Abstract
DNA triple helices offer new perspectives toward oligonucleotide-directed gene regulation. However, the poor stability of some of these structures might limit their use under physiological conditions. Specific ligands can intercalate into DNA triple helices and stabilize them. Molecular modeling and thermal denaturation experiments suggest that benzo[f]pyrido[3,4-b]quinoxaline derivatives intercalate into triple helices by stacking preferentially with the Hoogsteen-paired bases. Based on this model, it was predicted that a benzo[f]quino[3,4-b]quinoxaline derivative, which possesses an additional aromatic ring, could engage additional stacking interactions with the pyrimidine strand of the Watson–Crick double helix upon binding of this pentacyclic ligand to a triplex structure. This compound was synthesized. Thermal denaturation experiments and inhibition of restriction enzyme cleavage show that this new compound can indeed stabilize triple helices with great efficiency and specificity and/or induce triple helix formation under physiological conditions.
Triple helix formation recently has been the focus of considerable interest because of possible applications in developing new molecular biology tools as well as therapeutic agents (1), and because of the possible relevance of H-DNA structures in biological systems (2). In intermolecular structures, an oligopyrimidine⋅oligopurine sequence of DNA duplex is bound by a third-strand oligonucleotide in the major groove. Two main types of triple helices have been described, depending on the orientation of the third strand (3). The first reported triple-helical complexes involved pyrimidic third strands whose binding rests on Hoogsteen hydrogen bonds between a T⋅A base pair and thymine, and between a C⋅G base pair and protonated cytosine (4, 5). The (T,C)-containing oligonucleotide binds parallel to the oligopurine strand in the so-called pyrimidine motif. A second category of triple helices contains purines in the third strand, which is oriented antiparallel to the oligopurine strand. C⋅G×G and T⋅A×A base triplets are formed after a reverse Hoogsteen hydrogen bonding scheme (6, 7). Oligonucleotides containing T and G can also form triple helices whose orientation depends on base sequence (see ref. 8 and references therein).
The binding of a third-strand oligonucleotide to a target DNA duplex generally involves a thermodynamically weaker interaction than duplex formation itself (9, 10). Exceptions concern binding of some G-rich oligonucleotides in the purine motif (11). The requirement for protonation of cytosines in the pyrimidine motif makes these triple helices marginally stable at physiological pH. This has led to the design of chemical modifications that could increase the affinity of the third strand for its target. These modifications include the use of nonnatural bases (see ref. 3 for a review) and N3′ → P5′ phosphoramidate backbone (12), as well as oligonucleotide-intercalator conjugates (13).
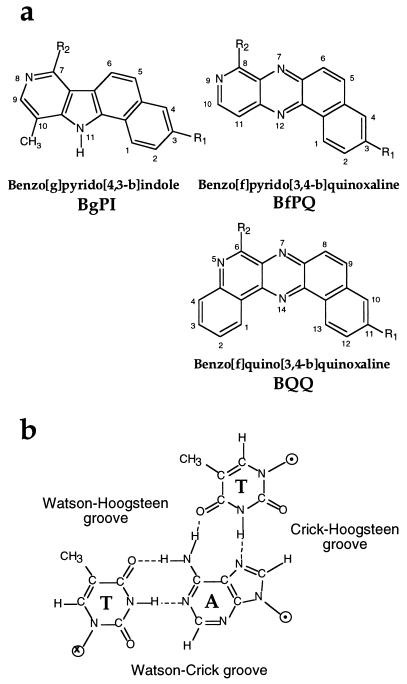
An alternative approach involves using nucleic acid ligands that selectively stabilize triple helices. For example, ethidium bromide has been shown to bind and stabilize a triple helix made of poly(dT)⋅poly(dA) × poly(dT), which contains only T⋅A×T triplets (14). However, this compound poorly stabilizes (or even destabilizes) the triple helices containing both T⋅A×T and C⋅G×C+ base triplets, probably as a result of electrostatic repulsion (15). Benzopyridoindole derivatives (BPI, Fig. 1a) were the first molecules reported to strongly stabilize this latter type of triple helices even though they have a preference for T⋅A×T stretches (16). Several other intercalators (17–20) as well as various DNA minor groove ligands (21–23) have also been shown to bind to DNA triple helices. Intercalators usually stabilize to a greater extent triple helices containing T⋅A×T triplets, whereas minor groove binders usually destabilize triplexes, except in a particular case where the triple helix involved an RNA strand (24). Because no structural data are available on triple helix–ligand complexes, not much is known about the interactions that direct specific intercalation into triple helices. BPI derivatives have been shown to intercalate between T⋅A×T base triplets by excitation fluorescence energy transfer from base triplets to ligands (16, 25) and by linear and circular dichroism (26). By comparing the effects of various substituents of BPI on triple helix stabilization, molecular modeling has proposed two models of intercalation for BePI and BgPI molecules in triple helices (27).
Figure 1.
(a) Structure of different triplex-specific ligands shown to intercalate into triple helices. In the present study, R1 = OCH3 and R2 = NH(CH2)3NH2. (b) Structure of a T⋅A×T base triplet, with the names given to the different grooves.
Benzo[f] pyridoquinoxalines (BfPQ, Fig. 1a) were synthesized as analogues of BPI derivatives, in which the indole nucleus is replaced by a quinoxaline heterocycle (28). These molecules have been shown to stabilize significantly triple helices whereas they bind poorly to double helices. In the present paper, we used experimental data and molecular modeling to propose a model for intercalation of BPQ molecules in triple helices whereby stacking of the four-aromatic-ring system involves nearly exclusively the Hoogsteen base pair. Based on this model, it is predicted that better ligands could be obtained by increasing stacking interactions with base triplets. A new family of ligands with a pentacyclic aromatic ring system was synthesized to optimize stacking interactions with the Watson strand of triple helices (i.e., the pyrimidine strand of the underlying double helix). The triple helix-binding properties of benzo[f]quino[3,4-b]quinoxaline derivatives (BQQ, Fig. 1a), which fulfill these expectations, are described in this paper.
MATERIALS AND METHODS
Molecular Modeling.
A DNA triplex structure was constructed by molecular modeling techniques by using coordinates that correctly take into account the sugar conformation of (T,C)-motif triple helices (29). This structure is closer to a B-form DNA as reported by NMR studies (30, 31) than the structure previously proposed by Arnott et al. (32) based on fiber x-ray diffraction. The jumna program allows us to construct DNA structures according to their helical parameters (33). An intercalation site can be easily created in the triplex by doubling the rise parameter for two adjacent T⋅A×T base triplets (rise = 6.8 Å), and subsequently decreasing the twist parameter between these two triplets from 34 to 16° to reduce bond distance constraints. Molecular structures for BfPQ and BQQ (see Fig. 1a) were constructed by using the builder module of the insight ii package (Molecular Simulations) and minimized by using the discover module. BfPQ and BQQ derivatives bearing an identical side chain (R2 = NH(CH2)3NH2) were docked into the triplex intercalation site. Then, energy minimization was performed to generate conformations of lowest energy. Energies were also calculated for the free ligand molecules and the triplex structure in the absence of ligand. Interaction energies were then estimated from these separate calculations. Solvent and counter-ions were not explicitly included in these calculations. Instead, a sigmoidal distance-dependent dielectric constant was used, and each phosphate group was assigned half an electronic charge.
Thermal Denaturation Experiments.
Triple helix stability was measured by using UV absorption spectroscopy. All thermal denaturation studies were carried out on a Uvikon 940 spectrophotometer, interfaced to an IBM-AT personal computer for data collection and analysis. Temperature control of the cell holder was achieved by a Haake D8 circulating water bath. The temperature of the water bath was decreased from 80 to 0°C and then increased back to 80°C at a rate of 0.1°C/min with a Haake PG 20 thermoprogrammer, and the absorbance at 260 nm was recorded every 10 min.
Inhibition of DraI Restriction Enzyme Cleavage by Oligonucleotides.
A plasmid (pLTR-HIV) containing about half of the HIV proviral genome was used as a substrate for the restriction enzyme DraI. The DraI digestion of 0.5 μg pLTR-HIV was performed at 37°C during 10 min with 10 units of DraI in a pH 7.5 buffer containing 10 mM Tris⋅HCl, 10 mM MgCl2, 50 mM NaCl, and 1 mM DTT. The products of the reaction were loaded on an ethidium bromide-stained agarose gel (0.8%).
RESULTS
Design of Benzo[f]quino[3,4-b]quinoxaline Ligands.
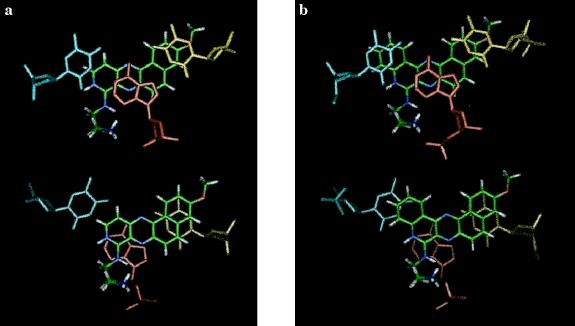
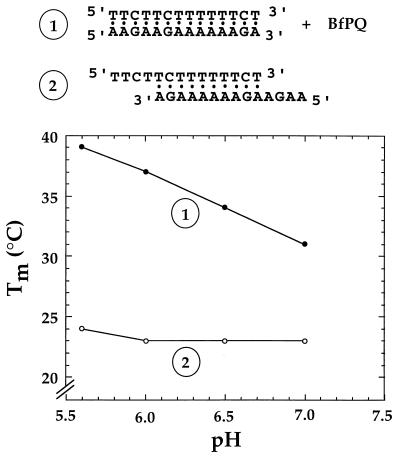
A molecular modeling method was used previously to propose a model for the interaction of BPI derivatives with triplex structures. This model was in agreement with experimental data obtained with BPI derivatives bearing different aminoalkylamine side chains (27). This combined computational and experimental method was used for the related BfPQ derivatives. BfPQ derivatives can be inserted readily into a triplex intercalation site. Their tetracyclic aromatic ring system provides reasonably good stacking interactions with base triplets. The aminoalkyl side chain (R2 in Fig. 1a) could be located either in the Watson–Crick groove or in the Watson–Hoogsteen groove of the triple helix (see Fig. 1b). In contrast to previous exhaustive studies of BPI derivatives, only a limited number of BPQ derivatives were synthesized and investigated. However, a study of the stabilization afforded by the available compounds indicated that the aminoalkyl side chain plays a similar role for BPQ and BPI analogues. Therefore, we propose that, as for their benzo[g]pyridoindole analogues (27) (see Fig. 1a), the aminoalkyl side chain at position 8 of BfPQ (R2 in Fig. 1a) is located in the Watson–Crick groove of triple helices (which corresponds to the minor groove of the target DNA duplex). The energy-minimized model shows that stacking interactions are rather weak with the pyrimidine strand of the Watson–Crick double helix (Watson strand) but strong with Hoogsteen base pairs (Fig. 2a). This preferred stacking with the Hoogsteen base pairs suggested that BfPQ could induce the formation of a Hoogsteen-paired parallel duplex in the absence of the Watson strand, as previously observed with BPI derivatives (34). Fig. 3 shows the melting temperature of the mixture of 5′-AAGAAGAAAAAAGA-3′ and 5′-TTCTTCTTTTTTCT-3′ as a function of pH in the absence and presence of BfPQ. The pH-independent duplex formed in the absence of BfPQ corresponds to an antiparallel Watson–Crick duplex involving the formation of 10 contiguous base pairs (Fig. 3, complex 2). In contrast, in the presence of BfPQ, a pH-dependent and more stable complex is observed. This result indicates that BfPQ, as BPI derivatives, is able to induce parallel Hoogsteen base pairing between these two oligonucleotides (Fig. 3, complex 1). Complete Hoogsteen base pairing requires protonation of cytosines in the pyrimidine sequence, hence the pH-dependent stabilization of the Hoogsteen duplex. Therefore, the experimental data support the model derived from molecular mechanics calculations, in which BfPQ interacts mainly with the Crick–Hoogsteen base pairs of the triple helix (Fig. 2a).
Figure 2.
Energy-minimized models of BfPQ (a) or BQQ (b) intercalated in a triple helix made of Hoogsteen T⋅A×T base triplets. The planar ring system is shown stacked with the T⋅A×T base triplet on its 5′ side (with respect to the purine strand) (Upper) and on its 3′ side (Lower). The Watson strand (pyrimidine strand) is shown in blue, the Crick strand (purine strand) is in red, and the Hoogsteen strand (third strand) is in yellow. The intercalated molecule is colored using standard colors for each atom (C in green, H in white, N in blue, and O in red).
Figure 3.
Melting temperature as a function of pH for the mixture of oligonucleotides (1 μM each), which can form either a 14-mer parallel Hoogsteen duplex in the presence of 15 μM BfPQ (1), or a 10-bp antiparallel Watson–Crick duplex in the absence of BfPQ (2). Experiments were carried out in a cacodylate buffer (10 mM) containing 0.1 M NaCl.
On the basis of the structure of the intercalated complex of BfPQ, we reasoned that it should be possible to further enhance the binding of triplex-specific ligands by increasing stacking interactions with the Watson strand. This could be achieved by adding an additional ring adjacent to the pyridine ring of BfPQ. An energy-minimized model of BQQ derivatives (Fig. 2b) showed that these molecules could engage better stacking interactions with both the base triplets adjacent to the intercalation site (including bases in the Watson strand) than the parental BfPQ molecules. The synthesis of BQQ derivatives will be described elsewhere. The BQQ derivative of Fig. 1a with R1 = OCH3 and R2 = NH(CH2)3NH2 was compared with the BfPQ derivative with the same substituents (28) for its ability to stabilize triple helices.
Stabilization of a (T,C)-Motif Triple Helix by BQQ.
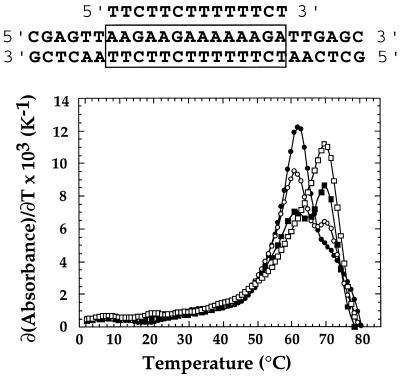
The ability of the newly synthesized BQQ compound to bind triple-helical DNA was characterized and compared with that of the related compounds BgPI and BfPQ by means of thermal denaturation experiments. Stabilization of the triple helix (Table 1) as well as that of the target duplex was studied by UV absorption spectroscopy. In this triple-helical system, a 14-mer oligonucleotide binds to a 26-bp DNA target. This sequence was already used to study the interactions of benzopyridoindole derivatives (16, 27). The triplex was formed by first mixing both strands of the duplex at 1.5 μM concentration of each strand and then adding 1.8 μM of the third strand in a pH 6.2 cacodylate buffer (10 mM) containing 0.1 M NaCl. Usually, a typical melting curve shows two transitions: the one observed at a lower temperature corresponds to the triplex-to-duplex transition (Tm3→2), whereas the other one at higher temperature is from the duplex-to-single strands transition (Tm2→1). As previously described, the triplex-to-duplex transition was not reversible, because of the slow kinetics of triplex formation, in the absence of any ligand (9). The apparent half-dissociation temperature obtained upon increasing temperature at a rate of 0.1°C/min was 18°C. With this rate of heating and cooling the width (δTm) of the hysteresis curve was 4°C. Triple helix-specific ligands were shown to stabilize this triple helix and abolish the hysteresis phenomenon (16, 27). The relative stabilization induced by various ligands was estimated from the difference between the melting temperatures in the presence of 10 μM of the compound and that of the triplex alone (ΔTm, Table 1). The half-dissociation temperature corresponding to the triplex-to-duplex transition was increased from 18 to 46°C (ΔTm3→2 = 28°C) for BgPI and 47°C (ΔTm3→2 = 29°C) for BfPQ, whereas the Tm of the duplex-to-single-strands transition was also increased from 58 to 74°C (ΔTm2→1 = 16°C) for BgPI and 60°C (ΔTm2→1 = 2°C) for BfPQ. In the presence of BQQ, the melting curve exhibited an unusual monophasic profile with Tm = 69°C. The absence of any transition at a lower temperature could imply that either the triple helix did not form in the presence of BQQ or that the triplex was so stable in the presence of BQQ that it directly dissociated into single strands. Fig. 4 shows the first derivatives of the thermal denaturation profile of the target duplex in the presence of increasing concentrations of third-strand oligonucleotide in the presence of 10 μM BQQ. As the ratio of the third-strand-to-duplex increased from 0 to 1, the intensity of the peak initially observed in the presence of the duplex alone was decreased whereas a new peak emerged at a higher temperature. This experiment confirmed that the monophasic transition observed at 69°C was due to melting of the triplex into single strands. Therefore, in the presence of 10 μM BQQ, a triplex with 14 base triplets has a higher melting temperature (Tm3→2 = 69°C, ΔTm3→2 = 51°C) than the 26-bp target DNA duplex (Tm2→1 = 62°C, ΔTm2→1 = 4°C) in a pH 6.2 buffer containing 0.1 M NaCl, whereas the triplex dissociates at 18°C (40°C lower than that of the 26-bp duplex) in the absence of BQQ. It should be noted that BQQ not only is the best triplex-stabilizing ligand investigated until now but also has the highest triplex vs. duplex specificity (Δ(ΔTm) = 47°C).
Table 1.
Melting temperatures of the triplex and the duplex (sequences shown at the top of Fig. 4) in the absence and presence of 10 μM BgPI, BfPQ, and BQQ molecules
| Ligands | Tm, ±1°C
|
ΔTm, °C
|
||
|---|---|---|---|---|
| Triplex Tm3→2 | Duplex Tm2→1 | Triplex ΔTm3→2 | Duplex ΔTm2→1 | |
| None | 18 | 58 | ||
| BgPI | 46 | 74 | +28 | +16 |
| BfPQ | 47 | 60 | +29 | +2 |
| BQQ | 69 | 62 | +51 | +4 |
Tm3→2 is the melting temperature for the dissociation of triplex into duplex + thrid strand, except for the result obtained in the presence of BQQ, in which the triplex dissociates directly into three single strands. ΔTm indicates the difference of Tm in the presence and absence of ligands. Δ(ΔTm) shows the difference of ΔTm for the 14-mer triplex and the 26-bp duplex. Experiments were carried out in a 10 mM cacodylate buffer (pH 6.2) containing 0.1 M NaCl.
Figure 4.
First derivatives of the melting curves for the mixture of a 26-bp duplex with increasing third-strand concentrations in the presence of 10 μM BQQ (sequences are shown at the top of the figure). The duplex concentration was 1.5 μM, and the third-strand-to-duplex ratio was 0:1 (•), 0.33:1 (○), 0.66:1 (▪), and 1:1 (□). Experiments were carried out in a 10 mM cacodylate buffer (pH 6.2) containing 0.1 M NaCl.
Inhibition of Restriction Enzyme Cleavage by a Short (9-mer) Triplex-Forming Oligonucleotide.
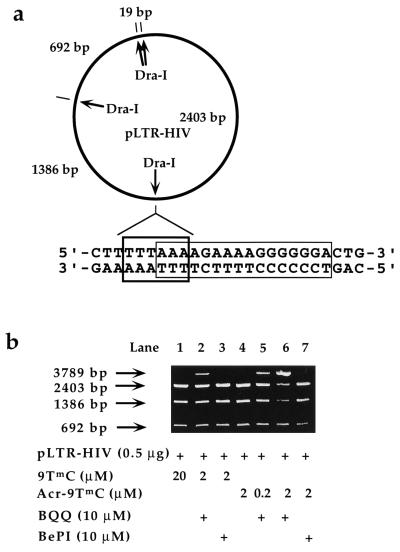
We further investigated triplex formation with the HIV 16-bp oligopyrimidine⋅oligopurine proviral DNA sequence corresponding to the polypurine tract (PPT) of HIV RNA. The restriction endonuclease DraI cleaves exactly at the junction of the triple helix site (5′-TTT↓AAA-3′) (Fig. 5), and triple helix formation inhibits DNA cleavage (35). A plasmid (pLTR-HIV) carrying the 16-bp oligopyrimidine⋅oligopurine sequence was used as a substrate for the DraI enzyme. The plasmid contains four DraI cleavage sites (Fig. 5a), and a complete digestion of the plasmid gave four fragments. One cleavage fragment was too short (19 bp) to be observed on nondenaturing agarose gels. Inhibition of cleavage at the junction of the 16-bp sequence should lead to the disappearance of the 1,386- and 2,403-bp cleavage products and to the appearance of a 3,789-bp fragment (Fig. 5b). It has been shown previously that the 16-bp PPT sequence can be targeted by a 15-mer (T,C,G)-containing oligonucleotide (5′-T4mCT4G6-3′, where mC is 5-methylcytosine) and by a 9-mer (5′-T4mCT4-3′) (12, 35). Because BQQ ligands are likely to bind to the two A4 tracts of the oligopurine sequence, we chose to assess the ability of ligand-assisted triple helix formation to interfere with the cleavage of DraI enzyme with the 9-mer (T,C)-containing oligonucleotide (9TmC) and its analog with an acridine tethered at the 5′ end (Acr-9TmC). Covalent attachment of acridine to the 5′ end of the 9-mer raised the Tm value from 8 to 26°C. The Tm3→2 of 9TmC is about 8°C at 1.5 μM concentration in the absence of ligand and under the conditions of the restriction enzyme assay (pH 7.5). This Tm3→2 value rose to 50°C in the presence of 10 μM BQQ. At 37°C, the 9-mer (9TmC) was not expected to inhibit DraI cleavage. Fig. 5b (lane 1) shows that no inhibition was observed at 20 μM concentration. Addition of 10 μM BQQ stabilized the complex sufficiently to observe partial inhibition in the presence of 2 μM of the 9-mer (Fig. 5b, lane 2) (50% inhibition was observed at about 5 μM of the 9-mer). With the acridine-conjugated 9-mer (Acr-9TmC), partial inhibition was observed with the presence of 10 μM BQQ at concentrations as low as 0.2 μM (Fig. 5b, lane 5). At 2 μM, 90% inhibition was obtained whereas no effect was observed in the absence of BQQ (Fig. 5b, lane 6). The efficiency of DraI inhibition depended on ligand concentration: the concentration of the oligonucleotide Acr-9TmC required to achieve 50% inhibition (IC50) raised from 0.4 to 2 μM when that of BQQ dropped from 10 to 2 μM (data not shown). Fig. 5b also shows that BQQ is a much more potent inducer of stable triple helix formation than BePI, which is the lead compound in this family of triplex-specific ligands, because 10 μM BePI failed to prevent DraI cleavage with 2 μM of 9TmC and Acr-9TmC (Fig. 5b, lanes 3 and 7).
Figure 5.
Inhibition of DraI restriction endonuclease cleavage by oligonucleotides in the presence of BQQ or BePI. (a) The plasmid (pLTR-HIV) contains four DraI cleavage sites that generate DNA fragments of 19, 692, 1,386, and 2,403 bp. One of these four sites corresponds to the junction of the 16-bp oligopyrimidine⋅oligopurine sequence (polypurine tract, PPT) of the HIV-1 provirus contained within the nef gene. (b) Agarose gel reveals the extent of DraI cleavage inhibition by showing the decreased 1,386- and 2,403-bp fragments and the increased 3,789-bp fragment. The intensity of the 692-bp fragment is not affected. The concentration of oligonucleotides and that of ligands are indicated (see Materials and Methods for experimental conditions).
DISCUSSION
Molecular modeling has suggested that BfPQ intercalates within a triple helix by stacking nearly exclusively with the Hoogsteen base pair of the base triplets. This mode of binding should promote the formation of parallel-stranded Hoogsteen-paired double helices, as experimentally observed. Based on this model, we have proposed that the extension of the four-membered ring to a five-membered ring with the appropriate shape should optimize stacking interaction with all three bases of the base triplets adjacent to the intercalation site. The synthesis of a BQQ derivative has confirmed this prediction. BQQ turns out to be the best triplex-stabilizing ligand that has been described until now. It binds tightly enough to a triplex with 14 base triplets (Table 1) to increase its melting temperature above that of a 26-bp duplex containing the 14-bp target sequence. However, the most interesting property is its discrimination between triplex and duplex structures. For the triplex with 14 base triplets described in Table 1, the triplex-to-duplex transition (Tm3→2) is shifted by 51°C whereas the duplex-to-single-strands transition (Tm2→1) is shifted by 4°C only. It should be noted that ΔTm3→2 measures a difference in binding for triplex vs. duplex structures and not the strength of binding to the triplex alone. Similarly, ΔTm2→1 measures a difference in binding to the duplex vs. single strands. In the absence of structural data, it is difficult to explain why BQQ and BfPQ do not bind strongly to double helices in contrast to BePI and BgPI derivatives. All BPI derivatives have been shown to self-associate, thus forming dimers and multimers (36). BfPQ and BQQ derivatives also self-associate. Both the chemical nature of the ring system and the shape of the molecule (dictated by pyridoquinoxaline of BfPQ vs. pyridoindole of BgPI) play a role in defining the stacking interactions that these molecules can engage with nucleic acid base pairs and base triplets, whereas electrostatic interactions take place between the terminal amino group of the side chain, which is protonated at pH 7, and the Watson–Crick groove of triple helices or the minor groove of double helices. Molecular modeling suggests that BfPQ and BQQ intercalate into duplexes with their long axis perpendicular to the base pairs. Therefore, parts of the hydrophobic ring system remain exposed to the solvent, resulting in unfavorable binding energy. Increased stacking interactions and a lower exposure to the solvent would make intercalation of these molecules into triplexes much more favorable.
The design of ligands that bind strongly to triple-helical structures and have a high discrimination between triplexes and duplexes opens new possibilities to control gene expression at the transcriptional level. Intermolecular triple-helical complexes formed by antigene oligonucleotides with target oligopyrimidine⋅oligopurine sequences could be stabilized by triplex-specific ligands, thereby leading to an enhanced biological response. Alternatively, intramolecular triple-helical structures (H-DNA) formed by polypyrimidine⋅polypurine sequences presenting a mirror symmetry could be induced by triplex-specific ligands and stabilized even under conditions where they would be converted back to a double helix during physiological processes. Experiments are underway to investigate these possibilities.
Acknowledgments
We thank O. Schwartz for the gift of the plasmid construction containing the HIV-nef gene. This research was supported by grants from the Agence Nationale de Recherche sur le SIDA. The work of C.E. was supported by a grant from Institut de Formation Supérieure Biomédicale.
ABBREVIATIONS
- BPI
benzopyridoindole
- BfPQ
benzo[f]pyridoquinoxalines
- BQQ
benzo[f]quino[3,4-b]quinoxaline
References
- 1.Thuong N T, Hélène C. Angew Chem Int Ed. 1993;32:666–690. [Google Scholar]
- 2.Mirkin S M, Frank-Kamenetskii M D. Annu Rev Biophys Biomol Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 3.Sun J S, Hélène C. Curr Op Struct Biol. 1993;3:345–356. [Google Scholar]
- 4.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout J L, Thuong N T, Lhomme J, Hélène C. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser H E, Dervan P B. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 6.Beal P A, Dervan P B. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 7.Pilch D S, Levenson C, Shafer R H. Biochemistry. 1991;30:6081–6087. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]
- 8.de Bizemont T, Duval-Valentin G, Sun J S, Bisagni E, Garestier T, Helene C. Nucleic Acids Res. 1996;24:1136–1143. doi: 10.1093/nar/24.6.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rougée M, Faucon B, Mergny J L, Barcelo F, Giovannangeli C, Garestier T, Hélène C. Biochemistry. 1992;31:9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- 10.Shindo H, Torigoe H, Sarai A. Biochemistry. 1993;32:8963–8969. doi: 10.1021/bi00085a030. [DOI] [PubMed] [Google Scholar]
- 11.Svinarchuk F, Bertrand J R, Malvy C. Nucleic Acids Res. 1994;22:3742–3747. doi: 10.1093/nar/22.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escudé C, Giovannangeli C, Sun J S, Lloyd D H, Chen J K, Gryaznov S M, Garestier T, Hélène C. Proc Natl Acad Sci USA. 1996;93:4365–4369. doi: 10.1073/pnas.93.9.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J S, François J C, Montenay-Garestier T, Saison-Behmoaras T, Roig V, Chassignol M, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1989;86:9198–9202. doi: 10.1073/pnas.86.23.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaria P V, Shafer R H. J Biol Chem. 1991;266:5417–5423. [PubMed] [Google Scholar]
- 15.Mergny J L, Collier D, Rougée M, Montenay-Garestier T, Hélène C. Nucleic Acids Res. 1991;19:1521–1526. doi: 10.1093/nar/19.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mergny J L, Duval-Valentin G, Nguyen C H, Perrouault L, Faucon B, Rougée M, Montenay-Garestier T, Bisagni E, Hélène C. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 17.Lee J S, Latimer L J P, Hampel K J. Biochemistry. 1993;32:5591–5597. doi: 10.1021/bi00072a014. [DOI] [PubMed] [Google Scholar]
- 18.Wilson W D, Tanious F A, Mizan S, Yao S, Kiselyov A S, Zon G, Strekowski L. Biochemistry. 1993;32:10614–10621. doi: 10.1021/bi00091a011. [DOI] [PubMed] [Google Scholar]
- 19.Fox K R, Polucci P, Jenkins T C, Neidle S. Proc Natl Acad Sci USA. 1995;92:7887–7891. doi: 10.1073/pnas.92.17.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latimer L J P, Payton N, Forsyth G, Lee J S. Biochem Cell Biol. 1995;73:11–18. doi: 10.1139/o95-002. [DOI] [PubMed] [Google Scholar]
- 21.Park Y W, Breslauer K J. Proc Natl Acad Sci USA. 1992;89:6653–6657. doi: 10.1073/pnas.89.14.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand M, Thuong N T, Maurizot J C. J Biol Chem. 1992;267:24394–24399. [PubMed] [Google Scholar]
- 23.Durand M, Thuong N T, Maurizot J C. J Biomol Struct Dyn. 1994;11:1191–1202. doi: 10.1080/07391102.1994.10508063. [DOI] [PubMed] [Google Scholar]
- 24.Pilch D S, Breslauer K J. Proc Natl Acad Sci USA. 1994;91:9332–9336. doi: 10.1073/pnas.91.20.9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilch D S, Waring M J, Sun J S, Rougée M, Nguyen C H, Bisagni E, Garestier T, Hélène C. J Mol Biol. 1993;232:926–946. doi: 10.1006/jmbi.1993.1440. [DOI] [PubMed] [Google Scholar]
- 26.Kim S K, Sun J S, Garestier T, Hélène C, Bisagni E, Rodger A, Norden B. Biopolymers. 1997;42:101–111. [Google Scholar]
- 27.Escude C, Nguyen C H, Mergny J L, Sun J S, Bisagni E, Garestier T, Helene C. J Am Chem Soc. 1995;117:10212–10219. [Google Scholar]
- 28.Nguyen C H, Fan E, Riou J F, Bisserey M C, Vrignaud P, Lavelle F, Bisagni E. Anticancer Drug Des. 1995;10:277–297. [PubMed] [Google Scholar]
- 29.Ouali M, Letellier R, Adnet F, Liquier J, Sun J S, Lavery R, Taillandier E. Biochemistry. 1993;32:2098–2103. doi: 10.1021/bi00059a030. [DOI] [PubMed] [Google Scholar]
- 30.Macaya R F, Schultze P, Feigon J. J Am Chem Soc. 1992;114:781–783. [Google Scholar]
- 31.Radhakrishnan I, Patel D J. Biochemistry. 1994;33:11405–11416. doi: 10.1021/bi00204a001. [DOI] [PubMed] [Google Scholar]
- 32.Arnott S, Bond P J, Selsing E, Smith P J C. Nucleic Acids Res. 1976;3:2459–2470. doi: 10.1093/nar/3.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 34.Escudé C, Mohammadi S, Sun J S, Nguyen C-H, Bisagni E, Liquier J, Taillandier E, Garestier T, Hélène C. Chem Biol. 1996;3:57–65. doi: 10.1016/s1074-5521(96)90085-x. [DOI] [PubMed] [Google Scholar]
- 35.Giovannangéli C, Montenay-Garestier T, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1992;89:8631–8635. doi: 10.1073/pnas.89.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilch D S, Martin M-T, Nguyen C H, Sun J S, Bisagni E, Garestier T, Hélène C. J Am Chem Soc. 1993;115:9942–9951. [Google Scholar]