Abstract
The most common base substitution arising from oxidative damage of DNA is a GC → AT transition. In an effort to determine the oxidized lesion(s) that gives rise to this mutation, the mutagenicity of three oxidized cytosines, 5-hydroxycytosine, 5-hydroxyuracil, and uracil glycol, were investigated in Escherichia coli. An M13 viral genome was constructed to contain a single oxidized cytosine at a specific site. Replication in vivo of the single-stranded genomes yielded mutation frequencies of 0.05%, 83%, and 80% for 5-hydroxycytosine, 5-hydroxyuracil, and uracil glycol, respectively. The predominant mutation observed was C → T. A model for C → T oxidative mutagenesis is suggested in which initial cytosine oxidation is followed by deamination to a poorly repaired uracil derivative that is strongly miscoding during replication.
Oxidative DNA damage results from exposure to both endogenous and exogenous oxidizing agents and has been implicated in a variety of degenerative disorders, including cancer and aging (1, 2). Reactive oxygen molecules, in the forms of superoxide, hydrogen peroxide, and hydroxyl radicals, are formed in vivo as byproducts of normal aerobic metabolism. Oxidizing species are also formed during the metabolism of organic carcinogens and following exposure to ionizing radiation. Although cells possess many defenses against oxidative damage, it has been estimated that the genome of a mammalian cell receives about 104–105 oxidative hits per day (3).
The most frequently occurring base substitution mutation observed in aerobic organisms is a GC → AT transition (4, 5). This substitution is also the most abundant genetic change induced as a consequence of oxidative DNA damage (6, 7). In search of the chemical precursor to these GC → AT transitions, early studies focused on 7,8-dihydro-8-oxoguanine (8-oxo-G), a frequently detected oxidized DNA lesion. Subsequent studies revealed, however, that 8-oxo-G gives rise to G → T transversions (8–10), leaving unidentified the genesis of GC → AT mutations.
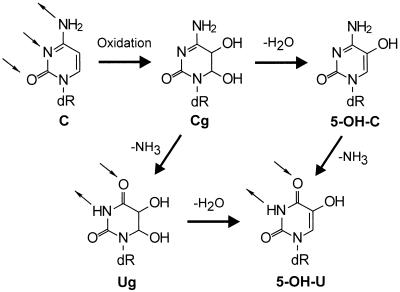
Recently, several studies have hinted that oxidized cytosines—5-hydroxycytosine (5-OH-C) and/or 5-hydroxyuracil (5-OH-U)—might contribute significantly to the GC → AT mutations observed in Escherichia coli (11–14). As shown in Fig. 1, oxidation of cytosine can give rise to 5,6-dihydroxy-5,6-dihydrocytosine (Cg), an unstable DNA lesion that can break down further to form 5-OH-C, 5-OH-U, and 5,6-dihydroxy-5,6-dihydrouracil (Ug) (15). The three lesions, 5-OH-C, 5-OH-U, and Ug, have been identified in both untreated DNA and DNA that has been treated with an oxidizing agent (15, 16).
Figure 1.
Oxidation of cytosine can result in the formation of the relatively unstable species, Cg, that can either dehydrate, deaminate, or undergo both reactions to form 5-OH-C, Ug, or 5-OH-U, respectively. (Small arrows denote hydrogen bond donors and acceptors.)
An important implication of this oxidative deamination pathway is that oxidized uracil species can form from the original cytosine. Thus, through deamination of the 4-amino group of cytosine, a cytosine-like base pairing moiety can be replaced by a thymine-like base pairing moiety. To test the in vivo consequences of 5-OH-C, 5-OH-U, and Ug, these lesions were separately inserted into viral DNA, introduced into E. coli, and the viral progeny sequenced to determine mutation frequency and specificity. Here we present definitive data showing that oxidized uracil derivatives (5-OH-U and Ug) efficiently cause GC → AT transitions in vivo. Furthermore, we demonstrate that 5-OH-C is only weakly mutagenic in E. coli. The results, in aggregate, suggest a model for oxidative mutagenesis in which initial cytosine oxidation is followed by deamination to a poorly repaired uracil derivative that is strongly miscoding during replication.
MATERIALS AND METHODS
Materials.
Restriction enzymes and bacteriophage T4 polynucleotide kinase were from New England Biolabs. Sephadex G-50 Quick Spin columns were from Boehringer Mannheim. Bacteriophage T4 DNA ligase and exonuclease III were from Pharmacia. Uracil DNA glycosylase was from GIBCO/BRL. 5-Bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) and isopropyl β-d-thiogalactopyranoside (IPTG) were from Gold Biotechnology (St. Louis). Deoxyadenosine 5′-[α-[35S]thio]triphosphate ([α35S]dATP) and Sequenase version 2.0 sequencing reagents were from Amersham. Plasmid preparation kits were from Qiagen (Chatsworth, CA). Bacteriophage M13mp7L2 was from C. W. Lawrence (17). The cell line used for transfection was DL7 (AB1157; (lac) ΔU169) (18), and for plating was GW5100 (JM103, P1−) from Graham Walker (Massachusetts Institute of Technology).
Synthesis of an Oligonucleotide Containing a Single 5-OH-C, 5-OH-U, or Ug Lesion.
Oligonucleotides 5′-CAXGCAG-3′ (where X is 5-OH-C, 5-OH-U, or Ug) were prepared as described (19). Oligonucleotides were characterized by digestion with nuclease P1 and calf intestinal phosphatase, and the corresponding nucleosides were analyzed by high-pressure liquid chromatography (HPLC). Molecular weights of the oligonucleotides were confirmed by electrospray mass spectrometry. The integrity of the oxidized cytosines in the ligated genome was confirmed by subjecting the 7-mer oligonucleotides containing either 5-OH-C, 5-OH-U, or Ug to the same conditions employed for genome construction. No degradation of the modified oligonucleotides was detected as measured by reversed-phase HPLC (data not shown).
Construction of a Single-Stranded (ss) M13 Genome Containing a Single 5-OH-C, 5-OH-U, or Ug.
5-OH-C- and 5-OH-U-containing oligonucleotides were synthesized from the corresponding phosphoramidites (19) and then purified by 23% polyacrylamide gel electrophoresis. M13mp7L2 ss DNA (1.05 pmol) was linearized by restriction enzyme digestion with EcoRI (31.5 units). To the linear ss DNA was annealed a 47-mer oligonucleotide scaffold (2.1 pmol) complementary to 20 bases on both the 5′ and 3′ ends of the linear DNA. The resulting circular M13 molecule (≈50% yield) contained a seven base gap that was complementary to the sequence 5′-CACGCAG-3′. Into the seven base gap was inserted an equimolar amount of a 5′-phosphorylated 7-mer oligonucleotide of the sequence 5′-CAXGCAG-3′, where X = oxidized cytosine. The oligonucleotide scaffold was removed by heating at 100°C for 2 min in the presence of a 2-fold molar excess of the 47-mer scaffold’s complement, resulting in the construction of a uniquely modified ss circular genome.
The Ug-containing oligonucleotide was prepared by oxidation of the corresponding uracil-containing oligonucleotide with OsO4 followed by HPLC purification (20). Because of the lability of the Ug lesion, a modified genome construction scheme was employed that involved the use of a uracilated 47-mer scaffold, and the concerted efforts of uracil DNA glycosylase (UDG) (5 units/pmol DNA) and exonuclease III (47 units/pmol DNA) for scaffold removal (21). (Ug was previously shown not to be a substrate for UDG.)
Singly modified genomes were constructed and characterized by restriction enzyme digestion to assess the amount of oligonucleotide fully ligated in the genome (21). The efficiencies of genome construction for cytosine, 5-OH-C, 5-OH-U, and Ug were determined to be 85, 80, 44, and 32%, respectively (data not shown).
The genomes were constructed such that the lesion was situated within a unique ApaLI restriction site. As described below, placement of the adduct within a restriction site provided a strategy for enrichment of mutant DNA.
Transformation of E. coli with Bacteriophage M13 Genomes.
Immediately after preparation, genomes were centrifuged for 4 min at 2,000 × g at 4°C through a Sephadex G-50 Quick Spin column preequilibrated in H2O. A 30 μl portion of each genome preparation mixture was examined by gel electrophoresis, and the presence of completely ligated genome was confirmed by comigration with ss circular DNA.
E. coli DL7 cells were prepared for transformation by electroporation as described (8, 22). A 190 μl portion of cell suspension was added to 5–10 μl of the DNA solution. The mixture was transferred to a cold Bio-Rad gene pulser cuvette (0.2 cm) and electroporations were performed with a BTX electro cell manipulator 600 system (San Diego) set at 50 mF and 129 Ω. The electroporation field strength optimal for cell survival and transformation efficiency was 12.5 kV/cm. Immediately after electroporation, 1 ml of SOC medium (23) was added, and a portion of the bacterial suspension was plated in the presence of GW5100 cells, X-Gal, and IPTG. The number of infective centers per transfection was determined by plaque counting. The mixture was incubated further for 2 hr at 37°C, after which the progeny phage were isolated from the supernatant.
Mutant Enrichment Protocol.
E. coli cells were transformed as described above with 5-OH-C, 5-OH-U, Ug, and control genomes. DNA used in the above transformations gave rise to dark-blue and colorless plaques. Dark-blue plaques resulted from restoration of the M13mp7L2 lacZ reading frame (which is out of frame by +2) upon ligation of the 7-mer oligonucleotide. The sequence context of the system was such that both wild-type and mutant progeny exhibited a dark-blue phenotype. As determined by sequence analysis, the colorless plaques resulted from undigested parental M13mp7L2 DNA.
For each lesion, the progeny phage were pooled from a total of 50,000 independent transformational events generated in at least 9 transformations. The pooled progeny phage were used to produce replicative form (RF) DNA by using the Qiagen midiplasmid purification system. The pool was enriched for mutants by digesting the RF progeny DNA with the restriction enzyme, ApaLI. As mentioned above, the lesion was situated within a unique ApaLI restriction sequence. If replication past the lesion did not result in mutation, the ApaLI restriction sequence would remain intact and, upon digestion with the enzyme, the DNA molecule would be linearized and biologically inactivated. However, if a mutation did occur, the DNA molecule would be refractory to cleavage by ApaLI, and hence retain its ability to replicate in E. coli and form a plaque.
Determination of Mutation Frequency.
Equimolar amounts of RF DNA (250 ng) were treated either in the presence of ApaLI (20 units/μl) or without the enzyme at 37°C for 4 hr in the appropriate buffer. The two fractions, labeled ApaLI+ and ApaLI−, respectively, were diluted to 100 μl and 10 μl (12 ng of DNA) were used to transform 190 μl of E. coli DL7 cells by electroporation as described above.
The mutation frequency was defined as the ratio of mutant to wild-type plus mutant progeny. The ratio of infective center plaques produced from the ApaLI+ fraction to the ApaLI− fraction gave the percentage not digested by ApaLI, the first approximation of mutation frequency. Individual plaques from the ApaLI+ fraction were sequenced to determine the mutational specificity of each lesion. Because the ApaLI+ fraction included some wild-type DNA that had evaded digestion (as determined by sequence analysis), the true mutation frequency was determined by multiplying the percentage not digested by ApaLI by the ratio of mutants sequenced.
RESULTS
The purpose of this study was to evaluate the mutagenic potency and specificity of the lesions 5-OH-C, 5-OH-U, and Ug by using a ss DNA system that served as a model for replication past an oxidized cytosine lesion.
Construction and Transfection into E. coli of a ss M13 Genome Containing a Single 5-OH-C, 5-OH-U, or Ug.
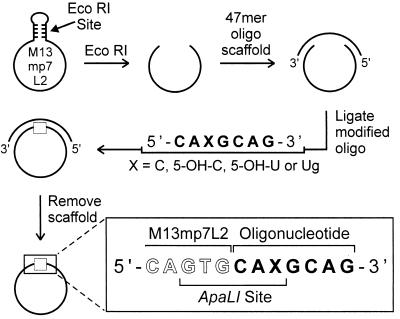
The viral genomes for this study were constructed from M13mp7L2 ss DNA as shown in Fig. 2 (21, 24). 5-OH-C- and 5-OH-U-containing oligonucleotides were constructed from chemically synthesized 5-OH-C and 5-OH-U phosphoramidites (19), and the Ug oligonucleotide was synthesized by selective OsO4 oxidation of the corresponding uracil-containing oligonucleotide followed by HPLC purification (20). In each case, the modified base was situated within a unique ApaLI recognition site.
Figure 2.
Construction of singly modified genomes as described. Briefly, a singly modified heptamer was ligated into the lacZ coding region of an M13mp7L2 bacteriophage genome. The three bases on the 3′ end of the M13 molecule combined with the three bases on the 5′ end of the oligonucleotide generate a unique recognition site for the restriction enzyme ApaLI, 5′-GTGCAC-3′.
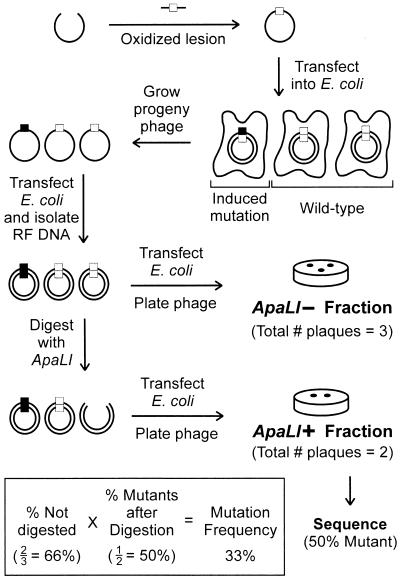
As shown in Fig. 3, the ligated genomes containing an oxidized lesion were transfected into wild-type E. coli cells by electroporation (25). Mutant enrichment was accomplished by taking advantage of the original placement of the adduct in a unique ApaLI recognition site; induced mutants were insensitive to cleavage by this enzyme, whereas wild-type progeny were linearized and rendered nonviable. Individual plaques not digested by ApaLI were sequenced to determine the types of mutations induced by each lesion. The mutation frequency of each lesion was calculated by multiplying the percentage of DNA not digested by ApaLI by the percentage of mutants in the undigested population (see equation in Fig. 3).
Figure 3.
Experimental design to evaluate the mutation frequency of 5-OH-C, 5-OH-U, and Ug. Mutation frequency was determined in two steps. First, a preliminary mutation frequency was determined by measuring the ratio of phage produced from RF DNA that was either treated with or without ApaLI. That ratio, or preliminary mutation frequency, was next adjusted by multiplying it by the fraction of phage in the ApaLI-resistant population that was verified to be mutant by DNA sequencing. This adjustment is necessary because some wild-type RF DNA escapes ApaLI selection. In the example shown, 66% of the progeny were in the pool that was resistant to digestion by ApaLI. Of these, one-half were true mutants as determined by sequencing. Thus, as shown, the mutation frequency was (66%)(50%) = 33%.
Mutation Frequency and Specificity of 5-OH-C, 5-OH-U, and Ug.
5-OH-C-, 5-OH-U-, and Ug-containing genomes were replicated within E. coli. As summarized in Table 1, the mutation frequencies of 5-OH-C, 5-OH-U, and Ug were determined in parallel with an unmodified cytosine. The mutation frequency of 5-OH-C was found to be quite modest (0.05%), although greater than the background frequency of the control (0.003%). The predominant mutation observed was a C → T transition, although a few C → G transversions were also detected.
Table 1.
Mutation frequencies and specificities of 5-OH-C, 5-OH-U, and Ug
| Lesion | Not digested by ApaLI, % | Mutants in ApaLI+, % | Lesion mutation frequency, % | Mutation specificity (no. of mutants) |
|---|---|---|---|---|
| Cytosine | 0.08 | 4 (3/79) | 0.003 | C → T (3) |
| 5-OH-C | 0.09 | 57 (79/139) | 0.05 | C → T (77) |
| C → G (2) | ||||
| 5-OH-U | 83 | 100 (95/95) | 83 | C → T (95) |
| Ug | 80 | 100 (92/92) | 80 | C → T (92) |
Data represent a total of three independent genome constructions and a total of nine transfections per lesion. The progeny phage from these transfections were combined. ApaLI digest data for cytosine, 5-OH-C, and 5-OH-U represent one experiment and for Ug represent three pooled digests. Sequence analysis was performed once.
In contrast to the low mutation frequency of 5-OH-C, the mutagenicities of 5-OH-U and Ug were strikingly high. The mutation frequencies of 5-OH-U and Ug were determined to be 83% and 80%, respectively, with 100% of the sequenced mutations derived from these two lesions being C → T transitions.
DISCUSSION
5-OH-U and Ug are oxidative deamination products of cytosine, and the extremely high mutation frequencies observed may result from deamination of the 4-amino group of the oxidized cytosine, altering the base pairing preference of the pyrimidine from guanine to adenine (see Fig. 1). Because 5-OH-U and Ug are present in oxidatively damaged DNA (16), the 83% and 80% mutation frequencies observed for these lesions implicate 5-OH-U and Ug as likely sources of the frequently observed C → T transitions in the mutational spectrum of oxidatively damaged DNA.
A modest mutation frequency (one order of magnitude above background) was observed for 5-OH-C. The predominant mutation observed for this lesion was a C → T transition, and several C → G transversions were also observed. The data presented here for 5-OH-C are in agreement with in vitro polymerase bypass and dNTP incorporation studies demonstrating that this lesion is processed without significant error by the Klenow fragment of DNA polymerase I (12, 13).
Although one previous study suggested that 5-OH-C was significantly mutagenic in E. coli (2.5% mutation frequency), the technique used to generate 5-OH-C in that study also allowed for the generation of other oxidized products (14). In that experiment, dCTP was oxidized with Fe2+ and H2O2, and the 5-OH-dCTP peak was isolated by HPLC. The putative cytosine oxidation product was incorporated into a viral genome by the low fidelity HIV reverse transcriptase. To cause a C → T mutation, the oxidized species had to be incorporated opposite template guanines. Given the high mutagenicity of the 5-OH-U and Ug discovered here, it is possible that interpretation of the results of the earlier study could be skewed considerably if only a small amount of a highly mutagenic oxidized uridine or readily deaminated oxidized cytosine species was present after HPLC purification. Because of the low fidelity of HIV reverse transcriptase, the possibility that an oxidized uracil or readily deaminated cytosine was incorporated opposite template guanines cannot be excluded.
The work presented here employed a direct chemical synthesis to generate 5-OH-C, thus minimizing contamination by other oxidized cytosine derivatives (19). Because the levels of 5-OH-C, 5-OH-U, and Ug in DNA are comparable, the biological significance of 5-OH-C relative to 5-OH-U and Ug may be minimal when one considers that the mutation frequency of 5-OH-C is three orders of magnitude smaller than that of 5-OH-U and Ug (Table 1). However, the contribution of these lesions to the overall observed spectra of C → T mutations cannot be assessed until further experiments addressing the rates of formation of these lesions are performed. More specifically, it would be of interest to examine the rates of cytosine glycol deamination and/or dehydration that lead to the formation of 5-OH-C, 5-OH-U, and Ug in cellular DNA. In addition, the relative contribution of these lesions compared with that of uracil arising from deamination of cytosine should be addressed.
The background mutation level of the experiment (0.003%) was defined by the observed mutation frequency of the control genome containing unmodified cytosine. A spontaneous mutation frequency of 0.003% corresponds to 3 mutational events per 100,000 bases or 3 × 10−5. In previous studies (26), the spontaneous mutation frequency of the lacI gene of E. coli was determined to be 2 × 10−6. The handling of the unmodified oligonucleotide (heating, air exposure) and the genetic engineering techniques used for genome construction could result in the slight elevation of the spontaneous mutation frequency as observed in this study. Although only 3 spontaneous mutants were observed, it is interesting to note that all were C → T transitions, identical to those observed for the oxidized cytosines, underscoring the role that these lesions may play in spontaneous DNA mutation.
Although the cells used in this study were fully repair proficient (wild type), the observed mutation frequencies for 5-OH-U and Ug clearly reflect the absence of significant cellular repair of these lesions in this system. This lack of repair possibly stems from the fact that the lesions are only briefly part of a double-stranded DNA template, thus circumventing the cell’s double-stranded DNA repair system. If true, the system presented here provides an in vivo mutation frequency that closely represents the inherent miscoding properties of oxidized cytosines. Thus, the data imply that if 5-OH-U and Ug are not repaired then these lesions are nearly 100% mutagenic. Precedent for such high inherent miscoding comes from the work of Moriya et al. (27), which showed 100% mutation frequency for 1,N2-(1,3-propano)-2′-deoxyguanosine lesions in ss vectors in vivo (27). The above line of reasoning (assuming that in this particular cellular system the lesions in question are not repaired) further implies that the miscoding potential of 5-OH-C is inherently low. It cannot be discounted, of course, that the 5-OH-C lesions in this study might have been repaired by proteins acting on ss DNA.
Given the apparent mutagenicity of 5-OH-U and Ug when not repaired in a wild-type E. coli host, and given the frequency of these oxidized, deaminated cytosines detected in DNA, it seems likely that naturally occurring 5-OH-U and Ug lesions in E. coli are repaired at the level of double-stranded DNA. It is anticipated that ongoing in vivo research will shed light on the ability of E. coli to repair double-stranded DNA containing 5-OH-U and Ug lesions, eventually identifying and isolating any specific bacterial repair proteins. E. coli proteins that have been implicated by in vitro studies in the removal of oxidized cytosine and uracil lesions from DNA include uracil DNA glycosylase (5-OH-U) (28), formamidopyrimidine DNA glycosylase (5-OH-C and 5-OH-U) (29), endonuclease III (5-OH-C, 5-OH-U, and Ug) (20, 29–31), and endonuclease VIII (5-OH-C and 5-OH-U) (32).
The adverse role that oxidized, deaminated cytosines may play in mammalian cells remains to be investigated. 5-OH-U and Ug have both been detected in mammalian tissues and in human cells at levels comparable to that of 8-oxo-G (16). For example, the levels of 5-OH-U, Ug, and 8-oxo-G in human leukocytes have been determined to be 7, 20, and 12 adducts per 107 bases, respectively. If mammalian polymerases misincorporate bases opposite 5-OH-U and Ug with similar frequency to that of E. coli polymerases, these lesions pose a serious threat to genomic integrity. Proteins that may be involved in diminishing the cellular effects of oxidized cytosines in mammalian cells include functional homologues of endonuclease III and TDG, a mismatch repair protein that specifically removes T and uracil from G/T and G/U mismatches (33–35).
Although the correlation between oxidative DNA damage and human disease is well established, the relationship between specific oxidative lesions and mutation has only recently begun to unfold. To understand this process fully, the exact types and biological significance of oxidative lesions formed in DNA must be determined. This work suggests that 5-OH-U and Ug lesions may be the main cause of cellular C → T transitions, and should help to focus attention on the possible mutagenic role of oxidized, deaminated cytosines in mammalian systems.
Acknowledgments
We thank A. Lee for technical assistance; E. A. Bailey, D. Mathur, M. Morningstar, and D. Wang for experimental suggestions and helpful discussions; and J. Barry, P. Vourous, and J. S. Wishnock for mass spectrometry characterization of the modified oligonucleotides. We are grateful to E. A. Wintner and D. Wang for critical readings of the manuscript. This work was supported in part by National Cancer Institute Grant CA52127 and by National Institute for Environmental Health Sciences Grant NIEHS07020.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: 8-oxo-G, 7,8-dihydro-8-oxoguanine; 5-OH-C, 5-hydroxycytosine; 5-OH-U, 5-hydroxyuracil; Cg, 5,6-dihydroxy-5,6-dihydrocytosine; Ug, uracil glycol; HPLC, high-pressure liquid chromatography; ss, single-stranded; RF, replicative form.
References
- 1.Mitra G, Pauly G T, Kumar R, Pei G K, Hughes S H, Moschel R C, Barbacid M. Proc Natl Acad Sci USA. 1989;86:8650–8654. doi: 10.1073/pnas.86.22.8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goth R, Rajewsky M F. Proc Natl Acad Sci USA. 1974;71:639–643. doi: 10.1073/pnas.71.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaaper R M, Danforth B N, Glickman B W. J Mol Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- 5.Schaaper R M, Dunn R L. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tkeshelashvili L K, McBride T, Spence K, Loeb L A. J Biol Chem. 1991;266:6401–6406. [PubMed] [Google Scholar]
- 7.Moraes E C, Keyse S M, Tyrrell R M. Carcinogenesis. 1990;11:283–293. doi: 10.1093/carcin/11.2.283. [DOI] [PubMed] [Google Scholar]
- 8.Wood M L, Dizdaroglu M, Gajewski E, Essigmann J M. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 9.Shibutani S, Takeshita M, Grollman A P. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 10.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 11.Roy-Burman S, Roy-Burman P, Visser D W. Biochem Pharmacol. 1970;19:2745–2756. doi: 10.1016/0006-2952(70)90101-2. [DOI] [PubMed] [Google Scholar]
- 12.Purmal A A, Kow Y W, Wallace S S. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purmal A A, Kow Y W, Wallace S S. Nucleic Acids Res. 1994;22:3930–3935. doi: 10.1093/nar/22.19.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feig D I, Sowers L C, Loeb L A. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dizdaroglu M, Holwitt E, Hagan M P, Blakely W F. Biochem J. 1986;235:531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner J R, Hu C C, Ames B N. Proc Natl Acad Sci USA. 1992;89:3380–3384. doi: 10.1073/pnas.89.8.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee S K, Borden A, Christensen R B, LeClerc J E, Lawrence C W. J Bacteriol. 1990;172:2105–2112. doi: 10.1128/jb.172.4.2105-2112.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasko D D, Harvey S C, Malaikal S B, Kadlubar F F, Essigmann J M. J Biol Chem. 1988;263:15429–15435. [PubMed] [Google Scholar]
- 19.Morningstar M L, Kreutzer D A, Essigmann J M. Chem Res Toxicol. 1997;10:1345–1350. doi: 10.1021/tx970052a. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Essigmann J M. Biochemistry. 1997;36:8628–8633. doi: 10.1021/bi970341y. [DOI] [PubMed] [Google Scholar]
- 21.Bailey E A, Iyer R S, Harris T M, Essigmann J M. Nucleic Acids Res. 1996;24:2821–2828. doi: 10.1093/nar/24.14.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey E A, Iyer R S, Stone M P, Harris T M, Essigmann J M. Proc Natl Acad Sci USA. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. DNA Cloning: A Practical Approach. Oxford, U.K.: IRL; 1985. pp. 109–135. [Google Scholar]
- 24.LeClerc J E, Borden A, Lawrence C W. Proc Natl Acad Sci USA. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messing J. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 26.Coulondre C, Miller J H. J Mol Biol. 1977;117:525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- 27.Moriya M, Zhang W, Johnson F, Grollman A P. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaruga P, Dizdaroglu M. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 30.Dizdaroglu M, Laval J, Boiteux S. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 31.Wagner J R, Blount B C, Weinfeld M. Anal Biochem. 1996;233:76–86. doi: 10.1006/abio.1996.0010. [DOI] [PubMed] [Google Scholar]
- 32.Melamede R J, Hatahet Z, Kow Y W, Ide H, Wallace S S. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 33.Wiebauer K, Jiricny J. Nature (London) 1989;339:234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- 34.Neddermann P, Jiricny J. Proc Natl Acad Sci USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallinari P, Jiricny J. Nature (London) 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]