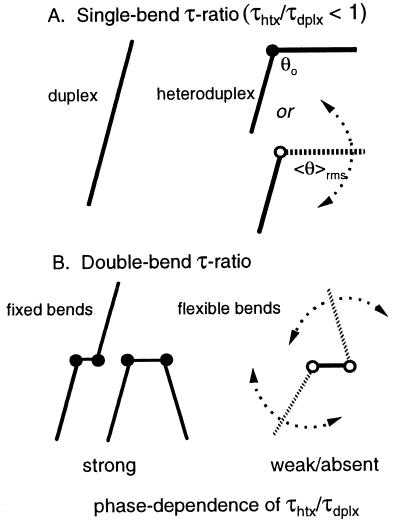
In the current work, we describe an experimental approach (“phased τ ratio”; Fig. 1) for quantifying long-range flexibility in RNA. The method exploits the sensitivity of the rotational (birefringence) decay times of helices possessing two variably spaced bends to the flexibilities of the individual bends (8, 9). The current approach is conceptually similar to gel-based studies of RNA molecules possessing two bulge-induced bends. Such studies have used the dependence of gel mobility on the phasing between the bends to estimate the helix repeat of RNA (10, 11); however, gel methods cannot at present be used to quantify bend flexibility. Although NMR methods are capable of providing evidence for internal dynamics/flexibility in RNA (12, 13), such information is generally difficult to quantify in terms of long-range flexibility. An alternative approach, time-resolved fluorescent resonance energy transfer, is capable of characterizing the long-range dynamics of nucleic acids (14, 15) but has not yet been applied to tRNA.
Figure 1.
Principle of the phased τ ratio approach. (A) RNA molecules (heteroduplex; htx) with single bends have shorter birefringence decay times (τ) than a linear (duplex; dplx) control, for either fixed (θo) or flexible (〈θ〉rms) interstem angles, because the heteroduplex is less extended in solution. The resulting single-bend τ ratio, τhtx/τdplx, does not generally provide a measure of the range (dispersion) of the angle (8). (B) RNA molecules with two bends in various torsional alignments (phasings) will display τ ratios that are dependent on the additional flexibility of the bends. Double-bend constructs can thus be used in conjunction with single-bend (mean) angles to estimate the effective bend dispersion. 〈θ〉rms = 〈θ2〉½, where θ is the total instantaneous angle.
In a typical transient electric birefringence experiment, the terminal rotational decay time (τhtx) of a heteroduplex RNA molecule possessing a single centrally located nonhelix element is compared with the corresponding decay time (τdplx) for a fully duplex RNA molecule of equal contour length (100–200 bp), the two molecules differing only by the presence of a central nonhelical element (e.g., bulge) in the heteroduplex (8, 9). The resulting τ ratio (= τhtx/τdplx) is a sensitive quantitative measure of the angle between the helix stems that flank the nonhelix element (8). Such angles are “apparent” in that they may arise from fixed bends and/or points of increased flexibility; both types of flexure will reduce the rotational decay time of the heteroduplex (Fig. 1A). However, a distinction can be made if one examines heteroduplex RNA molecules that possess two bends with varied interbend spacing (Fig. 1B). If both bend angles are essentially fixed, the entire range of τ ratios (for all two-bend species) is specified by the individual (single bend) angles. For flexible bends, the phase variation of the τ ratios is diminished in proportion to the added flexibility of the nonhelix elements as phase coherence is lost (8). Application of the phased τ ratio method to yeast tRNAPhe (as unmodified RNA) reveals that, in the presence of magnesium ions, the global flexibility of the native tRNA core is not significantly greater than the flexibility of RNA helix. However, in the absence of divalent cations, the core becomes quite flexible under conditions where much of the secondary structure is likely to be retained.
MATERIALS AND METHODS
Production of Two-Element Heteroduplex RNA Molecules.
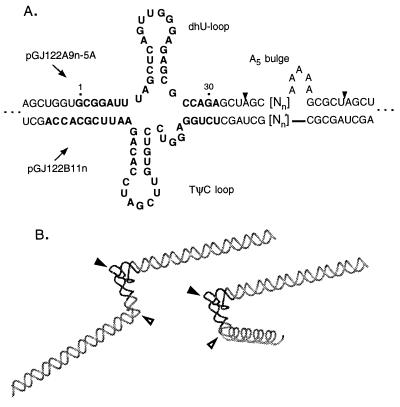
The secondary structures of the two-element RNA molecules used in the current work are depicted in Fig. 2A. The constructs are identical to the extended (E) [tRNAPhe] species as described (4), except for the insertion of oligomers for the A5 bulge and phasing segments (Nn) into the NheI site of the parent plasmids, pGJ122A9 and pGJ122B11. To create the 21 expression plasmids (3 plasmids for each of the following n values: n = 0, 2, 4, 6, 8, 10, and 12), the oligomers CTAGCNnA5GCG and CTAGCNnGCG (representing duplex oligodeoxynucleotides with 5′-CTAG overhangs, shown in italic type) were inserted into pGJ122A9 and pGJ122B11, respectively. The Nn phasing segments are as follows: N2, GC; N4, GAN2; N6, CTN4; N8, AGN6; N10, GCN8; N12, TAN10. The resultant plasmids were designated pGJ122A9n-5A and pGJ122B11n. T7 transcription of pairs of plasmids (for each n) and subsequent annealing of transcript pairs yields the desired heteroduplexes, designated E[tRNAPhe:A5]. A third parent plasmid, pGJ122A12, contains the same insert as pGJ122B11 but with the insert in the opposite orientation. Insertion of CTAGCNnGCG oligomers into pGJ122A12 creates a set of plasmids, pGJ122A12n, whose transcripts provide linear controls when annealed to transcripts from pGJ122B11n.
Figure 2.
Double-bend RNA constructs used in this work. (A) The secondary structures of the yeast tRNAPhe-A5 constructs, designated E[tRNAPhe:A5]. Dashed extensions represent an additional 66 bp of RNA helix (4). Numerals 1 and 30 correspond to the numbering of native tRNA. (▾) Centers of the NheI sites created by the phasing/A5 bulge insertions. Nn:N′n refer to n additional (phasing) base pairs. Plasmids giving rise to specific transcripts are indicated to the left. (B) Phosphate backbone ribbon representations of the tertiary arrangements of two of the two-bend constructs; the darker ribbon represents the tRNA backbone (4, 16); the bulge is represented by an additional segment of backbone docked to two adjacent helices using insight (Molecular Simulations, Waltham, MA) for illustrative purposes; no attempt was made to find an energetically correct conformation. The centers of the tRNA and bulge bends are represented by solid and open arrowheads, respectively.
Transcription reactions contained SmaI-digested plasmid DNA (0.06 mg/ml) in 40 mM Tris⋅HCl, pH 8.1/100 μM spermidine hydrochloride/5 mM DTT/0.001% Triton X-100/all four rNTPs (each at 2.5 mM)/20 mM MgCl2/T7 RNA polymerase (5 μg/ml protein). Equimolar amounts of transcripts from pGJ122A9n-5A and pGJ122B11n were annealed to produce E[tRNAPhe:A5] constructs (0 ≤ n ≤ 12); full-duplex controls were produced by annealing transcripts from pGJ122B11n and pGJ122A12n. Annealing reactions were performed in 100 mM Tris⋅HCl, pH 6.5/100 mM NaCl/5 mM EGTA/10 mM Na2EDTA, by heating to 95°C for 5 min, followed by cooling to room temperature over 30 min. Annealed RNA molecules were gel-purified as described (4, 17). In previous work, it was shown that the E[tRNAPhe] heteroduplex folds into the appropriate tertiary arrangement required for site-specific lead cleavage of the dhU-loop (17) (for further details of the purification and characterization of the E[tRNAPhe] species, see refs. 4 and 17).
Hydrodynamic Model.
A detailed description of the hydrodynamic approach used for the current computations, including an analysis of the τ ratio approach, has been presented (8, 18). The current structural model is identical to the model described (4), except for the addition of a second bend (A5 bulge). For the hydrodynamic computations, the heteroduplex constructs are represented by an equivalent RNA helix with a bend (tRNA) at bp 85 (4), and a second bend at bp (106 + n). The bend separations of (21 + n) bp include the transit distance through the tRNA core, expressed as equivalent base pairs (4). Thus, computed τ ratios represent the ratio of computed (terminal) decay times for RNA helices with point bends at bps 85 and (106 + n) to the computed decay times of the corresponding (182 + n)-bp duplex controls. Experimentally derived interstem angles are specified in Table 1 (17, 19). For computations involving altered flexural and/or torsional rigidities, in this work confined to the tRNA core, the effective persistence length (Peff) is altered uniformly over a region along the axial contour of the tRNA core between the inner aspects of the acceptor and anticodon stems (bps 7⋅66 and 27⋅37, respectively). The effective torsional elastic constant (Ceff) is altered over the region lying between the core vertex and bp 27⋅37. Reference helix parameters are as follows: twist, 12 bp per turn; rise, 2.8 Å per bp; hydrodynamic radius, 13 Å; helix (reference) persistence length (Phelix), 700 Å (20, 21); helix (reference) torsional elastic constant (Chelix), 3.0 × 10−19 erg⋅cm (1 erg = 0.1 μJ). Note that the τ ratios are essentially independent of the choice of helix parameters provided that the same parameters are used for both heteroduplex and duplex species. Each computed τ ratio represents 5 × 103 chains for the heteroduplex, and an equal number for the corresponding linear control; this ensemble size reduces the standard error of the single-point computation to less than 0.5%.
Table 1.
Interstem angles from previous single-bend measurements
| Element | Interstem angle, degrees
|
Ref. | |
|---|---|---|---|
| Mg2+ (2 mM) | No Mg2+ | ||
| E[tRNAPhe] | 70 ± 4 | 154 ± 5 | 17 |
| A5 bulge | 106 ± 3 | 105 ± 3 | 19 |
Interstem angles subtend the axes of the helices flanking the bend element; they are thus supplements to the previously reported bend angles. For E[tRNAPhe], the angle subtends the extended acceptor and anticodon stems.
Transient Electric Birefringence Measurements.
Experimental (transient electric birefringence) methods have been described in detail elsewhere (4, 19, 22). For the current measurements, the following pulse characteristics were used: pulse width, 1.0 μs; field strength, 10 kV/cm; repetition frequency, 1 Hz. Decay curves are averages of 128 individual decay transients. The following solution conditions were used: 9 μg of RNA per 35-μl cell volume; buffers are as specified in figure legends. Gel analysis of the RNA samples after transient electric birefringence measurements demonstrates the absence of strand separation or degradation.
Data Analysis.
Decay times for each birefringence decay curve are obtained from double-exponential fits using the Levenberg–Marquardt method (23). Individual τ ratio data points (see Fig. 5) are determined by averaging τ values from 15 decay curves; the resultant data points are associated with a standard deviation (σ) of 0.026. Tests of statistical significance for departures of computed curves from the experimental τ ratio values were performed by using the χ2 statistic,
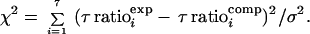 |
Optimal fits were obtained by minimizing χ2; significance at the 1% level is specified by χ2 > 16.8 (degrees of freedom = 6).
Figure 5.
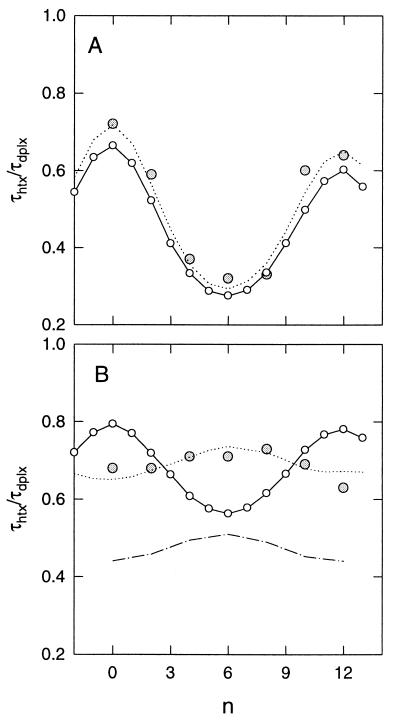
Phased τ ratio plots for the E[tRNAPhe:A5] species in the presence (A) or absence (B) of magnesium ions. τ ratios, τhtx/τdplx, are plotted as a function of the number (n) of phasing base pairs (see Fig. 2). For experimental τ ratios (shaded circles; diameter ≈ σ for 15 measurements), the reference (duplex) helix length is (182 + n) bp; for computed τ ratios (n = −2 to 13), the bend centers are separated by (21 + n) bp. In both panels, the continuous curves (○) represent τ ratio values computed from the angles in Table 1 and standard helix parameters; they are independent of the experimental data shown, except that both curves are phase-aligned to the experimental curve in A. The absolute bend directions were adjusted to provide phase coherence with the experimental data and should be considered arbitrary. The dashed lines represent global best fits to the experimental data by allowing the angles, Peff, Ceff, and phasing (for B) to vary. In B, dashed lines and dashed and dotted lines represent reductions in Ceff and Peff, respectively, to yield phase amplitudes that are approximately equal to the experimental amplitude.
RESULTS
The Electrophoretic Mobilities of the tRNA-A5 Bulge Constructs Are Strongly Influenced by the Torsional Phasing of the Two Bends.
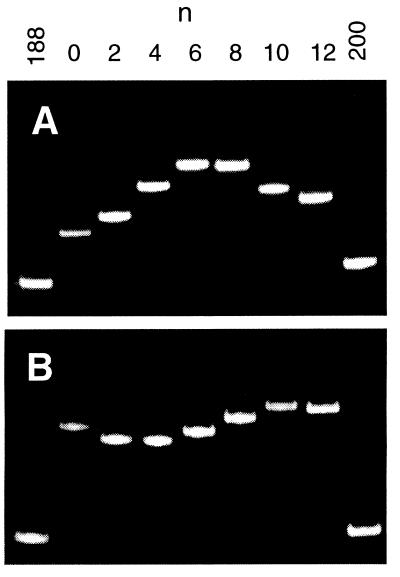
To apply the phased τ ratio method to yeast tRNAPhe, a set of RNA heteroduplexes was constructed in which the tRNA core is variably phased with an A5 bulge (Fig. 2). The electrophoretic mobilities of these constructs display a dramatic dependence on the phasing of the tRNA core and A5 bulge, indicative of a significant “fixed-bend” character for each element (Fig. 3). This phase dependence is particularly strong in the presence of magnesium ions (Fig. 3A). Although the relative mobilities cannot be used to quantify the bend angles, the dramatic reduction in the phase dependence of the mobilities in the absence of magnesium ions (Fig. 3B) suggests that the tRNA and/or bulge elements possess additional flexibility under the latter conditions. Moreover, μmin has shifted from n ≈ 6 to n ≈ 10, suggesting that there is a torsional realignment of one or both of the bends. Finally, it should be noted that all heteroduplex bands in Fig. 3 are narrow singlets, suggesting that conformers arising from bend angle dispersion must be equilibrating on a time scale that is short compared with the time of the electrophoresis experiment (for further discussion of this point, see ref. 24).
Figure 3.
Electrophoretic behavior of the E[tRNAPhe:A5] species in the presence of 2 mM Mg2+ (A) and in the absence of magnesium ions (B). In each gel image, the two outside lanes contain duplex control RNA molecules, with lengths of 188 bp and 200 bp as indicated; the remaining seven lanes contain heteroduplex molecules with n phasing base pairs (Fig. 2; n = 0 to 12). Electrophoresis was performed on nondenaturing 6% polyacrylamide gels (acrylamide/N,N′-methylenebisacrylamide ratio, 29:1) at room temperature; gel buffer was 10 mM sodium phosphate, pH 7.2/2 mM MgCl2 (A) or 10 mM sodium phosphate, pH 7.2/0.125 mM Na2EDTA (B).
Birefringence Decay Profiles for the Two-Bend Constructs Provide Evidence for Both Increased Bend Dispersion and Altered Torsional Phasing in the Absence of Magnesium Ions.
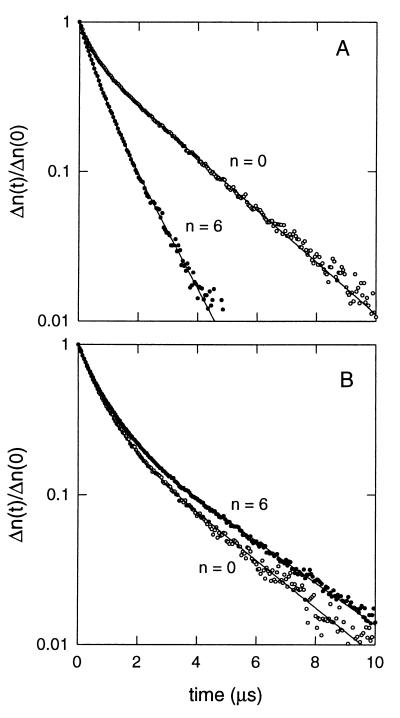
For representative birefringence decay curves for the n = 0 and n = 6 constructs are displayed in Fig. 4. A distinctive feature of both decay profiles in Fig. 4B (no magnesium ions) is that the fast-decay amplitudes (60–70%) are much larger than their counterparts in Fig. 4A (30–40%). Although we have not analyzed this characteristic further, the larger fast-decay components are consistent with an increase in bend dispersion (18, 24). Two additional features of the curves in Fig. 4B are the reduced separation between the n = 0 and n = 6 profiles (relative to the corresponding profiles in Fig. 4A), and the inversion of the profiles (τn = 6 > τn = 0 in Fig. 4B). Both features are consistent with the gel results: the reduced separation with reduced mobility variation and the profile inversion with a shift in the torsional phasing of the two bends.
Figure 4.
Representative birefringence decay curves for two E[tRNAPhe:A5] species, n was as indicated. (A) Presence of 2 mM Mg2+. (B) Absence of magnesium ions. Solid lines represent double-exponential fits to the experimental data by using the Levenberg–Marquardt method (23); terminal (slower) decay times were used for the τ ratio analysis. Buffers are as specified in the legend to Fig. 3, except for the sodium phosphate concentration, which was 5 mM.
τ Ratio Measurements Indicate That, in the Presence of Millimolar Magnesium Ions, the tRNA Core Lacks Additional Flexibility.
As indicated above, a powerful test for the absence of additional flexibility within the single-bend loci is quantitative agreement between the set of experimental τ ratios for the two-bend constructs and the curve predicted from the separately determined angles for each bend. It is evident from Fig. 5A that there is good agreement between the experimental data for the two-bend constructs and the curve predicted on the basis of the single-bend data in Table 1. This comparison can be quantified by considering two features of the τ ratio curve, namely, its amplitude and its mean, defined operationally as (τ ration=0 − τ ration=6) and [(τ ration=0 + τ ration=6)/2], respectively. In Fig. 5A, the curve means (predicted, 0.47; experimental, 0.51) and amplitudes (predicted, 0.39; experimental, 0.42) are both in good agreement. The experimental curve was obtained by a global (least-squares) fit to the two-bend τ ratio data, allowing both angles to vary. The resultant “experimental” curve yields interstem angles of 80° (tRNA core) and 111° (A5 bulge). The optimal angles, which are essentially independent of variations in either Peff or Ceff (see below), suggest that the tRNA interstem angle is more likely to lie in the range of 75–80°.
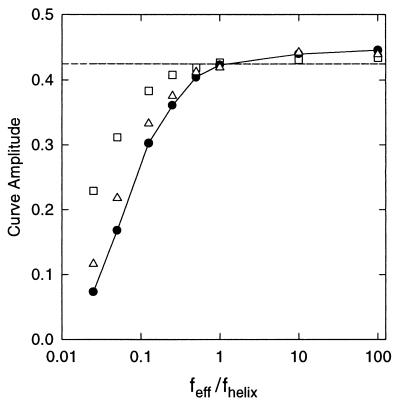
An upper bound for any additional dispersion of the anticodon–acceptor interstem angle can be estimated by assuming that no additional flexibility exists within the bulge element and by uniformly reducing Peff and/or Ceff within the tRNA core until the computed τ ratio curve differs significantly from the experimental data. For this analysis, the tRNA core is represented by a span of once-bent RNA helix with dimensions approximating those of the core region. For yeast tRNAPhe, this span is equivalent to about 13 bp, and extends from bp 7⋅66 to bp 27⋅37 (ref. 4 and see Materials and Methods). The results of this analysis are presented in Fig. 6. It is noteworthy that a 10% reduction in the amplitude of the experimental curve (Fig. 5A; i.e., from 0.42 to 0.38) is significant at the 1% level, irrespective of the nature of the added flexibility (i.e., torsional, bending, or both). Moreover, this result is not sensitive to the choice of the starting curve (optimal vs. predicted) in Fig. 5A.
Figure 6.
Dependence of the computed amplitude (≡ τ ration=0 − τ ration=6) of the phased τ ratio curve on the flexibility of the tRNA core, using the optimal experimental amplitude (dashed line) in Fig. 5A. Amplitudes are plotted as a function of feff/fhelix, where f = P (□), C (○), or either P or C (•), for constant P/C (= Phelix/Chelix). For feff/fhelix = 1, the tRNA core possesses no additional bending (P) or torsional (C) flexibility relative to an equivalent length of RNA helix. The horizontal dashed line represents the experimental amplitude for E[tRNAPhe:A5] in 2 mM Mg2+.
For a uniform increase in flexibility within the tRNA core, the 10% reduction in curve amplitude is exceeded for ∼4-fold reductions in either Ceff or Peff, or for a combined ∼1.6-fold reduction in Peff and Ceff. For the combined reduction (constant Poisson’s ratio ≡ kTP/C − 1), the 1% significance level corresponds to an increase of the rms fluctuations† for the polar and twist angles per base pair of only 25%, equivalent to an increase in the rms bend fluctuation from 5° to 7° for native RNA helix. Thus, in the presence of magnesium ions, the experimental data are consistent with the absence of any additional flexibility within the tRNA core. Additional flexibility, if present, is likely to be modest, with equivalent rms polar and/or torsional fluctuations that are within a factor of 1.5–2.0 of the corresponding fluctuations within the RNA helix.
In the foregoing analysis, we have assumed that the angles in Table 1 represent the fixed components of the interstem angles, with added angle dispersion (reduced Ceff or Peff) leaving the apparent angles unchanged. However, this solution is not necessarily unique; that is, differing combinations of fixed and stochastic components may give rise to the same apparent mean angle (8, 18). Therefore, to test for uniqueness, we have determined which combinations of fixed interstem angle (±20° of the optimal angle of 80°) and Peff/Phelix (≥0.05) yield apparent angles that are within the range of 75–85°. No fixed angle less than 75° or greater than 90° falls within the specified range for any value of Peff/Phelix. Moreover, even for 5° shifts toward the optimal tRNA interstem angle (e.g., 90° fixed to 85° apparent; Peff/Phelix = 0.08; 85° fixed to 80° apparent; Peff/Phelix = 0.13), the predicted curve amplitudes were unacceptably small (0.28 and 0.34, respectively) at the 1% significance level. Thus, the current solution is unique and confirms the earlier observation that for angles in the 60–100° range, added angle dispersion results in relatively little skewing of the apparent angle (8, 18). This uniqueness does not necessarily hold for much larger interstem angles, as will be seen in the following section.
The Flexibility of the tRNA Core Is Increased in the Absence of Magnesium Ions.
In the absence of magnesium ions, the experimental τ ratio curve (Fig. 5B) has a much smaller amplitude (∼0.08) than predicted (∼0.22) from the single-bend data in Table 1, although the curve means differ by less than 5% (0.69, experimental; 0.66, predicted). This result represents strong evidence for the presence of additional flexibility of one (or both) bends. Further support for added flexibility derives from the large fast amplitudes of the birefringence decay profiles in Fig. 4B, and more specifically, from the large fast amplitude for the decay profile of the individual tRNA element in the absence of magnesium ions (17). It should also be noted that the torsional phasing of the two bends is shifted substantially relative to the phasing observed in Fig. 5A. This shift is anticipated from the gel results in Fig. 3, where the species with minimum mobility has been shifted by Δn ∼ 4 (∼120°). A least-squares analysis of the data in Fig. 5B indicates that the shift in free solution is closer to 180° (Δn ∼ 6), although a precise determination of the phase shift for the latter is precluded by the small curve amplitude.
Despite clear evidence for the presence of additional flexibility, the magnitude of the added flexibility is more difficult to quantify, because reductions in Peff and Ceff influence the curve amplitude and mean in different ways. We can, however, draw some conclusions as to the bounds for Peff and Ceff with the current (tRNA core) model.
Reduced Ceff.
For pure reductions in Ceff (greater torsional dispersion), the optimal fit to the experimental data (minimum χ2) yields angles of 160° (tRNA) and 110° (A5 bulge) and Ceff/Chelix = 0.02. These optimal angles differ by only 4–5% from the angles in Table 1. In addition, Ceff/Chelix varies by less than 2-fold for a range of angles that are within ±5° of the angles reported in Table 1. Finally, bulge angles ≤100° are all rejected at the 1% level irrespective of the value of Ceff/Chelix. Although an approximately 50-fold reduction in Ceff (∼7-fold increase in average angle dispersion) is formally possible, it is unlikely that such a reduction would occur in the absence of a coordinate reduction in Peff; thus, we consider the other limiting model as well.
Reduced Peff.
By using the angles specified in Table 1, a pure reduction in Peff/Phelix sufficient to yield the experimental amplitude (0.08) leads to a substantial downward displacement of the curve, because of the effects of polar weighting of the angle (8). However, in the presence of added flexibility of the tRNA core, the reported angle (154°; Table 1) is likely to represent a skewed average of any additional polar dispersion, with a larger (i.e., more linear) fixed angle component (see discussion in ref. 8). For example, for a linear core (180° angle), reduction of Peff/Phelix to 0.25 would reduce the apparent interstem angle to the observed value of 154°, although this limiting case predicts a zero amplitude for the τ ratio curve. For a fixed tRNA angle of 10° and a Peff/Phelix of 0.4, both the amplitude and mean of the experimental curve are recovered; and for Peff/Phelix > 0.4, reductions in Ceff/Chelix are also required to match the experimental amplitude.
Thus, although the experimental curve in Fig. 5B cannot be explained without invoking additional flexibility of the tRNA core, the added flexibility may take more than one form within the limits of the current model, which assumes a uniform elastic response within the core. In particular, the curve can be reproduced through modest reductions in bending rigidity with attendant increases in the interstem angle, more substantial reductions in torsional rigidity, or a combination thereof.
DISCUSSION
In the presence of magnesium ions, the amplitudes of the experimental and computed τ ratio phasing curves are in close agreement (Fig. 5A), suggesting that the tRNA core is not significantly more flexible than an equivalent length of (bent) RNA helix. Thus, for a mean angle of 70–80° between the anticodon and acceptor stems (Table 1; current analysis), the angle dispersion is expected to have a SD in the range of 15–18°, the expected dispersion for a bent RNA helix of roughly the same dimensions as the tRNA core. The “equivalent-helix” model used herein is designed to account for the intrinsic flexibility of the native RNA helix; the τ ratio approach itself accounts for the flexibility of the helix extensions. The current results are entirely consistent with the angle dispersion observed in a previous electron micrographic analysis of the extended tRNA species (5, 21) and provide a physical basis for the dispersion observed in the electron micrographic investigation.
The agreement between the experimental and predicted curves in Fig. 5A allows one to draw two additional conclusions: (i) The A5 bulge is likely to be relatively rigid because the amplitude of the τ ratio curves reflects the combined effects of both bends; any increase in bulge flexibility would further restrict the permissible range of motion for the tRNA core. (ii) The helix extensions do not appreciably influence the mean interstem angles, even for the cis phasing of the bends (τ ratio minimum), where the extensions suffer their closest approach. This result is of general importance for single-bend studies in which the interstem angles may be quite acute; it argues that the derived single-bend angles are valid.
Two features of the experimental data obtained in the absence of magnesium ions (Fig. 5B) also deserve further comment. (i) The mean of the experimental data is near the mean of the computed curve, again indicating that there is little angle bias created by the helix extensions, even under conditions where at least one bend is highly flexible. (ii) The maximum in the τ ratio curve near n = 6 suggests that there is a substantial shift in the phase relationship between the two bends. There does appear to be a difference in the extent of the phase shift between the gel and solution results. The origin (and significance) of this difference is unclear at this point but may be related to the influence of the gel matrix on the two-bend constructs. In this regard, the phase relationship between the bends is based on a positive tilt at the bulge center and a positive or negative roll for the tRNA bend center (Fig. 5 A or B, respectively), with an assumed (average) helix repeat of 12 bp per turn between the bend centers. The absolute directions of the bends should be considered arbitrary until the direction of the bulge-induced bend has been defined, both in the presence and absence of magnesium ions. However, neither the absolute bend directions nor the interbend phasing has any effect on either the curve amplitudes or conclusions regarding flexibility.
There are two further implications of the amplitude plots in Fig. 6. (i) Elements of tertiary structure that are more rigid than the surrounding helix (Peff/Phelix > 1) will have little influence on the overall spatial distribution of the flanking helices, a consequence of the intrinsic flexibility of the latter. (ii) Slight increases in flexibility (e.g., Peff/Phelix ∼ 0.5) will have relatively minor effects on the overall spatial distribution of the arms for the tRNA core in the presence of magnesium ions, also due to the intrinsic flexibility of the RNA helix. This latter point is important from a mechanistic standpoint, because it bears on the issue of the entropic free energy change accompanying a change in global conformation (for further discussion, see ref. 25).
The current computational analysis has used a simplifying model in which the core of the tRNA is represented by an approximately 13-bp segment of helix with flexibility parameters (Peff and Ceff) that are uniformly varied. The elastic response within the core is unlikely to be uniform (1, 2, 6, 7), and the altered core flexibility (e.g., in response to withdrawal of magnesium ions) is unlikely to be uniformly distributed. Nevertheless, the current approach can be used to characterize the intrinsic global flexibility of the tRNA core (as well as other nonhelix elements). In particular, it can be used to examine the influence of specific tertiary interactions on both the interstem angles themselves and the stabilization of the core to structural deformation. It is also assumed that the span of the central core region (equivalent to about 13 bp) is unchanged in the absence of magnesium ions. We cannot rule out some increase in this span with the loss of tertiary structure. However, any such increase would shift the mean of the experimental curve upward relative to the mean of the computed curve; in fact, the means of those curves differ by less than 5% (Fig. 5B). In any case, failure of this assumption would not significantly affect the conclusions regarding additional flexibility, which are based principally on the amplitude of the phasing curve. This issue is analyzed in detail elsewhere (8).
Finally, it is interesting to speculate on the nature of the transition to the more open flexible core upon the removal of magnesium ions. Under the conditions used in the current analysis (2 mM Mg2+ and 5–10 mM sodium phosphate at 4°C), much of the secondary structure within the tRNA core appears to be preserved. In particular, the current measurements were performed at temperatures well below the thermally induced hyperchromic transition of the extended tRNA moiety, and the transition to the open form is essentially isochromic (ref. 17 and references and further discussion therein). Furthermore, interstem stacking is preserved within the T acceptor and D anticodon domains (12, 26). Thus, the transition itself may involve the loss of one or more critically coordinated magnesium ions, believed to exist in the 8–12 turn and between the D and T loops.
CONCLUSION
A modified form of transient electric birefringence experiment (phased τ ratio method) has been used to demonstrate that in the presence of magnesium ions the core of yeast tRNAPhe does not possess additional flexibility relative to an equivalent span of RNA helix. However, upon removal of divalent cations, the tRNA core gains flexibility as its tertiary structure is disrupted. The self-consistency of the single-bend and double-bend τ ratio measurements provides a strong argument against the concern that either direct or indirect interactions between the helix extensions are influencing the interstem angles. Thus, the current approach should be broadly applicable to the study of both intrinsic and protein-induced bending of RNA and DNA.
Acknowledgments
We thank Dr. M. Zacharias for useful discussions, Dr. M. Olmsted for program modifications, Drs. M. Yarus and T. Cech for useful comments, and J. Mills for help with the production of the DNA oligonucleotides. This work was supported by a grant from the National Institutes of Health (GM 52557 to P.J.H.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: E, extended.
〈δθ〉rms ≡ (〈θ〉2 − θo2)½, where θ is the total instantaneous polar angle per base pair, θo is the fixed component of the angle, and 〈θ〉rms = 〈θ2〉½. Equivalent definitions apply to fluctuations, 〈δϕ〉rms, in the twist angle.
References
- 1.Holbrook S R, Sussman J L, Warrant R W, Kim S-H. J Mol Biol. 1978;123:631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- 2.Hingerty B, Brown R S, Jack A. J Mol Biol. 1978;124:523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- 3.Stout C D, Mizuno H, Rubin J, Brennan T, Rao S T, Sundaralingam M. Nucleic Acids Res. 1976;3:1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friederich M W, Gast F-U, Vacano E, Hagerman P J. Proc Natl Acad Sci USA. 1995;92:4803–4807. doi: 10.1073/pnas.92.11.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T M, Wang Y-H, Zaug A J, Griffith J D, Cech T R. EMBO J. 1995;14:4849–4859. doi: 10.1002/j.1460-2075.1995.tb00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tung C-S, Harvey S C, McCammon J A. Biopolymers. 1984;23:2173–2193. doi: 10.1002/bip.360231106. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura S, Doi J. Nucleic Acids Res. 1994;22:514–521. doi: 10.1093/nar/22.3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vacano E, Hagerman P J. Biophys J. 1997;73:306–317. doi: 10.1016/S0006-3495(97)78071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagerman P J. Curr Opin Struct Biol. 1996;6:643–649. doi: 10.1016/s0959-440x(96)80031-5. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya A, Murchie A I H, Lilley D M J. Nature (London) 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 11.Tang R S, Draper D E. Biochemistry. 1990;29:5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- 12.Heerschap A, Haasnoot C A G, Hilbers C W. Nucleic Acids Res. 1983;11:4483–4499. doi: 10.1093/nar/11.13.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen L X, Tinoco I., Jr J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 14.Eis P S, Millar D P. Biochemistry. 1993;32:13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- 15.Yang M, Millar D P. Biochemistry. 1996;35:7959–7967. doi: 10.1021/bi952892z. [DOI] [PubMed] [Google Scholar]
- 16.Sussman J L, Holbrook S R, Warrant R W, Church G M, Kim S-H. J Mol Biol. 1978;123:607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- 17.Friederich M W, Hagerman P J. Biochemistry. 1997;36:6090–6099. doi: 10.1021/bi970066f. [DOI] [PubMed] [Google Scholar]
- 18.Zacharias M, Hagerman P J. Biophys J. 1997;73:318–332. doi: 10.1016/S0006-3495(97)78072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zacharias M, Hagerman P J. J Mol Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 20.Kebbekus P, Draper D E, Hagerman P J. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- 21.Hagerman P J. Annu Rev Biophys Biomol Struct. 1997;26:139–156. doi: 10.1146/annurev.biophys.26.1.139. [DOI] [PubMed] [Google Scholar]
- 22.Hagerman K R, Hagerman P J. J Mol Biol. 1996;260:207–223. doi: 10.1006/jmbi.1996.0393. [DOI] [PubMed] [Google Scholar]
- 23.Press W H, Vetterling W T, Teukolsky S A, Flannery B P. Numerical Recipes in Fortran: The Art of Scientific Computing. Cambridge, U.K.: Cambridge Univ. Press; 1992. pp. 678–683. [Google Scholar]
- 24.Shen Z, Hagerman P J. J Mol Biol. 1994;241:415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- 25.Zacharias M, Hagerman P J. J Mol Biol. 1996;257:276–289. doi: 10.1006/jmbi.1996.0162. [DOI] [PubMed] [Google Scholar]
- 26.Hilbers C W, Heerschap A, Haasnoot C A G, Walters J A L I. J Biomol Struct Dyn. 1983;1:183–207. doi: 10.1080/07391102.1983.10507434. [DOI] [PubMed] [Google Scholar]