Abstract
Osteoclasts, the multinucleated cells that resorb bone, develop from hematopoietic cells of monocyte/macrophage lineage. Osteoclast-like cells (OCLs) are formed by coculturing spleen cells with osteoblasts or bone marrow stromal cells in the presence of bone-resorbing factors. The cell-to-cell interaction between osteoblasts/stromal cells and osteoclast progenitors is essential for OCL formation. Recently, we purified and molecularly cloned osteoclastogenesis-inhibitory factor (OCIF), which was identical to osteoprotegerin (OPG). OPG/OCIF is a secreted member of the tumor necrosis factor receptor family and inhibits osteoclastogenesis by interrupting the cell-to-cell interaction. Here we report the expression cloning of a ligand for OPG/OCIF from a complementary DNA library of mouse stromal cells. The protein was found to be a member of the membrane-associated tumor necrosis factor ligand family and induced OCL formation from osteoclast progenitors. A genetically engineered soluble form containing the extracellular domain of the protein induced OCL formation from spleen cells in the absence of osteoblasts/stromal cells. OPG/OCIF abolished the OCL formation induced by the protein. Expression of its gene in osteoblasts/stromal cells was up-regulated by bone-resorbing factors. We conclude that the membrane-bound protein is osteoclast differentiation factor (ODF), a long-sought ligand mediating an essential signal to osteoclast progenitors for their differentiation into osteoclasts. ODF was found to be identical to TRANCE/RANKL, which enhances T-cell growth and dendritic-cell function. ODF seems to be an important regulator in not only osteoclastogenesis but also immune system.
Hemopoietic precursor cells differentiate into osteoclasts at bone-resorbing sites under the control of osteotropic hormones and local factors produced in the microenvironment (1–4). We previously established a cell culture system to produce osteoclast-like cells (OCLs) using cocultures of spleen cells and osteoblasts or bone marrow stromal cells (5, 6). In the cocultures, OCLs are formed from spleen cells in the presence of such stimulators of bone resorption as interleukin 6 (IL-6), IL-11, parathyroid hormone (PTH), prostaglandin E2 (PGE2), and 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] (1–3, 5, 6). These stimulators are classified into three categories in terms of signal transduction pathways: vitamin D receptor-mediated signals [1,25(OH)2D3]; protein kinase A-mediated signals (PTH and PGE2); and gp130-mediated signals (IL-6 and IL-11). They appear to transduce the signals that induce osteoclastogenesis to osteoblasts/stromal cells (1–3). No OCLs are formed, even in the presence of 1,25(OH)2D3, when the cocultures of osteoblasts/stromal cells and spleen cells are separated with a membrane filter (5–7). These observations indicated that osteoblasts/stromal cells are essential for in vitro osteoclastogenesis through a cell-to-cell interaction (1–3). Thus, we proposed a hypothetical membrane-bound factor(s) that is expressed on osteoblasts/stromal cells in response to the stimulators of bone resorption and induces osteoclastogenesis by signaling to osteoclast progenitors. We named the factor “osteoclast differentiation factor (ODF)” (1–3).
Recently, we reported the purification of osteoclastogenesis-inhibitory factor (OCIF) from conditioned medium of human fibroblasts (8). OCIF specifically inhibits in vitro OCL formation elicited through the three distinct signaling pathways stimulated by 1,25(OH)2D3, PTH, or IL-11 (8). The nucleotide sequence analysis of OCIF cDNA (9) has demonstrated that OCIF is identical to osteoprotegerin (OPG) (10), which was initially identified as a novel secreted member of the tumor-necrosis factor (TNF) receptor family in an expressed sequence tag cDNA project (10). The analyses of transgenic mice overexpressing OPG/OCIF and animals injected with OPG/OCIF have demonstrated that OPG/OCIF suppresses bone resorption associated with osteoclast development (9, 10). OPG/OCIF lacks apparent transmembrane domain (9, 10) and seems to act as a soluble inhibitory receptor in osteoclastogenesis.
A mouse bone marrow-derived stromal cell line, ST2, is known to support OCL formation from mouse spleen cells in the presence of 1,25(OH)2D3 and dexamethasone (Dex) (6). We have recently reported that OPG/OCIF specifically binds to ST2 cells treated with 1,25(OH)2D3 and Dex, but not to untreated ST2 cells (8, 9). When the binding sites on the treated ST2 cells were occupied by OPG/OCIF, the cells failed to support OCL formation from spleen cells (9). Cross-linking study using [125I]OPG/OCIF revealed that a 40-kDa protein on the treated ST2 cells binds to OPG/OCIF (9). These results raised the possibility that the binding protein of 40 kDa is a ligand for OPG/OCIF and identical to ODF. Thus, we screened a cDNA expression library of ST2 cells treated with 1,25(OH)2D3 and Dex to isolate the cDNA encoding the OPG/OCIF binding protein. Here we report that the protein is a member of the membrane-associated TNF ligand family and has characteristics that satisfy the major criteria of ODF.
MATERIALS AND METHODS
Cell Culture.
ST2 cells (Riken Cell Bank, Tsukuba) were cultured in α-minimal essential medium (MEM) (GIBCO/BRL) containing 10% fetal calf serum (FCS). COS-7 cells (Riken Cell Bank) were cultured in Dulbecco’s modified eagle’s medium (DMEM; GIBCO/BRL) containing 10% FCS. C7 cells, which were a generous gift from S.-I. Hayashi (Tottori University, Yonago), were cultured in α-MEM containing 10% FCS and 1 ng/ml of human macrophage colony-stimulating factor (M-CSF) (Green Cross, Osaka).
cDNA Cloning.
A cDNA expression library in pcDL-SRα296 (11) was constructed using Great Lengths cDNA synthesis kit (CLONTECH) from poly(A)+ RNA of ST2 cells cultured in the presence of 10−8 M of 1,25(OH)2D3 and 10−7 M of Dex for 5 days. The resulting ST2 expression library was plated onto 24-well plates at a density of ≈100 bacterial colonies per well. Subpools of plasmid DNA were prepared from each bacterial plate using a Qiawell plasmid purification system (Qiagen, Chatwsworth, CA) and were transfected into COS-7 cells grown in 24-well plates using Lipofectamine (GIBCO/BRL). Transfectants were cultured for 2 days and incubated with 3 × 105 cpm of [125I]OPG/OCIF at 37°C for 1 h as described (9). A positive pool obtained was partitioned into smaller subpools until a single cDNA clone, designated pOBM291, was isolated. The cDNA was sequenced using AmpliTaq DyeDeoxy Terminator Cycle Sequencing on an ABI 377 sequencer (Perkin–Elmer). For the binding analysis using [125I]OPG/OCIF, COS-7 cells transfected with pOBM291 (the ODF expression vector) or pcDL-SRα296 (the empty vector) were cultured for 2 days, and then incubated with 1 nM of [125I]OPG/OCIF (1 × 106 cpm) at 37°C for 1 h in the presence or absence of 400 nM of unlabeled OPG/OCIF as described (9).
Preparation of a Soluble ODF (sODF).
sODF was prepared by fusing the extracellular domain of ODF (Asp76–Asp316) to the C-terminal end of thioredoxin using ThioFusion Expression System (Invitrogen). A DNA fragment encoding the extracellular domain of ODF was prepared by digesting pOBM291 with BamHI and EcoRI. PCR was performed using ODF cDNA as a template to amplify the 3′ portion of the cDNA with the following two primers: 5′-ATCAGAAGACAGCACTCACT-3′ and 5′-GGGGTCGACCTAGGACATCCATGCTAATGTTCC-3′. The PCR product was then digested with SalI and EcoRI and a 160-bp fragment was isolated. The BamHI–EcoRI and SalI–EcoRI fragments and pTrxFus vector digested with BamHI and SalI were ligated to yield pTrx-ODF. An Escherichia coli strain GI724 was transformed with pTrx-ODF. sODF was purified from the soluble cytoplamic fraction of the transformants by affinity chromatography on an OPG/OCIF-immobilized column and by gel filtration chromatography.
Expression and Molecular Characterization of Full-Length ODF.
COS-7 cells transfected with the ODF expression vector or the empty vector were cultured for 2 days. The cells were pulse-labeled with 0.2 mCi/ml (1 Ci = 37 Gbq) Express Protein Labeling Mix (NEN) for 4 h in cysteine-methionine-free medium, washed twice with PBS, and lysed in 10 mM of Tris⋅HCl, pH 8.0, containing 1% Triton X-100, 0.14 M of NaCl, 0.1% bovine hemoglobin, and protease inhibitors (12). After removal of cell debris by centrifugation, the lysates (100 μl) were incubated with or without 1 μg of OPG/OCIF for 1 h at 4°C. Five micrograms of anti-OPG/OCIF or mouse immunogloblin (IgG) was added to the mixture and the complexes formed were bound to protein A–Sepharose gel. OPG/OCIF-associated immunoprecipitates were analyzed on a 12.5% SDS/polyacrylamide gel under reducing conditions.
OCL Formation Assay.
COS-7 cells transfected with the ODF expression vector or the empty vector were cultured for 2 days on coverslips in a 24-well plate, fixed in PBS containing 1% paraformaldehyde for 8 min at room temperature, and washed four times with PBS. Mouse spleen cells (7 × 105 cells) obtained from 6- to 15-week-old male ddY mice (6) were cultured for 6 days either on the fixed cells in α-MEM containing 10% FCS and 10 ng/ml of human M-CSF or in the same medium in the presence or absence of various concentrations of sODF in a 24-well plate. After treatment, the cells were subjected to tartrate-resistant acid phosphatase (TRAP) staining, calcitonin binding, and autoradiography. For TRAP staining, the cells were fixed and stained for acid phosphatase in the presence of 50 mM of sodium tartrate (6). For calcitonin binding, the cells were incubated with 0.25 nM of [125I]salmon calcitonin (Amersham) for 1 h at 37°C in the presence or absence of 100 nM of unlabeled calcitonin. For autoradiography, the cells cultured in LaboTech chamber slides (Nunc) were incubated with [125I]calcitonin, stained for TRAP, dipped in NTB-2 emulsion (Kodak), and subjected to autoradiography. For OCL formation from C7 cells, the cells (5 × 104 cells) were cultured for a week in the presence of 10 or 20 ng/ml of M-CSF and various concentrations of sODF, and stained for TRAP.
Dentine Resorption Assay.
Spleen cells (2 × 107 cells) were cultured in α-MEM containing 10% FCS, 10 ng/ml of M-CSF, and 30 ng/ml of sODF for 6 days in a 10-cm dish. The cultured cells were recovered from the culture with a scraper and were collected by centrifugation. The cells (1 × 105 cells) were cultured for additional 3 days on dentine slices in the same medium. After removal of the cells, the slices were stained with Mayer’s hematoxylin to visualize resorption pits (13).
Binding Analysis of [125I]sODF to C7 Cells.
Radioiodination of sODF with [125I]Bolton–Hunter Reagent (NEN) was performed as described (14). C7 cells (1 × 105 cells) were cultured in a 24-well plate. Binding analysis was performed by incubating C7 cells with 1.0 × 106 cpm of [125I]sODF for 1.5 h at 37°C in the presence or absence of 400-fold excess unlabeled sODF as described previously (9).
Northern Blot Analysis.
Isolation of total RNA and hybridization were done as described (15). Poly(A)+ RNA was isolated from total RNA using an oligo(dT) cellulose spin column. A blot contained 2 μg per lane of poly(A)+ RNA from trabecular bone and bone marrow of mouse femur (see Fig. 4C). Mouse multitissue Northern blots (OriGene Technologies, Rockville, MD) contained 2 μg per lane of poly(A)+ RNA (Fig. 4C). Other blots contained 20 μg per lane of total RNA (Fig. 4 A and B). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control.
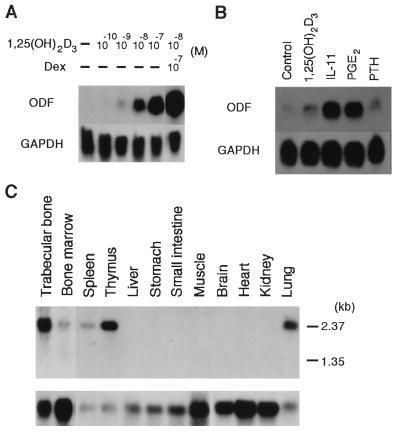
Figure 4.
Expression of ODF gene. (A) A blot loaded 20 μg of total RNA from ST2 cells cultured for 4 days in the presence or absence of various concentrations of 1,25(OH)2D3 and Dex as indicated was probed with ODF or GAPDH cDNA. (B) A blot loaded 20 μg of total RNA from primary osteoblasts cultured for 6 days in the absence or presence of 10−8 M of 1,25(OH)2D3, 10−8 M of IL-11, 10−6 M of PGE2, or 5 × 10−8 M of PTH was probed with ODF or GAPDH cDNA. (C) Blots of mouse tissue poly(A)+ RNA (2 μg) were probed with ODF cDNA (Upper) or GAPDH cDNA (Lower).
Preparation of Primary Osteoblasts.
Primary osteoblasts were prepared from calvaria of newborn ddY mice as described (16). The osteoblasts were cultured in α-MEM containing 10% FCS in the presence of 10−8 M of 1,25(OH)2D3, 10−8 M of IL-11, 10−6 M of PGE2, or 5 × 10−8 M of PTH for 6 days and subjected to the isolation of RNA.
RESULTS
Cloning of Mouse ODF cDNA.
By screening a cDNA expression library of ST2 cells treated with 1,25(OH)2D3 and Dex using [125I]OPG/OCIF, a cDNA clone, designated pOBM291, was isolated from ≈5 × 104 colonies. pOBM291 contained a 1.65-kb insert with an ORF encoding 316 amino acids (Mr 36K) (Fig. 1A). Hydropathy analysis showed the absence of a signal sequence and the presence of an internal 24-residue hydrophobic domain, which presumably represents a transmembrane domain. This structure is typical of a type II transmembrane protein with an extracellular C-terminal region. A homology search of the GenBank sequence database revealed that the C-terminal 165 residues of the protein had a significant homology to the extracellular domains of the TNF ligand family members (17–19) (data not shown). The protein satisfied the major criteria for ODF, the predicted mediator of osteoclastogenesis (1, 2), in the light of its biological activity and its gene expression regulated by bone resorbing factors (see below). We renamed the protein as ODF. Almost simultaneously, a new member of the TNF ligand family designated as TRANCE (20) or RANKL (21) was independently cloned. Comparison of the predicted amino-acid sequences of ODF and TRANCE/RANKL revealed identity between the two proteins.
Figure 1.
ODF is an OPG/OCIF-binding protein. (A) Schematic structure of ODF. Domains: C, cytoplasmic; TM, transmembrane; E, extracellular. Arrowhead represents the N terminus (Asp76) of ODF, which is fused to the C-terminal end of thioredoxin. (B) Binding analysis of [125I]OPG/OCIF to COS-7 cells transfected with the ODF expression vector (COSODF) or COS-7 cells transfected with the empty vector (COSVec). Data are expressed as mean ± SD of three cultures. (C) Coimmunoprecipitation of ODF-OPG/OCIF complexes with anti-OPG/OCIF antibody (Ab).
Specific Binding of OPG/OCIF to ODF.
[125I]OPG/OCIF was incubated with COS-7 cells transfected with the ODF expression vector or an empty vector. As shown in Fig. 1B, OPG/OCIF specifically bound to COS-7 cells expressing ODF (COSODF), but not to control COS-7 cells transfected with the empty vector (COSVec). To determine molecular weight of ODF, we pulse-chase labeled COSODF cells and co-immunoprecipitated the cell lysates with anti-OPG/OCIF antibody in the presence of OPG/OCIF. The results demonstrated that ODF is a 40-kDa protein (Fig. 1C).
Biological Function of ODF.
To examine whether ODF mediates a cell-to-cell signal responsible for osteoclastogenesis, we carried out an in vitro OCL formation assay by evaluating TRAP activity and calcitonin binding, a combination of which is unique to osteoclasts (1, 22). When COSODF or COSVec cells were fixed with paraformaldehyde and then mouse spleen cells were cultured on the fixed cells for 6 days in the presence of M-CSF, TRAP-positive mononuclear and multinucleated cells appeared on the COSODF cells but not on the COSVec cells (Table 1). In addition, [125I]calcitonin specifically bound to the cells cultured on the COSODF cells. Concurrent addition of OPG/OCIF to the cultures inhibited both the TRAP-positive cell formation and the calcitonin binding in a dose-dependent manner (Table 1). These results suggest that ODF mediates the cell-to-cell signaling essential for osteoclastogenesis.
Table 1.
OCL formation from spleen cells by fixed ODF-expressing COS cells
| Cells | M-CSF, ng/ml | OPG/OCIF, ng/ml | TRAP-positive cells, n/well
|
[125I]calcitonin binding, cpm/well | |
|---|---|---|---|---|---|
| Mononuclear | Multinucleated | ||||
| COSODF | 0 | 0 | 0 | 0 | ND |
| 0 | 515 ± 87 | 32 ± 9 | 2212 ± 397 | ||
| 10 | 10 | 109 ± 60 | 6 ± 5 | 85 ± 25 | |
| 100 | 0.5 ± 0.8 | 0 | 29 ± 13 | ||
| COSVec | 10 | 0 | 0.8 ± 0.9 | 0 | 0 ± 21 |
Multinucleated cells were defined as cells containing three or more nuclei. The results are expressed as the means ± SD of six cultures. ND, not determined.
sODF Induces OCL Formation From Spleen Cells in the Absence of Osteoblasts/Stromal Cells.
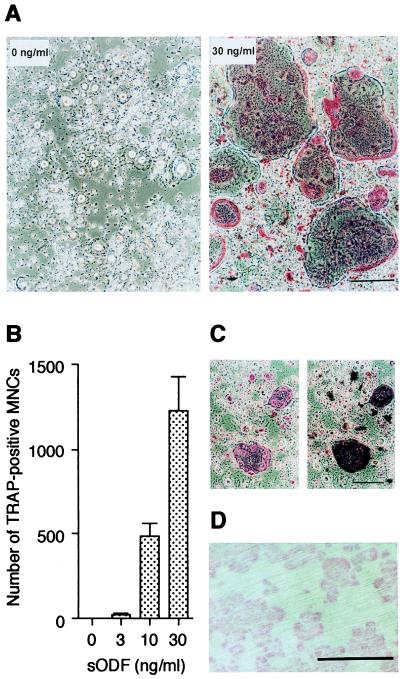
The results from the experiments using COSODF cells did not exclude the possibility that other proteins co-expressed on COSODF cells might be involved in the signaling for osteoclastogenesis. Therefore, we next examined the effect of sODF, which is produced by fusing the extracellular domain of ODF to thioredoxin, on in vitro OCL formation. When mouse spleen cells were cultured in the presence of 10 ng/ml of M-CSF and various concentrations of sODF, TRAP-positive multinucleated cells were formed in a dose-dependent manner (Fig. 2 A and B). In the cocultures, giant TRAP-positive cells were formed in the presence of 30 ng/ml of sODF and the cells with numerous nuclei ranged in size up to 500 μm in diameter (Fig. 2A). Strong TRAP stain was observed in the periphery of the nuclei. The morphological features of the TRAP-positive multinucleated cells were quite similar to those formed in cocultures of spleen cells and stromal cells. Autoradiography using [125I]calcitonin confirmed the presence of calcitonin receptors on the induced TRAP-positive cells (Fig. 2C). OPG/OCIF negated the effect of sODF in a dose-dependent manner (data not shown). Furthermore, when these OCLs were cultured on dentine slices for 3 days in the presence of M-CSF and sODF, numerous resorption pits were formed on the slices (Fig. 2D).
Figure 2.
sODF induces OCL formation from spleen cells in the absence of osteoblasts/stromal cells. (A-C) Mouse spleen cells were cultured in the presence of 10 ng/ml of M-CSF and various concentrations of sODF for 6 days. (A) Morphology of OCLs induced from spleen cells in the presence or absence of 30 ng/ml sODF. (B) The number of TRAP-positive multinucleated cells induced by sODF. Data are expressed as mean ± SD of three cultures. (C) Autoradiography of [125I]calcitonin bound to OCLs induced by 30 ng/ml of sODF. The cells were incubated with [125I]calcitonin, stained for TRAP (Left), and processed for autoradiography (Right). (D) Resorption pits formed on a dentine slice by the induced OCLs. [Bars = 200 μm (A, C, and D).]
sODF Acts Directly on Osteoclast Progenitors.
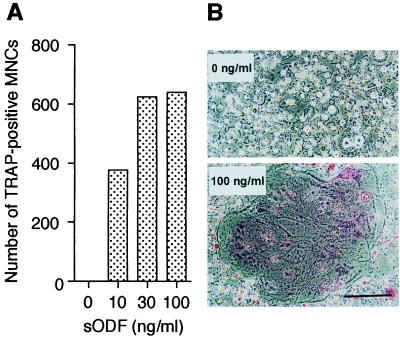
Mature monocytes and alveolar macrophages as well as several cell lines of the macrophage lineage can differentiate into OCLs, when cocultured with stromal cells in the presence of 1,25(OH)2D3 (23–25). A macrophage cell line, C7, is also capable of differentiating into OCLs in such a coculture system (26). Because spleen cells are a mixture of hematopoietic cell and stromal cell populations, we used C7 cells to examine whether ODF acts directly on osteoclast progenitors. sODF induced the formation of TRAP-positive multinucleated cells from C7 cells in a dose-dependent manner in the presence of M-CSF (Fig. 3A). The TRAP-positive multinucleated cells formed from C7 cells were morphologically similar to those formed from spleen cells in the presence of M-CSF and sODF (Fig. 3B; see also Fig. 2A) and produced numerous resorption pits on dentine slices (data not shown). Binding analysis showed that [125I]sODF specifically bound to C7 cells [7663 ± 125 cpm (minus unlabeled sODF) vs. 2735 ± 158 cpm (plus unlabeled sODF), mean ± SD].
Figure 3.
sODF induces OCL formation from C7 cells. (A) The number of TRAP-positive multinucleated cells induced by sODF. C7 cells were cultured in the presence of 10 ng/ml of M-CSF and various concentrations of sODF for 6 days. Data are expressed as means of two cultures. (B) Morphology of OCLs induced from C7 cells in the presence of 20 ng/ml of M-CSF and 0 or 100 ng/ml of sODF. [Bar = 200 μm.]
Expression of ODF Gene in Stromal Cells, Osteoblasts, and Tissues.
Northern blot analysis using a 1.0-kb ODF cDNA probe showed that a single mRNA transcript of approximately 2.4 kb was present in ST2 cells treated with 1,25(OH)2D3 and Dex, but not in the untreated cells (Fig. 4A). The results are consistent with the observation that OPG/OCIF binds to ST2 cells treated with 1,25(OH)2D3 and Dex (9). Expression of the ODF gene was found to be strikingly up-regulated by 1,25(OH)2D3 in a dose-dependent manner in a range of 10−10 to 10−7 M (Fig. 4A). Dex alone did not show any effects on the expression of the ODF gene (data not shown). The combination of 10−8 M of 1,25(OH)2D3 and 10−7 M of Dex, which is known to show a maximal effect on OCL formation in the cocultures of spleen cells and bone marrow stromal cells (6), gave the highest up-regulation of the ODF gene expression (Fig. 4A). Up-regulation of the ODF gene expression was also observed in mouse primary osteoblasts cultured in the presence of 1,25(OH)2D3, IL-11, PGE2, or PTH (Fig. 4B). The up-regulation of ODF expression was consistent with the observation that the binding of [125I]OPG/OCIF to osteoblasts increased when the cells were treated with each stimulator (data not shown). OPG/OCIF inhibited OCL formation elicited by each stimulator in the cocultures of spleen cells and osteoblasts (data not shown).
Analysis of tissue distribution of the ODF transcript showed that the gene was strongly expressed in trabecular bone, thymus, and lung, and weakly expressed in spleen and bone marrow (Fig. 4C).
DISCUSSION
During the past decade, we demonstrated that a cell-to-cell contact between osteoblasts/stromal cells and osteoclast progenitors is important for osteoclastogenesis (1–3, 5, 6). In the present study we molecularly cloned a mouse OPG/OCIF-binding protein and identified it as ODF (1–3), the long-sought-after factor essential for osteoclastogenesis. The following evidence demonstrates that ODF is expressed on the membrane of osteoblasts/stromal cells in response to three distinct bone-resorbing signals and mediates an essential signal to osteoclast progenitors for their differentiation into osteoclasts: (i) the fixed COSODF cells induced the OCL formation from spleen cells; (ii) sODF induced the OCL formation from spleen cells or C7 cells in the absence of osteoblasts/stromal cells, and these OCLs were capable of forming numerous pits on dentine slices; (iii) the three distinct signals, through vitamin D receptor, protein kinase A, and gp130, independently up-regulated the expression of ODF gene in osteoblasts and induced ODF on the membrane; and (iv) the specific binding of [125I]sODF to C7 cells suggested the direct action of ODF on osteoclast progenitors of the monocyte/macrophage lineage.
Because ODF is identified as an OPG/OCIF binding protein, it is important to understand the relationship between ODF and OPG/OCIF. In the present study, we demonstrated that OPG/OCIF specifically abolishes the effect of both membrane-bound- and soluble-form ODF. OPG/OCIF also inhibited the OCL formation elicited through the three distinct signaling pathways in cocultures of spleen cells and osteoblasts, as it did in the culture of unfractionated bone cells or cocultures of spleen cells and stromal cell lines (8). These results strongly suggest that OPG/OCIF inhibits in vitro osteoclastogenesis by directly binding to ODF on osteoblasts/stromal cells and eventually interrupting ODF-mediated signaling from osteoblasts/stromal cells to osteoclast progenitors. On the analogy of the TNF ligand family that interacts with a parallel TNF receptor family (17–19), we suspect that OPG/OCIF acts as a soluble form competitor of ODF receptor presumably expressed on osteoclast progenitors (see below).
M-CSF produced by osteoblasts/stromal cells appears to be essential for the proliferation and differentiation of osteoclast progenitors (27–31). This is consistent with our observation that M-CSF was indispensable for ODF-mediated OCL formation. The concentration of M-CSF seems to be crucial for ODF-mediated OCL formation. A high concentration of M-CSF (≥40 ng/ml) suppressed OCL formation from sODF-stimulated spleen cells (N.S., unpublished observation). Perkins and Kling (32) also reported that addition of exogenous M-CSF to the cocultures of ST2 cells and spleen cells causes a dose-dependent decrease in the number of OCLs accompanied by an increase in the number of macrophage. These results suggest that not only ODF expression but also local concentrations of M-CSF influence osteoclast differentiation. We previously reported that the expression of OPG/OCIF gene in ST2 cells is down-regulated by 1,25(OH)2D3 and up-regulated by calcium ions (9). In addition, the level of OPG/OCIF expression in trabecular bones of ovariectomized rats decreased to 60% of that of sham-operated rats (H.Y., unpublished observation). These results imply a possible role of OPG/OCIF as a local regulator for osteoclastogenesis. Northern blot analysis revealed that ODF gene was highly expressed in not only osseous tissues but also nonosseous tissues such as thymus and lung. At present, we cannot explain why osteoclasts are not seen in nonosseous tissues expressing a high level of ODF mRNA. The microenvironment suitable for osteoclastogenesis seems to require a balance in the local concentrations of M-CSF and OPG/OCIF, presence of osteoclast progenitors, and ODF expression on osteoblast/stromal cells.
During the preparation of this manuscript, a new member of the TNF ligand family designated TRANCE (20) or RANK ligand (RANKL; ref. 21) was independently cloned. Analysis of the predicted amino-acid sequence revealed that ODF is identical to TRANCE/RANKL. TRANCE was identified as an activator of c-Jun N-terminal kinase in T-cells. TRANCE is abundantly expressed in T cells but not in B cells. RANKL was identified as a ligand for RANK, a new member of the TNF receptor family derived from dendritic cells. RANKL activates NF-κB and enhances T cell growth and dendritic cell function. Together, TRANCE/RANKL/ODF seems to be an important regulator of T cells, dendritic cells, and osteoclasts. It should be noted that both dendritic cells and osteoclasts are generated from hematopoietic cells of the monocyte–macrophage lineage (33). Recently, Iotsova et al. (34) reported that NF-κB1 and NF-κB2 double-knockout mice develop osteopetrosis because of a defect of osteoclast differentiation. This report suggests that the activation of NF-κB is important for osteoclastogenesis. Taken all together, these observations imply the identity of RANK with ODF receptor. The mechanism by which ODF mediates signals to osteoclast progenitors through a possible ODF receptor, RANK, should be elucidated. We found that the expression of the TRANCE/RANKL/ODF gene is up-regulated by osteotropic factors, including 1,25(OH)2D3. Vitamin D and other osteotropic factors may also be involved in the TRANCE/RANKL/ODF-mediated immune responses. Our findings appear to give an insight into the understanding of the potential immunological roles of vitamin D (35).
The present study indicates that osteoblasts/stromal cells play an essential role in osteoclastogenesis through the expression of ODF on the membrane (Fig. 5). Identification of ODF as TRANCE/RANKL and as a ligand for OPG/OCIF raises the possibility that ODF and OPG/OCIF are involved in the regulation of not only osteoclastogenesis but also immune system. Further characterization of TRANCE/RANKL/ODF, OPG/OCIF, and the possible ODF receptor, RANK, will shed light on osteoclast biology and immunology.
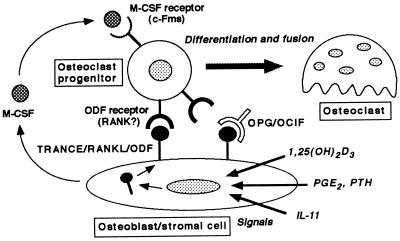
Figure 5.
A model illustrating a mechanism by which osteoblasts/stromal cells regulate osteoclastogenesis. Three distinct signals stimulated by 1,25(OH)2D3, PGE2/PTH, and IL-11 induce TRANCE/RANKL/ODF expression on osteoblasts/stromal cells. ODF mediates a signal for osteoclastogenesis through ODF receptor (RANK?) expressed on osteoclast progenitors. OPG/OCIF inhibits osteoclastogenesis by interrupting the binding of ODF and ODF receptor. M-CSF produced by osteoblasts/stromal cells is also indispensable for proliferation and differentiation of osteoclast progenitors.
Acknowledgments
We thank F. Kobayashi and C. Mashiyama for technical assistance, and M. Hosono, T. Kawaguchi, T. Nakakarumai, H. Kawahara, and S. Ishida for the preparation of recombinant human OPG/OCIF.
ABBREVIATIONS
- 1
25(OH)2D3, 1,25-dihydroxyvitamin D3
- Dex
dexamethasone
- OCIF
osteoclastogenesis-inhibitory factor
- OCL
osteoclast-like cell
- ODF
osteoclast differentiation factor
- OPG
osteoprotegerin
- PGE2
prostaglandin E2
- PTH
parathyroid hormone
- sODF
soluble osteoclast differentiation factor
- TRAP
tartrate-resistant acid phosphatase
- TNF
tumor necrosis factor
- M-CSF
macrophage colony-stimulating factor
- MEM
α-minimal essential medium
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB008426).
References
- 1.Suda T, Takahashi N, Martin T J. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- 2.Suda T, Takahashi N, Martin T J. In: Endocrine Review Monographs. Bikle D D, Negrovilar A, editors. Vol. 4. Bethesda: Endocrine Soc.; 1995. pp. 266–270. [Google Scholar]
- 3.Suda T, Udagawa N, Nakamura I, Miyaura C, Takahashi N. Bone. 1995;17:87S–91S. doi: 10.1016/8756-3282(95)00185-g. [DOI] [PubMed] [Google Scholar]
- 4.Roodman G D. Endocr Rev. 1996;17:308–332. doi: 10.1210/edrv-17-4-308. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi N, Akatsu T, Uadagawa N, Sasaki T, Yamaguchi A, Moseley J M, Martin T J, Suda T. Endocrinology. 1988;123:2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- 6.Udagawa N, Takahashi N, Akatsu T, Sasaki T, Yamaguchi A, Kodama H, Martin T J, Suda T. Endocrinology. 1989;125:1805–1813. doi: 10.1210/endo-125-4-1805. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita T, Asano K, Takahashi N, Akatsu T, Udagawa N, Sasaki T, Martin T J, Suda T. J Cell Physiol. 1990;145:587–595. doi: 10.1002/jcp.1041450327. [DOI] [PubMed] [Google Scholar]
- 8.Tsuda E, Goto M, Mochizuki S, Yano K, Kobayashi F, Morinaga T, Higashio K. Biochem Biophys Res Commun. 1997;234:137–142. doi: 10.1006/bbrc.1997.6603. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda H, Shima N, Nakagawa N, Mochizuki S, Yano K, Fujise N, Sato Y, Goto M, Yamaguchi K, Kuriyama M, Kanno T, Murakami A, Tsuda E, Morinaga T, Higashio K. Endocrinology. 1998;139:1329–1337. doi: 10.1210/endo.139.3.5837. [DOI] [PubMed] [Google Scholar]
- 10.Simonet W S, Lacey D L, Dunstan C R, Kelley M, Chang M-S, et al. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 11.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 13.Tamura T, Takahashi N, Akatsu T, Sasaki T, Udagawa N, Tanaka S, Suda T. J Bone Miner Res. 1993;8:953–960. doi: 10.1002/jbmr.5650080808. [DOI] [PubMed] [Google Scholar]
- 14.Bolton A E, Hunter W M. Biochem J. 1973;133:529–539. doi: 10.1042/bj1330529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuda H, Imai E, Shiota A, Fujise N, Morinaga T, Higashio K. Hepatology. 1996;24:636–642. doi: 10.1053/jhep.1996.v24.pm0008781336. [DOI] [PubMed] [Google Scholar]
- 16.Akatsu T, Tamura T, Takahashi N, Udagawa N, Tanaka S, Sasaki T, Yamaguchi A, Nagata N, Suda T. J Bone Miner Res. 1992;7:1297–1306. doi: 10.1002/jbmr.5650071109. [DOI] [PubMed] [Google Scholar]
- 17.Beutler B, van Huffel C. Science. 1994;264:667–668. doi: 10.1126/science.8171316. [DOI] [PubMed] [Google Scholar]
- 18.Smith C A, Farrah T, Goodwin R G. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 19.Gruss H-J, Dower S K. Blood. 1995;85:3378–3404. [PubMed] [Google Scholar]
- 20.Wong B R, Rho J, Arron J, Robinson E, Orlinick J, Chao M, Kalachikov S, Cayani E, Bartlett F S, III, Frankel W N, Lee S Y, Choi Y. J Biol Chem. 1997;272:25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 21.Anderson D A, Maraskovsky E, Billingsley W L, Dougall W C, Tometsko M E, Roux E R, Teepe M C, DuBose R F, Cosman D, Galibert L. Nature (London) 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 22.Chambers T J, Hall T J. Vitamins Hormones. 1991;46:41–86. doi: 10.1016/s0083-6729(08)60682-2. [DOI] [PubMed] [Google Scholar]
- 23.Udagawa N, Takahashi N, Akatsu T, Tanaka H, Sasaki T, Nishihara T, Koga T, Martin T J, Suda T. Proc Natl Acad Sci USA. 1990;87:7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers T J, Owens J M, Hattersley G, Jat P S, Noble M D. Proc Natl Acad Sci USA. 1993;90:5578–5582. doi: 10.1073/pnas.90.12.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin J H, Kukita A, Kazunori O, Katsuki T, Kohashi O. Endocrinology. 1995;136:4285–4292. doi: 10.1210/endo.136.10.7664646. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto A, Kunisada T, Hemmi H, Yamane T, Yasuda H, Miyake K, Yamazaki H, Hayashi S-I. Biochem Biophys Res Commun. 1998;242:703–709. doi: 10.1006/bbrc.1997.8046. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz L D, Nishikawa S. Nature (London) 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi N, Uadagawa N, Akatsu T, Tanaka H, Isogai Y, Suda T. Endocrinology. 1991;128:1792–1796. doi: 10.1210/endo-128-4-1792. [DOI] [PubMed] [Google Scholar]
- 29.Hattersley G, Owens J, Flanagan A M, Chambers T J. Biochem Biophys Res Commun. 1991;177:526–531. doi: 10.1016/0006-291x(91)92015-c. [DOI] [PubMed] [Google Scholar]
- 30.Kodama H, Nose M, Niida S, Yamasaki A. J Exp Med. 1991;173:1291–1294. doi: 10.1084/jem.173.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka S, Takahashi N, Udagawa N, Tamura T, Akatsu T, Stanley E R, Kurokawa T, Suda T. J Clin Invest. 1993;91:257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins S L, Kling S J. Am J Physiol. 1995;269:E1024–E1030. doi: 10.1152/ajpendo.1995.269.6.E1024. [DOI] [PubMed] [Google Scholar]
- 33.Akagawa K S, Takasuka N, Nozaki Y, Komuro I, Azuma M, Ueda M, Naito M, Takahashi K. Blood. 1996;88:4029–4039. [PubMed] [Google Scholar]
- 34.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Nature Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 35.Lemire J. In: Vitamin D. Feldman D, Glorieux F H, Pike J W, editors. San Diego: Academic; 1997. pp. 1167–1181. [Google Scholar]