Abstract
A cluster of four tRNA genes in Escherichia coli is co-transcribed with an adjacent gene encoding elongation factor Tu. The resultant transcript that specifies both structural (tRNA) and informational (mRNA) RNA may not be an uncommon occurrence and has interesting regulatory implications.
In Escherichia coli, where regulation of transcription has been most extensively studied, the RNAs synthesized are generally divided into two categories—the class of informational or mRNAs that are metabolically unstable, and the class of structural RNAs such as rRNA and tRNA that do not show appreciable turnover. The two classes are also distinguishable by their regulation pathways. In particular, stable RNA is subject to a coordinate regulation that is responsive to the availability of amino acids for protein synthesis. In general, this type of control only affects informational RNAs encoding proteins required in protein synthesis.
Transfer RNA genes are distributed throughout the E. coli genome as part of ribosomal RNA operons or as clusters of tRNA genes containing either different tRNA genes or repeats of the same gene (for review see ref. 1). We report here a novel transcription unit composed of a tRNA cluster and mRNA and suggest that the co-transcription of structural and informational RNA may not be an uncommon occurrence. Possible implications of these dual function transcripts are also discussed.
Organization and structure of the tyrU gene cluster
The tyrU gene cluster is situated between the rrnB ribosomal operon and the tufB gene at 89 min (ref. 2). The tyrU locus encodes the major tyrosine accepting species, , which occurs as a single copy within a cluster of three other tRNA genes3–5. The tufB locus codes for elongation factor (EF)-Tu, which promotes the binding of aminoacyl-tRNA to ribosomes. In addition to its key role in translation, EF-Tu functions as a structural component of Qβ replicase (see ref. 6 for review).
Figure 1 shows the DNA sequence of the tyrU gene cluster and the 5′ end of the adjacent tufB gene, which is located only 114 base pairs (bp) downstream of the tRNA cluster. The tyrU promoter (as identified by hybridization analyses presented below) is located directly upstream of the first tRNA gene in the cluster and shares two features with the tyrT promoter region: a region of G-C bias flanking the Pribnow box (Fig. 1 and ref. 7), and wo dyad symmetries, one involving the −35 region and the other centred near the Pribnow sequence (Fig. 1 and ref. 8). These sequence features are present in the promoter regions of other stringently controlled genes and may therefore have a role in their coordinate synthesis. The Pribnow box participates in a dyad symmetry in the ribosomal protein operons9 and the G + C-rich sequence following the Pribnow heptamer has been noted for rRNA and ribosomal protein operons10.
Fig. 1.
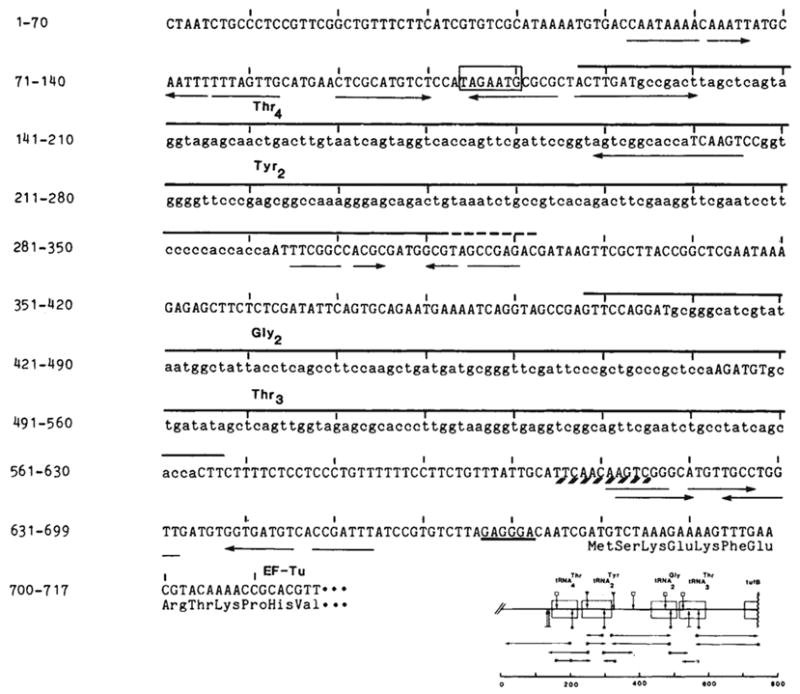
Nucleotide sequences of the tyrU cluster and proximal tufB region. The DNA strand with the sequence of the RNA transcript is shown. The source of DNA was the transducing phage λ rifd 18 (ref. 51; supplied by M. Nomura or N. Fiil). DNA sequencing was by the method of Maxam and Gilbert52; *, 5′-labelled end of the restriction fragments; →, direction and extent of the sequence obtained; the scale is in base pairs. Complement and overlap were obtained for all new sequences reported here with the exception of the sequence upstream of the gene. The sequence is identical to that reported by An and Friesen16 with the exception of an additional base pair they place at position 317. Structural tRNA sequence (lower case) and the two dimeric precursors are overlined. The immer is 170 bases. The precise 3′ terminus (---) of (~200 bases) is not known. It is located in a region of dyad symmetry (→ ←) as is the site between the tRNA cluster and the tufB gene at which processing or termination occurs (positions 608–618; see text). Transcription from the presumptive promoter would initiate at the A 7 bases upstream of as inferred from the results of Taylor and Burgess14. The Pribnow box (□) is preceded by a −35 sequence (TTGCAT) similar to the consensus sequence TTGACA of Rosenberg and Court53 and also byan A+T-rich region (88% A-T from −63 to −38). A potential ribosome binding site54, located 6 bases upstream of tufB, is underlined. The first 12 amino acids of tubB (ref. 19) are listed below their respective codons.
A co-transcript of tRNA and mRNA
Knowledge of the organization and structure of the tyrU cluster and adjacent tufB gene has facilitated analyses of the in vivo transcripts and processing intermediates. The methods developed by Alwine and co-workers11 were used to detect RNA species that comprise a small fraction of the total cellular RNA. The experimental approach involved fractionating denatured RNA by gel electrophoresis, transferring the RNA to diazo-benzyloxymethyl (DBM) paper and probing the RNA blots with radiolabelled restriction fragments isolated from the tyrU region (as detailed in Fig. 2 legend). Detection of RNA precursors was maximized by using E. coli strains deficient in RNA processing. Precursor tRNAs accumulate in these strains as a result of mutations blocking and/or delaying their processing12.
Fig. 2.
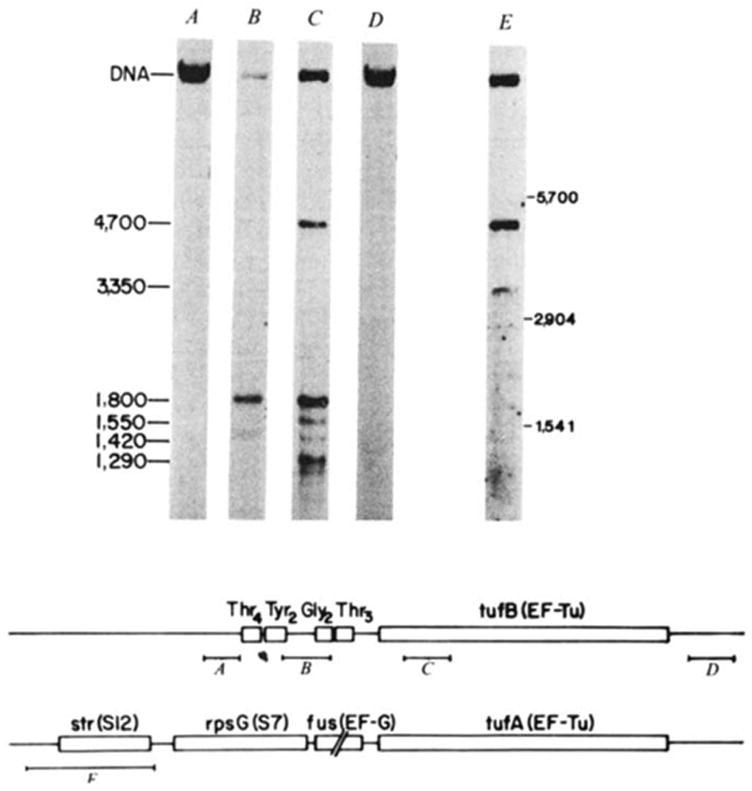
Characterization of the transcript specifying the tyrU cluster and tufB. E. coli strain ABL-1 (RNase III− RNase Pts; ref. 55; gift of W. McClain) was grown for several generations at 30 °C, and then incubated at 42 °C for 30 min to inactivate RNase P. RNA was extracted according to Gegenheimer and Apirion55 as detailed elsewhere30, denatured with glyoxal56, electrophoresed in 1.5% agarose gels with recirculation of the 10 mM sodium phosphate, pH 6.5, 1 mM EDTA running buffer, and transferred to DBM paper11. The RNA blots were probed with DNA fragments (A-E) which were nick translated57 with [α-32P]dCTP (NEN; 2–3,000 Ci mmol−1) to a specific activity of ~5 × 107 c.p.m. per μg. Hybridizations were carried out in the presence of 10% dextran sulphate as detailed by Alwine et al.11 but omitting carrier DNA. Blots were washed first in 0.3 M NaCl, 0.03 M sodium citrate, 0.1% SDS at 23 °C for 20 min, and then in 15 mM NaCl, 1.5 mM sodium citrate, 0.1% SDS for 45 min at 50–55 °C. The 32P-labelled probes isolated from the region of λ rifd18 depicted in the diagram were: A, HhaI-145; B, HinfI-190; C, HincII-SmaI-187; D, BstNI-HaeIII-185. Probe E is a HaeIII-530 fragment encompassing the S12 ribosomal protein gene, which was isolated from λ fus3 (obtained from J. Yates and M. Nomura). Molecular weight markers were 16S (1,541 bases), 23S (2,904 bases) and 30S RNA (~5,700 bases). Lanes B, C and E have ~2 μg RNA per lane, and lanes A and D have 30 μg per lane. The scale is in bases. Structural sequences for tRNAs and tufB are boxed.
Figure 2 shows results of this analysis for selected probes. The tRNA-specific probe (Fig. 2 B) hybridized to one band of 1,800 bases (transcripts smaller than ~700 bases were detected on the higher composition gels shown in Fig. 5). The tufB probe (Fig. 2 C) also hybridized to the 1,800-base transcript, as well as to four other transcripts sized at 4,700, 1,550, 1,420 and 1,290 bases. As detailed below, the 4,700-base transcript contained sequences from the other gene encoding EF-Tu, tufA (ref. 13), and the lower molecular weight transcripts probably represent tufA and tufB processing intermediates. The probes immediately upstream (Fig. 2 A) or downstream (Fig. 2 D) of the tRNA-tufB region did not hybridize to the 1,800-base transcript. This collection of probes thus defines a transcript of 1,800 bases which contains sequences from both the tRNA gene cluster and the tufB gene.
Fig. 5.
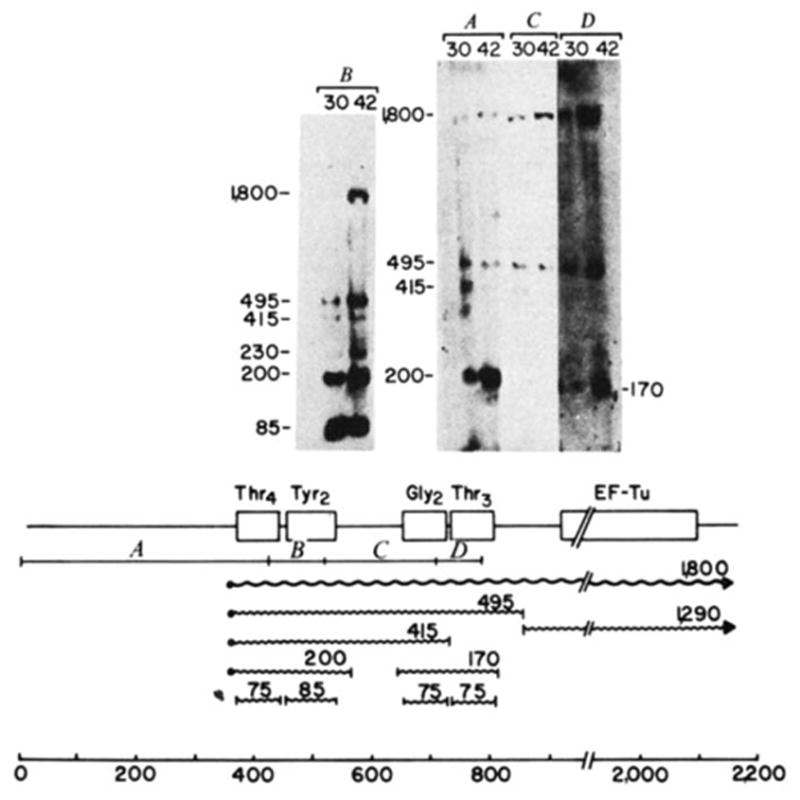
Processing intermediates of the tyrU cluster. RNA was extracted from E. coli strain A49 (RNase Pts; ref. 59; gift of W. McClain) grown at either 30 or 42 °C, electrophoresed on 2% agarose gels at 30 μg RNA per slot and blotted onto DBM paper. Transfer conditions were similar to those described for Fig. 2 except that glyoxal was removed with a lower concentration of sodium hydroxide (10 mM compared with 50 mM for Fig. 2). Molecular weight markers were 5′-end-labelled HaeIII and HincII fragments of ΦX174. The gel shown in B was electrophoresed for a shorter time to retain mature tRNA. The 32P-labeIled probes isolated from the regions of λ rifd18 illustrated in the diagram were: A, HinfI-420; B, HinfI-93; C, HinfI-190; D, HinfI-82. The putative primary transcript (
 ), processing intermediates and mature tRNAs (
), processing intermediates and mature tRNAs (
 ) are labelled by their respective sizes (in bases) on denaturing gels. The probe containing the structural tyrosine tRNA sequence (B) hybridized to a 230-base transcript that also hybridized with a
upstream probe and must therefore be a tyrT transcript. Probe C contained only 18 bases of structural tyrosine tRNA sequence, which severely limited its binding to the 200- or 230-base tyrosine transcripts in the stringent washing conditions used in these experiments. In the blot shown, the hybridization of probe C to the 170-base transcript did not photograph; in other blots this probe detectably bound the 170-base transcript. The processing scheme is discussed in the text.
) are labelled by their respective sizes (in bases) on denaturing gels. The probe containing the structural tyrosine tRNA sequence (B) hybridized to a 230-base transcript that also hybridized with a
upstream probe and must therefore be a tyrT transcript. Probe C contained only 18 bases of structural tyrosine tRNA sequence, which severely limited its binding to the 200- or 230-base tyrosine transcripts in the stringent washing conditions used in these experiments. In the blot shown, the hybridization of probe C to the 170-base transcript did not photograph; in other blots this probe detectably bound the 170-base transcript. The processing scheme is discussed in the text.
The 1,800-base transcript was further characterized by sequential hybridizations using probes covering different portions of the tyrU region. Figure 3 summarizes the hybridization results. Eight probes isolated from regions of structural tRNA or tufB sequence displayed hybridization to the 1,800-base transcript. Importantly, the sum of the restriction fragments that hybridized to the co-transcript corresponds to the molecular weight of the transcript (1,800 bases) determined on denaturing gels. Probes that failed to hybridize to the 1,800-base transcript defined the outer limits of the transcript: the 5′-proximal probe that produced negative results was separated from the structural sequence by 12 bp and the 3′-proximal probe began 85 bp downstream of tufB structural sequence. Thus, this series of probes specified a transcript encompassing both the tRNA cluster and the tufB gene.
Fig. 3.
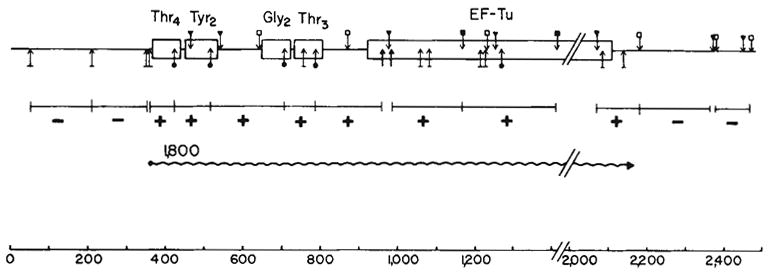
Organization and transcription of the tRNA-tufB region. The locations of the restriction sites were obtained from the sequence shown in Fig. 1 or ref. 16. Hybridization to the 1,800-base transcript (
 ) is denoted by a + in the appropriate restriction fragment (from λrifd18) and lack of hybridization by −. The HinfI-82 probe from the
region hybridized strongly to 16S RNA in addition to the 1,800-base transcript. This result is consistent with the presence of a number of 12–16-bp homologies between 16S RNA58 and
sequences. Due to the hybridization with 16S RNA, the HinfI-82 probe could not be used to determine whether the smaller tuf transcripts (1,550 and 1,420 bases shown in Fig. 2) were tufA or tufB specific. The scale is in bases. Structural sequences for the tRNAs and tufB are boxed.
) is denoted by a + in the appropriate restriction fragment (from λrifd18) and lack of hybridization by −. The HinfI-82 probe from the
region hybridized strongly to 16S RNA in addition to the 1,800-base transcript. This result is consistent with the presence of a number of 12–16-bp homologies between 16S RNA58 and
sequences. Due to the hybridization with 16S RNA, the HinfI-82 probe could not be used to determine whether the smaller tuf transcripts (1,550 and 1,420 bases shown in Fig. 2) were tufA or tufB specific. The scale is in bases. Structural sequences for the tRNAs and tufB are boxed.
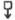 , BstI; ↓, HaeIII; ↥, HhaI;
, BstI; ↓, HaeIII; ↥, HhaI;
 HincII;
HincII;
 HinfI;
HinfI;
 SmaI.
SmaI.
To establish these unexpected results unequivocally, control experiments were carried out to ensure that: (1) failure to detect hybridization with the negative probes was not due to insufficient radioactivity, and (2) the observed hybridization with the positive probes was not the result of either contamination (with other labelled DNA) or the result of fortuitous homologies. The ability of each nick-translated probe to produce a strong hybridization signal to its respective band and to no other fragments of a digest of λ rifd 18 DNA was tested. As shown in Fig. 4 for selected probes and detailed in the legend, each probe hybridized only to its corresponding λ rifd18 restriction fragment.
Fig. 4.
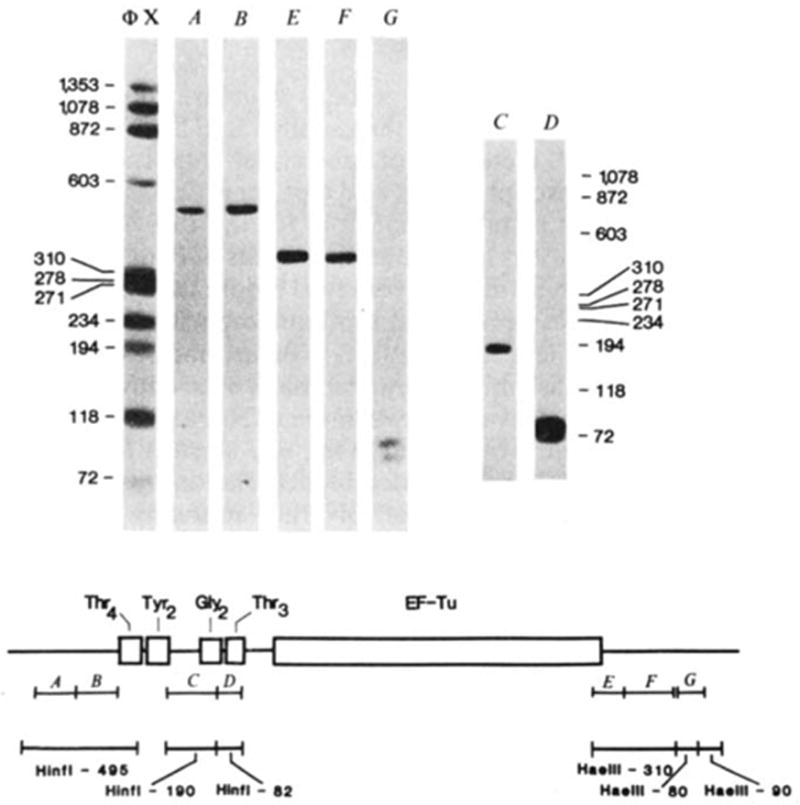
Hybridization of probes to λ rifd18 DNA blotted onto DBM paper. The BamHI-EcoRI-5,500 fragment containing the entire tRNA-tufB region was digested with either HinfI (lanes A–D) or HaeIII (lanes E–G) and 0.1 μg of each digest per slot was electrophoresed on a composite gel (5% acrylamide, 0.7% agarose) cross-linked with N, N′-diallyltartardiamide as detailed by Alwine et al.11. The gel was soaked in 2% periodic acid to dissolve the cross-links before blotting. Blots were hybridized to the nick-translated probes illustrated in the diagram and washed as described in Fig. 2 legend: A, HhaI-150; B, HhaI-145; C, HinfI-190; D, HinfI-82; E, HaeIII-BstNI-125; F, BstNI-HaeIII-185; G, BstNI-103. Probes A and B both hybridized only to the HinfI-495 fragment (depicted below the probe) from which they can be isolated by HhaI digestion. Similarly, probes E and F each hybridized to the HaeIII-310 fragment from which they can be isolated by BstNI digestion. The two structural tRNA probes, C and D, hybridized only to their respective HinfI-190 or HinfI-82 bands. Importantly, the -specific probe (C) did not hybridize to the fragment with which it has limited homology. Probe G hybridized to two HaeIII bands ~80 and 90 bp, consistent with the existence of one HaeIII site within the BstNI-103 fragment and a HaeIII site closely flanking each end (see Fig. 3). No hybridization was observed to additional bands when higher concentrations of DNA (0.5 μg per slot) or longer exposures were used. On the basis of the exposure times and DNA concentrations used in Fig. 3, the nick-translated probes can be expected to detect as little as 0.1 ng RNA. All the fragments (118–1,353 bp) of the ΦX174 HaeIII digest (molecular weight marker) exhibited comparable band intensities when probed with 32P-end-labelled ΦX174.
The hybridization results place the promoter for the tRNA-tufB co-transcript directly before the gene and the terminator closely following the tufB structural sequence. Note that these assignments are not precise, because a probe could share as much as 30 bases of homology with the transcript and fail to hybridize. Genetic evidence corroborates the location of the tufB promoter directly upstream of the tRNA cluster60. In addition, Taylor and Burgess14 found that a restriction fragment containing the presumptive tRNA-tufB promoter (labelled HaeIII-890 by Taylor and Burgess14 and HaeIII-700 by Rossi et al.15) formed filter-retainable complexes with RNA polymerase. Finally, inspection of the DNA sequence reveals a promoter region situated directly before the gene (see Fig. 1), a location consistent with the analyses of the functional promoter. The location of a possible termination sequence 34 bases following tufB (ref. 16) is also consistent with our hybridization results (Fig. 3). That termination occurs at this site in vivo is also suggested by the presence of gene U, which is transcribed from its own promoter ~200 bases downstream of tufB (refs 16,17).
Other EF-Tu transcripts
EF-Tu is encoded by duplicate genes that contribute approximately equivalent amounts of EF-Tu to the cell18,19. The two proteins differ only in the carboxy-terminal amino acid19, suggesting that they are functionally equivalent. The other gene for EF-Tu, tufA, is located at 72 min as the terminal gene of the str ribosomal protein operon13. The str-rpsG-fus-tufA genes are transcribed from a single promoter13 located 80 bp upstream of str structural sequence20, which could yield a primary transcript with an estimated size of 4,800 bases.
Probes prepared from tufB structural sequences hybridize to both tufA and tufB transcripts, as the DNA sequences of tufA and tufB differ at only 13 of 1,181 bp16,21. The largest tuf transcript was suspected to be tufA specific, as probes immediately upstream and downstream of the tRNA-tufB region failed to hybridize to it (Fig. 2). A probe isolated from the 5′ end of the str operon was used to prove that the 4,700-base transcript encoded the str operon. The str probe hybridized to the 4,700-base transcript that was also detected with the tufB structural probe (Fig. 2 E), indicating that the 4,700-base transcript corresponds to a full-sized str operon mRNA. This same probe also hybridized to a 3,350-base message that did not hybridize with tufB structural probes and could therefore encode the first three genes of the str operon (str-rpsG-fus). Our present data do not indicate whether the three smallest RNAs (1,290, 1,420, 1,550 bp (originate from tubB or tufA or both. As determined by densitometric tracings, these small RNAs are less abundant than the major 1,800-base transcript (by 2-, 22-, and 6-fold for 1,290-, 1,420- and 1,550-base transcripts respectively).
Modes of EF-Tu synthesis
The presence of one copy of EF-Tu in a ribosomal protein operon and the other in a tRNA operon may be important in coordinating the synthesis of these three components of the translational machinery. The intracellular concentration of EF-Tu does, in fact, parallel that of tRNAs and ribosomes during changing growth conditions in E. coli22,24. Moreover, EF-Tu synthesis from both loci is subject to stringent control in response to amino acid starvation23,25–28.
In addition to transcripts corresponding to full-size str operon (4,700 bases) and tRNA-tufB operon (1,800 bases), there are three smaller tuf messages (1,290, 1,420 and 1,550 bases) (see Fig. 2). The 1,290-base message corresponds to the non-tRNA cleavage product of the 1,800-base primary transcript as shown in Fig. 5. In addition, an RNA of this size could result from transcription initiating at a promoter site (TGATGTC) located 32 bases upstream of tufB. Indeed, some promoter activity has been observed in this region in vivo60. The intergenic region also contains a possible termination sequence (CAACAA; refs 29, 30) that overlaps a sequence (TTCAACAAGTC) postulated to be involved in processing because of its homology with the characterized tyrT processing site (ref. 31; see also Fig. 6). Due to this overlap the alternative, non-mutually exclusive pathways that could uncouple EF-Tu synthesis from that of the tRNA cluster (processing versus termination and re-initiation of transcription) cannot be distinguished. However, the presence of large amounts of the 1,800-base transcript (Fig. 2) indicates that termination following the tRNA cluster (if it occurs at all) cannot be very effective in these growth conditions.
Fig. 6.
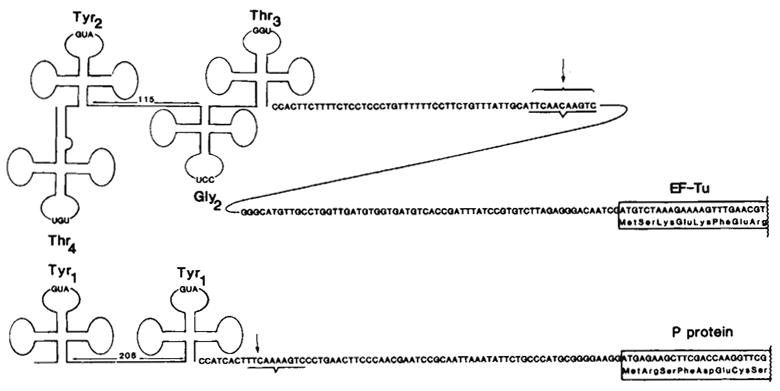
Comparison of the tyrT and tyrU primary transcripts. The tyrT transcript was characterized by Rossi et al.30 using RNA extracted from an RNase Pts E. coli strain infected with (doublet). The site conserved between the tyrT and tyrU gene clusters and known to be a 3′-processing site for tyrT transcripts31 is underlined. For tufB (ref. 19) and for the protamine-like protein (P protein; see text), the first eight amino acids are indicated. Translation of P protein also initiates at the methionine located 4 codons upstream of the start site shown, as indicated by the in vitro production of both a 29- and 33-amino acid basic protein that map to the first repeat61.
A similar potential for uncoupling exists at the tufA locus. The 3,350-base str-rpsG-fus message (Fig. 2) could result from either a termination or processing event or both. Analogous to the tufB region, promoter activity of DNA segments directly upstream of tufA has been observed in vivo (ref. 21 and J. D. Friesen and co-workers, personal communication). As noted below, the processing site conserved between the tyrT and tyrU transcripts is also present in the fus-tufA intergenic region, a finding that favours processing as a mode of uncoupling. Consistent with the suggested uncoupling of tufA synthesis, Nomura and co-workers find that translational repression of str operon by S7 protein does not inhibit EF-Tu synthesis in vitro32,33 or in vivo (M. Nomura, personal communication).
Processing intermediates of the tyrU cluster
Figure 5 gives the tRNA processing intermediates of the 1,800-base primary transcript. Each probe hybridized to mature tRNA (75–85 bases), the 495- and the 1,800-base transcripts. Only or probes hybridized to a 200-base transcript; similarly, only or probes hybridized to a 170-base transcript. The data suggest a processing scheme (depicted in the lower portion of Fig. 5) in which the primary transcript is initially processed to a 495-base transcript containing the tRNA cluster and a tufB mRNA of ~1,300 bases (Fig. 2). The tRNA precursor is subsequently processed to two dimeric precursors, a 200-base transcript containing and a 170-base transcript containing . Both these dimers are very close in size to those previously isolated from an E. coli strain with thermolabile RNase P activity34–36.
A secondary pathway for processing the tRNA-tufB transcripts may exist in RNase P-deficient strains. A 415-base transcript was apparent in lanes probed with A, B and C but not D, suggesting that this transcript consists of a trimer. This trimer may result from cleavage of either the 1,800- or 495-base transcript between the and structural genes.
The positions of the and dimers, based on the size of the processing intermediates (Fig. 5) or obtained from the sequence of the dimer34, are shown in Fig. 1. The long spacer separating the two dimers is similar to that found in tyrT (ref. 37) and supB–supE (ref. 38) and may be a common feature of tRNA clusters. The 114-base spacer separating the tRNA cluster and tufB is characterized by a 34-base C + T-rich region directly following the tRNAs.
The endonucleases responsible for cleaving the 1,800-base primary transcript have not been identified. One candidate was RNase III; however, there was no significant difference in the transcripts isolated from strains with or without functional RNase III (ABL-1 compared with A49). Other candidates include the tRNA processing endonuclease(s) partially purified by several laboratories36,39–41. It is of interest that the sequence conserved between the tyrT and tyrU primary transcripts, which is apparently the site for cleaving the tRNA cluster31 from the distal message (see Fig. 5), is also present in the fus-tufA intergenic region21 (TACCAAAGTC matches in 8 of 10 positions). The existence of a conserved processing site suggests that a single endonuclease may coordinately control EF-Tu processing from the tufA and tufB transcripts.
Finally, we note that in cells wild type for processing enzymes (CA274), the 1,800-base transcript is the only tyrU precursor detectable; none of the tRNA processing intermediates can be seen. Furthermore, as the amount of 1,800-base transcript in CA274 is comparable with that observed in cells deficient for processing (RNase Pts or the double mutant RNase III− RNase Pts), we suggest the tRNA sequences on the 1,800-base transcript are matured and function as tRNA in the cell.
Generality of dual function transcripts
The presence of tRNA on the same transcript as mRNA may not be rare in prokaryotes. An analysis of the in vivo transcripts of the gene in E. coli has revealed a 650-base transcript which, in addition to containing two sequences, can code for a small arginine-rich protein30. The putative basic protein is encoded by the first of the three repeated units that follow the structural gene37. That the tyrT transcript actually encodes protein is suggested by the in vitro production of a low molecular weight, basic protein that maps to the first repeat61.
Transcripts of the tyrT and tyrU gene clusters share several interesting features (Fig. 6): (1) the 5 ′ portion of both transcripts consists of two regions of cloverleaf secondary structure that are separated by a long spacer (115–208 bases), (2) the tyrosine structural sequences present on each transcript are almost identical, differing by only two bases in the variable loop region42, and (3) following the tRNA region there is an intergenic spacer of 67–114 bases containing a conserved sequence (TTCAAAAGTC; located in a region of dyad symmetry) which seems to be the site for processing the tRNA cluster from the mRNA portion of the transcript (ref. 31 and Fig. 5). Finally, the proteins encoded by the two transcripts may share the property of being produced in large quantities by the cell. EF-Tu represents as much as 5% of the total cell protein in E. coli22,24,43,44. The basic protein encoded in the tyrT transcript, if actually a DN A-binding protein as suggested by its resemblance to the basic protamines61, might also be expected to be relatively abundant.
Co-transcription of tRNA and mRNA has also been observed in other systems. In mitochondria, where transcription initiates at a unique site, almost every protein coding sequence is directly flanked by a tRNA gene45–47. Although the co-transcript seems to be very unstable, this arrangement may provide a requisite economy of space in the genome and/or conveniently placed processing signals in the nascent RNA45,47. Another system is bacteriophage T4, where a cluster of eight tRNAs is transcribed from the ipI promoter located 1,000 bases upstream40,48.
Physiological implications
The tRNA secondary and/or tertiary structure could stabilize adjacent mRNA by retarding the usually rapid 5′ to 3′ degradation of message49. Detection of substantial quantities of intact dual function transcript in cells wild type for processing enzymes is consistent with a stabilization role for the tRNA cluster. If mRNA stabilization is important, the results of Pedersen et al.50 imply that the tRNAs must be removed before translation. They observed that decay of protein synthetic capacity for tufA and tufB differed by less than 20 (ref. 50). In these experiments, RNA synthesis was inhibited by rifampicin or streptolygidin and it is possible that cleavage between the tRNA and tufB mRNA was also impaired. This would mask any storage form of the transcript that could be activated for translation by cleavage of the proximal tRNA. Alternatively (not mutually exclusive), co-transcription of tRNA and mRNA may be viewed as a mechanism for coordinately controlling the synthesis of key proteins with those of stable RNAs, thereby regulating the production of a particular protein with the growth requirements of the cell.
Conclusions
Analyses of the structure and transcription of the tyrU locus have revealed a new class of tRNA gene. Transfer RNA genes can be categorized by their proximity and co-transcription with either rRNA genes, other tRNA genes and/or genes encoding protein. It is tempting to speculate that the tRNA secondary and/or tertiary structure provides an additional level of post-transcriptional control for dual function transcripts. The precise role of the tRNA molecules on the co-transcript remains to be defined.
Acknowledgments
We thank E. Johnson and P. Smith for technical assistance, N. Fiil, W. McClain and J. Yates for supplying strains, and M. Nomura, J. Friesen and S. Altman for the communication of unpublished results. This work was supported by grants from the NIH (CA11208) and the American Cancer Society; J.R. was a NIH postdoctoral fellow (AI05245) and A.L. an ACS professor.
References
- 1.Ozeki H. In: RNA Polymerase, tRNA and Ribosomes; Their Genetics and Evolution. Osawa S, Ozeki H, Uchida H, Yura T, editors. Tokyo University Press; 1980. pp. 173–183. [Google Scholar]
- 2.Lindahl L, et al. J molec Biol. 1977;109:23–47. doi: 10.1016/s0022-2836(77)80044-2. [DOI] [PubMed] [Google Scholar]
- 3.Orias E, Gartner TK, Lannan JE, Betlach MJ. Bact. 1972;109:1125–1133. doi: 10.1128/jb.109.3.1125-1133.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Squires C, Konrad B, Kirschbaum J, Carbon J. Proc natn Acad Sci USA. 1973;70:438–441. doi: 10.1073/pnas.70.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi J, Landy A. Cell. 1979;16:523–534. doi: 10.1016/0092-8674(79)90027-8. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal T, Carmichael GG. A Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 7.Berman ML, Landy A. Proc natn Acad Sci USA. 1979;76:4303–4307. doi: 10.1073/pnas.76.9.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekiya T, Khorana HG. Proc natn Acad Sci USA. 1974;71:2978–2982. doi: 10.1073/pnas.71.8.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Post LE, Arfsten AE, Davis GR, Nomura M. J biol Chem. 1980;255:4653–4659. [PubMed] [Google Scholar]
- 10.Travers AA. J Bact. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alwine JC, et al. Meth Enzym. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- 12.Smith JD. Prog Nucleic Acid Res molec Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- 13.Jaskunas SR, Lindahl L, Nomura M, Burgess RR. Nature. 1975;257:458–462. doi: 10.1038/257458a0. [DOI] [PubMed] [Google Scholar]
- 14.Taylor WE, Burgess RR. Gene. 1979;6:331–365. doi: 10.1016/0378-1119(79)90073-8. [DOI] [PubMed] [Google Scholar]
- 15.Rossi J, Ross W, Egan J, Lipman DJ, Landy A. J molec Biol. 1979;128:21–47. doi: 10.1016/0022-2836(79)90307-3. [DOI] [PubMed] [Google Scholar]
- 16.An G, Friesen JD. Gene. 1980;12:33–39. doi: 10.1016/0378-1119(80)90013-x. [DOI] [PubMed] [Google Scholar]; J Bact. 1980;144:904–916. doi: 10.1128/jb.144.3.904-916.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto M, Nomura M. J Bact. 1979;137:584–594. doi: 10.1128/jb.137.1.584-594.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Meide PH, Borman TH, Van Kimmenade AMA, Van de Putte P, Bosch L. Proc natn Acad Sci USA. 1980;77:3922–3926. doi: 10.1073/pnas.77.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arai K, et al. Proc natn Acad Sci USA. 1980;77:1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Post L, Nomura M. J biol Chem. 1980;255:4660–4666. [PubMed] [Google Scholar]
- 21.Yokota T, Sugisaki H, Takanami M, Kaziro Y. Gene. 1980;12:25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- 22.Furano A. Proc natn Acad Sci USA. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyajima A, Kaziro Y. J Biochem. 1978;83:453–462. doi: 10.1093/oxfordjournals.jbchem.a131932. [DOI] [PubMed] [Google Scholar]
- 24.Pedersen S, Bloch PL, Reeh S, Neidhardt FC. Cell. 1978;14:179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 25.Furano AV, Wittel FP. J biol Chem. 1976;251:898–901. [PubMed] [Google Scholar]
- 26.Reeh S, Pedersen S, Friesen JD. Molec gen Genet. 1976;149:279–289. doi: 10.1007/BF00268529. [DOI] [PubMed] [Google Scholar]
- 27.Blumenthal RM, Lemaux PG, Neidhardt FC, Dennis PP. Molec gen Genet. 1976;149:291–296. doi: 10.1007/BF00268530. [DOI] [PubMed] [Google Scholar]
- 28.Shibuya M, Kaziro Y. J Biochem. 1979;86:403–411. doi: 10.1093/oxfordjournals.jbchem.a132539. [DOI] [PubMed] [Google Scholar]
- 29.Calva E, Burgess R. J biol Chem. 1981;255:11017–11022. [PubMed] [Google Scholar]
- 30.Rossi J, Egan T, Hudson L, Landy A. Cell. 1981;26:305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- 31.Sekiva T, Contreras R, Takeya T, Khorana G. J biol Chem. 1979;254:5802–5816. [PubMed] [Google Scholar]
- 32.Nomura M, Yates JL, Dean D, Post LE. Proc natn Acad Sci USA. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean D, Yates JL, Nomura M. Cell. 1981;24:413–419. doi: 10.1016/0092-8674(81)90331-7. [DOI] [PubMed] [Google Scholar]
- 34.Chang S, Carbon J. J biol Chem. 1975;250:5542–5555. [PubMed] [Google Scholar]
- 35.Ilgen C, Kirk LL, Carbon J. J biol Chem. 1976;251:922–929. [PubMed] [Google Scholar]
- 36.Sakano H, Shimura Y. J molec Biol. 1978;123:287–326. doi: 10.1016/0022-2836(78)90082-7. [DOI] [PubMed] [Google Scholar]
- 37.Egan J, Landy A. J biol Chem. 1978;253:3607–3622. [PubMed] [Google Scholar]
- 38.Nakajima N, Ozeki H, Shimura Y. Cell. 1981;23:239–249. doi: 10.1016/0092-8674(81)90288-9. [DOI] [PubMed] [Google Scholar]
- 39.Schedl P, Roberts J, Primakoff P. Cell. 1976;8:581–594. doi: 10.1016/0092-8674(76)90226-9. [DOI] [PubMed] [Google Scholar]
- 40.Goldfarb A, Daniel V. Nature. 1980;286:418–420. doi: 10.1038/286418a0. [DOI] [PubMed] [Google Scholar]; Nucleic Acids Res. 1980;8:4501–4516. doi: 10.1093/nar/8.19.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]; J molec Biol. 1981;146:393–412. doi: 10.1016/0022-2836(81)90039-5. [DOI] [PubMed] [Google Scholar]
- 41.Ray BK, Apirion D. Eur J Biochem. 1981;114:517–524. doi: 10.1111/j.1432-1033.1981.tb05175.x. [DOI] [PubMed] [Google Scholar]
- 42.Goodman HM, Abelson JM, Landy A, Zadrazil S, Smith JD. Eur J Biochem. 1970;13:461–483. doi: 10.1111/j.1432-1033.1970.tb00950.x. [DOI] [PubMed] [Google Scholar]
- 43.Jacobsen GR, Rosenbusch JP. Nature. 1976;261:23–26. doi: 10.1038/261023a0. [DOI] [PubMed] [Google Scholar]
- 44.Ivell R, Fasano D, Crechet JB, Parmeggiani A. Biochemistry. 1981;20:1355–1361. doi: 10.1021/bi00508a049. [DOI] [PubMed] [Google Scholar]
- 45.Battey J, Clayton D. J biol Chem. 1980;255:11599–11606. [PubMed] [Google Scholar]
- 46.Anderson S, et al. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 47.Montova J, Ojala D, Attardi G. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 48.Fukada K, Gossens L, Abelson J. J molec Biol. 1980;137:213–234. doi: 10.1016/0022-2836(80)90326-5. [DOI] [PubMed] [Google Scholar]
- 49.Lim L, Kennell D. J molec Biol. 1979;1980;135141:369–390. 227–233. doi: 10.1016/0022-2836(79)90442-x. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen S, Reeh S, Friesen J. Molec gen Genet. 1978;166:329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- 51.Kirschbaum JB, Konrad EB. J bact. 1973;116:517–526. doi: 10.1128/jb.116.2.517-526.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxam AM, Gilbert W. Proc natn Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenberg M, Court D. A Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 54.Shine J, Dalgarno L. Proc natn Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gegenheimer P, Apirion D. Cell. 1978;15:527–539. doi: 10.1016/0092-8674(78)90021-1. [DOI] [PubMed] [Google Scholar]
- 56.McMaster GK, Carmichael GG. Proc natn Acad Sci USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rigby P, Dieckmann M, Rhodes C, Berg P. J molec Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- 58.Brosius J, Palmer ML, Kennedy PJ, Noller HF. Proc natn Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schedl P, Primakoff P. Proc natn Acad Sci USA. 1973;70:2091–2096. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JS, An G, Friesen JD, Fiil NP. Cell. 1981;25:251–258. doi: 10.1016/0092-8674(81)90250-6. [DOI] [PubMed] [Google Scholar]
- 61.Altman S, Model P, Dixon G, Wosnick M. Cell. doi: 10.1016/0092-8674(81)90198-7. (in the press) [DOI] [PubMed] [Google Scholar]


