Abstract
Activation of phospholipase D1 (PLD1) by Arf has been implicated in vesicle transport and membrane trafficking. PLD1 has also been shown to be associated with the small GTPase RalA, which functions downstream from Ras in a Ras–RalA GTPase cascade that facilitates intracellular signal transduction. Although PLD1 associates directly with RalA, RalA has no effect upon the activity of PLD1. However, PLD1 precipitated from cell lysates with immobilized glutathione S-transferase–RalA fusion protein is active. This suggests the presence of an additional activating factor in the active RalA–PLD1 complexes. Because Arf stimulates PLD1, we looked for the presence of Arf in the active RalA–PLD1 complexes isolated from v-Src- and v-Ras-transformed cell lysates. Low levels of Arf protein were detected in RalA–PLD1 complexes; however, if guanosine 5′-[γ-thio]triphosphate was added to activate Arf and stimulate translocation to the membrane, high levels of Arf were precipitated by RalA from cell lysates. Interestingly, deletion of 11 amino-terminal amino acids unique to Ral GTPases, which abolished the ability of RalA to precipitate PLD activity, prevented the association between RalA and Arf. Brefeldin A, which inhibits Arf GDP–GTP exchange, inhibited PLD activity in v-Src- and v-Ras-transformed cells but not in the nontransformed cells, suggesting that the association of Arf with RalA is required for the increased PLD activity induced by v-Src and v-Ras. These data implicate Arf in the transduction of intracellular signals activated by v-Src and mediated by the Ras–RalA GTPase cascade. Because both Arf and PLD1 stimulate vesicle formation in the Golgi, these data raise the possibility that vesicle formation and trafficking may play a role in the transduction of intracellular signals.
Phospholipase D (PLD) activity is elevated in response to extracellular stimuli (1–6), in response to oncogenic signals (7–12), stress fiber formation (13), and in membrane trafficking and vesicular transport (14–16). PLD hydrolyzes phosphatidylcholine (PC) to phosphatidic acid (PA). Second messenger function for PA has been proposed where PA has been reported to activate signaling molecules such as the GTPase activating proteins for the small GTPases Ras, Rac, and Arf (17–19), phosphatidylinositol-4-kinase (20), and Raf (21). Another suspected role for PA is facilitating vesicle budding and transport (19). This hypothesis is based largely on the observation that PLD activity is stimulated by the small GTPase ARF (22, 23), which is required for vesicle budding in the Golgi (24). Consistent with a role for PA and PLD in vesicular transport, the addition of PLD to Golgi membranes stimulated vesicle formation in the absence of Arf (25–27). Thus, whereas PLD activity is apparently important for both intracellular signal transduction and vesicle transport, a relationship between these two cellular phenomena is not clear.
The activation of PLD by the oncogenic tyrosine kinase v-Src is mediated by a GTPase cascade of Ras and RalA (9, 10). An active PLD can be precipitated from cell lysates with immobilized GST-RalA fusion protein (10). Subsequent studies demonstrated that the PLD associated with RalA is PLD1 (28) and that this interaction is direct (29). Purified PLD1 has been shown to be inactive in the absence of other factors such as Arf (28, 30, 31). These data suggested that RalA may be an activator of PLD1 or that there is an activator of PLD associated with the active RalA–PLD complex precipitated from cell lysates. In this report, we have investigated the mechanism of PLD activation in RalA–PLD complexes.
MATERIALS AND METHODS
Cells and Cell Culture Conditions.
Normal and v-Src- and v-Ras-transformed BALB/c 3T3 and NIH 3T3 cells were maintained in DMEM supplemented with 10% newborn calf serum (HyClone). Cell cultures were made quiescent to reduce background PLD activity by growing to confluence and then replacing with fresh media containing 0.5% newborn calf serum for 1 day.
Materials.
GTP[γS] and brefeldin A (BFA) were purchased from Sigma; [3H]myristate and [3H]phosphatidylcholine were obtained from New England Nuclear. Precoated silica 60A thin layer chromatography plates were from Scientific Products.
Assay of PLD Activity.
Cell cultures were grown to confluence, at which time the media were replaced with fresh media containing 0.5% newborn calf serum for 1 day to reduce background PLD activity. Cells were treated with lysis buffer (25 mM Hepes, pH 7.2/100 mM KCl/10 mM NaCl/0.5 mM EGTA/0.5 mM EDTA/1 mM DTT) containing 1% Triton X-100. Protease inhibitors (12 μg/ml leupeptin/20 μg/ml aprotinin/0.1 mM phenylmethylsulfonyl fluoride) were included just before lysis (30 min at 4°C). Lysates were scraped from the plates with a rubber policeman and centrifuged for 10 min at 1,500 rpm; the supernatant was then centrifuged for 45 min at 30,000 rpm, the supernatant was recovered, and the protein concentration was determined using the Bio-Rad assay. Cell lysates (600 μg protein) were treated with immobilized GST-RalA fusion proteins as described previously (10). After 1.5 h at 4°C, the GST-RalA fusion proteins were separated from the lysate by microcentrifugation for 30 sec and washed three times with cold lysis buffer. A liposome-based in vitro PLD assay based largely on strategies used by Brown et al. (22) was used to assay PLD activity associated with Ral fusion proteins. PLD activity was determined by examining the ability to convert [3H]phosphatidylcholine in prepared liposomes to phosphatidylethanol in the presence of exogenously provided ethanol. Liposomes were prepared by mixing chloroform solutions of phospholipids containing 1 μCi/reaction of [3H]phosphatidylcholine (42 Ci/mM) and drying under a stream of nitrogen followed by resuspension with sonication for 10 min at 45°C in lysis buffer. The liposomes consist of phosphatidylethanolamine/phosphatidylinositol-4,5-bisphosphate/PC in a molar ratio of 16:1.4:1 with PC suspended to a final concentration of 8.6 μM. The reaction buffer consisted of lysis buffer plus 5 mM MgCl2, 0.16 mM CaCl2, and 1% ethanol in a total volume of 0.2 ml. The reaction mixtures were incubated for 15 min at 37°C and terminated by the addition of 2 ml of CHCl3/MeOH/HCl (1.0:1.0:0.006). Phase separation was achieved by adding 1 ml of 0.1 N HCl, 1 mM EGTA. The resolution of PLD products by thin layer chromatography and quantification was as described previously (32). Alternatively, the activity of purified PLD1 was determined using the choline release assay as described previously (28, 30).
In vivo PLD activity was determined by the transphosphatidylation reaction in the presence of 0.8% butanol as described (32). Cells in 35-mm culture dishes were prelabeled with [3H]myristate, 3 μCi (40 Ci/mmol) for 4–6 h in 2 ml of DMEM containing 0.5% newborn calf serum.
RalA Proteins.
The construction of the RalA mutants and the loading of immobilized Ral with guanine nucleotides was as described previously (10, 33).
Arf Proteins.
The Arf protein used to stimulate RalA-associated PLD activity was a generous gift of Paul Sternweis and Alex Brown. The Arf preparation was prepared as described previously by Brown et al. (34) from porcine brain. It was approximately 50% pure and contained a mixture of Arf1 and Arf3. Arf proteins were pretreated with GTP[γS] before use as described (34). Arf protein used to stimulate purified PLD1 was affinity-purified from sf9 insect cells infected with baculovirus expressing a Glu-tagged recombinant human Arf1 using immobilized mAb for the Glu tag (30).
Western Analysis of Arf.
Extraction of proteins from cultured cells was performed as previously described (10). Equal amounts of protein were subjected to SDS/PAGE using an 8% acrylamide separating gel. After transferring to nitrocellulose, the filters were incubated with an antibody raised against Arf (1D9), which recognizes all Arf isoforms (35).
RESULTS
RalA Does Not Activate PLD1.
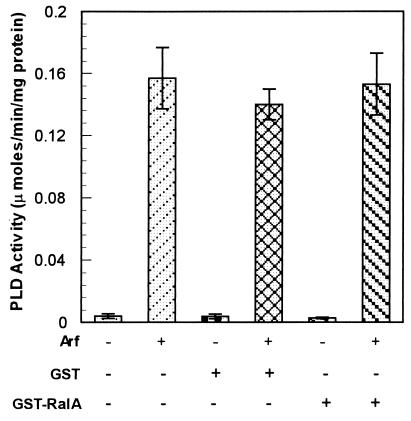
We previously reported that immobilized GST-RalA fusion protein precipitates an active PLD1 from v-Src and v-Ras-transformed cell lysates (10, 29). However, purified PLD1 is inactive in the absence of additional factors such as Arf, protein kinase C (PKC) α, or Rho family GTPases (28, 31). To determine whether the activation of the PLD1 associated with RalA was due to RalA, we examined whether RalA had any effect on purified PLD1. As shown in Fig. 1, RalA had no effect upon PLD1 activity or the PLD1 activity that was stimulated by Arf. RalA also had no effect upon the PLD activity stimulated by either Rho or PKC α (data not shown). These data indicate that RalA does not by itself activate PLD activity. However, as the PLD1 precipitated by RalA from cell lysates is active, it is likely that the PLD1 complex precipitated by RalA from cell lysates has, in addition to PLD1, a factor(s) that is essential for activating PLD1.
Figure 1.
RalA does not activate PLD1. Purified PLD1a (30) was treated with nothing, GST, or GST-RalA as shown, and PLD activity was determined in the absence or presence of GTP[γS]-activated Arf (5 μM). The assay volume was 100 μl and contained 10 ng of purified PLD1 and 1 μg of either GST or GST-RalA. The assay time was 30 min. The PLD activity is expressed in μmol/min per mg of PLD1 protein. Error bars represent the SD for triplicate determinations.
GTP-Dependent Association between RalA and Arf.
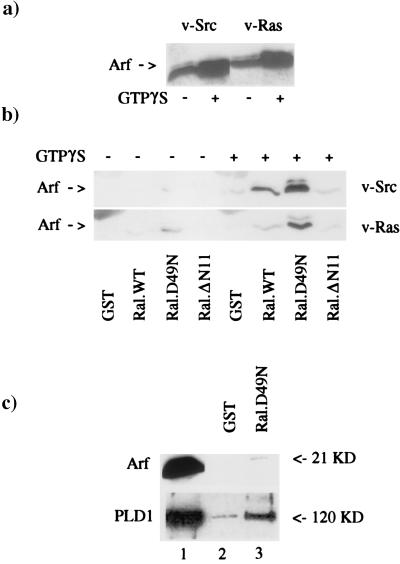
The data in Fig. 1 suggest the presence of a PLD1-activating factor in RalA–PLD1 complexes. We previously demonstrated that the PLD precipitated by RalA from cell lysates could be further stimulated by the addition of exogenously provided Arf (29). We therefore examined for the presence of Arf in the RalA precipitates from v-Src- and v-Ras-transformed cell lysates. Initially, we looked at the level of Arf in GST-RalA precipitates from total cell lysates; although Arf could be detected by Western blot analysis, the level of Arf in the RalA precipitates was only slightly above background (data not shown). We then examined the ability of GST-RalA to precipitate Arf from lysates of the membrane fraction, which is where most of the RalA-associated PLD activity is found (unpublished results). Isolated membranes were mixed with the cytosolic fraction in the presence and absence of GTP[γS], which activates Arf and stimulates membrane association (36). Including GTP[γS] with the cytosol increased the level of Arf that fractionated with membranes (Fig. 2A), suggesting that the GTP[γS] treatment was activating some of the cytosolic Arf. We then examined the ability of immobilized GST-RalA and two mutant GST-RalA proteins to precipitate Arf from membrane lysates. Wild-type RalA and the two mutant RalA proteins precipitate different levels of PLD activity. An effector domain mutant of RalA (D49N) precipitated more PLD activity than wild-type RalA, and a deletion of 11 amino-terminal Ral-specific amino acids (ΔN11) precipitated very little PLD activity (10). On examination of Arf levels in the RalA precipitates of the membrane fractions, we found that although Arf was easily detectable above background in the precipitates for the D49N RalA mutant, the levels of Arf associated with wild-type RalA were very low. However, if GTP[γS] was included, there was a dramatic increase in the ability of the GST-RalA fusion proteins to precipitate Arf (Fig. 2B). GTP[γS] increased the level of Arf that associated with RalA by greater than 10-fold for the D49N RalA mutant, which bound Arf the strongest. Interestingly, the amount of Arf associated with the three RalA proteins correlated well with the PLD activity that was precipitated by the RalA proteins (10), with the effector domain mutant (D49N) binding the most Arf and the amino-terminal deletion mutant (ΔN11) binding the least Arf. These data suggest a GTP-dependent association between Arf and RalA involving the N terminus of RalA.
Figure 2.
RalA precipitates Arf protein from cell lysates in a GTP-dependent manner. (A) Membrane and cytosolic fractions were isolated from v-Src- and v-Ras-transformed cells. The membrane fraction (250 μg protein) was mixed with cyotosol (750 μg protein) for 10 min at 30°C either in the absence or the presence of GTP[γS] (100 μM). The membranes were then recovered by centrifugation and lysed, and the level of Arf in the membrane fraction before and after GTP[γS] treatment was determined by Western blot analysis using mAb 1D9, which recognizes all Arf proteins (35). (B) Lysed membranes were treated with immobilized GST, GST-RalA, and the GST-RalA mutants D49N and ΔN11. The levels of Arf precipitated by GST and the GST-RalA fusion proteins were then determined by Western blot analysis. (C) Partially purified Arf (0.8 μg Arf protein; 0.035 nmol) used previously to activate the RalA-associated PLD (29) was treated with immobilized GST and GST-RalA D49N (0.70 nmol) in the presence of GTP[γS] (50 μM). The immobilized GST and GST-RalA proteins were recovered by centrifugation and washed three times with PBS. The precipitates were subjected to Western blot analysis using the 1D9 anti-Arf antibody. In lane 1 is the Arf preparation before treatment with either GST or GST-RalA proteins. The ability of GST-RalA D49N to precipitate PLD1 from an affinity-purified PLD1 preparation as previously reported (29) is shown for comparison.
We previously demonstrated that the interaction between RalA and PLD1 is direct by showing that purified GST-RalA could precipitate PLD1 from a purified preparation of PLD1 (29). We therefore investigated whether GST-RalA could precipitate Arf from the partially purified Arf preparation used to activate the RalA-associated PLD. As shown in Fig. 2C, GST-RalA D49N, which associated most strongly with Arf in the lysate precipitations, could not precipitate significant levels of Arf from the partially purified Arf that had been treated with GTP[γS]. The ability of RalA to precipitate PLD1 from a purified PLD1 preparation as reported previously (10) is shown for comparison. Additionally, the amount of Arf precipitated by GST-RalA D49N from membranes (Fig. 2, compare A, lanes 2 and 4, with B, lane 7) is substantially higher than the amount of Arf precipitated by GST-RalA D49N from the purified source (Fig. 2C, compare lanes 1 and 3). From this comparison, it is clear that the D49N RalA mutant precipitates Arf much more efficiently from the crude source than from the purified source. These data indicate that the interaction between RalA and Arf is indirect, suggesting that an additional factor is likely involved. As discussed below, the factor is not likely PLD1.
Differential Ability of RalA Mutants to Precipitate PLD Activity Is Not Caused by Differences in the Level of PLD1.
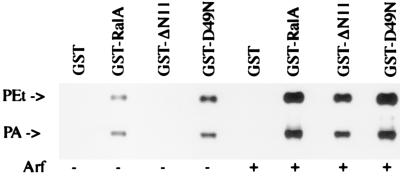
The data presented in Fig. 2 suggest that the differences in PLD activity associated with RalA and the RalA mutants are caused by differences in the level of a PLD activator rather than differences in the levels of PLD1 associated with the wild-type RalA and the RalA mutants. Because PLD1 is expressed at very low levels in mammalian cells (28–30), we have not been able to detect PLD1 protein in either cell lysates or RalA precipitates using standard Western blot analysis. Therefore, to determine the levels of PLD1 associated with RalA and the RalA mutants, we used an indirect assay that exploited the ability to observe PLD activity associated with RalA. We demonstrated previously that the PLD1 precipitated by GST-RalA could be further stimulated by the addition of Arf (29). If the PLD activity associated with RalA is caused by the level of Arf and not due to the level of PLD1, then exogenously provided Arf should reveal PLD activity associated with the ΔN11 mutant. In Fig. 3A, the differential precipitation of PLD activity by RalA and the RalA mutants is shown as reported previously (10). As predicted, when Arf was provided, PLD activity was detected in association with the ΔN11 RalA mutant. In fact, the level of PLD activity associated with the ΔN11 RalA mutant was increased in the presence of Arf almost to the level seen with Arf-treated wild-type RalA (Fig. 3). These data suggest that the differences in PLD activity associated with the RalA mutants is not caused by differences in PLD1 protein associated with these mutants but rather to differences in the levels of Arf seen in Fig. 2B.
Figure 3.
Differential precipitation of PLD activity by RalA mutants is lost in the presence of Arf. Lysates from v-Src-transformed murine fibroblasts were treated with GST, GST-RalA (wt-RalA), or GST fused to the RalA mutants D49N or ΔN11. The GST and GST-RalA fusion proteins were recovered, and the PLD activity, as measured by the production of PEt in the presence of 1% EtOH, in the pellets was determined in the presence and absence of Arf (40 nM) as shown. A representative experiment that was repeated three times is shown.
BFA Blocks the Elevation of PLD Activity in v-Src- and v-Ras-Transformed Cells.
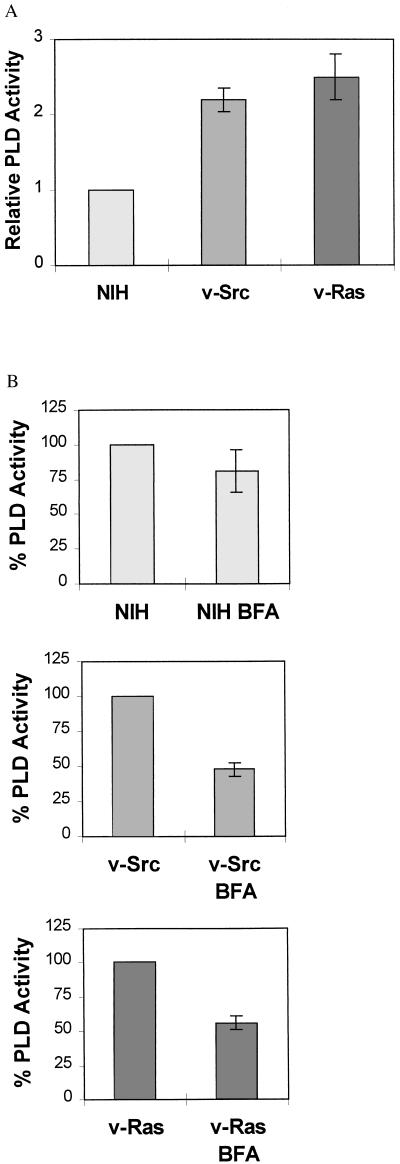
The data presented above suggest that there is a GTP-dependent association between Arf and RalA–PLD1 complexes that facilitates the activation of PLD1 in response to v-Src and v-Ras. The strong stimulation of RalA–Arf association by GTP[γS] suggests that Arf must be activated for the association to occur. To investigate whether Arf activation was required for the activation of PLD1 by v-Src and v-Ras, we examined the effect of BFA, which inhibits GDP–GTP exchange upon Arf (37, 38), on the PLD activity in v-Src- and v-Ras-transformed cells. PLD activity was examined by examining PLD-catalyzed transphosphatidylation of PC in the presence of butanol in cells prelabeled with [3H]myristate, which is incorporated into PC species utilized by the v-Src-induced PLD (32). As shown in Fig. 4A and reported previously (9, 10), there is elevated PLD activity in the v-Src- and v-Ras-transformed cells relative to the parental NIH 3T3 cells. BFA treatment substantially reduced PLD activity in both v-Src- and v-Ras-transformed cells, having much less of an effect upon the background PLD activity observed in the NIH 3T3 cells (Fig. 4B). The reduced effect of BFA on the PLD activity in the parental cells may be because BFA is added only 10 min before the addition of butanol (for detection of PLD transphosphatidylation), and therefore only acutely activated PLD is severely inhibited. The basal PLD activity may not be so dependent upon the continual activation of Arf and may also represent a substantial amount of PLD2 activity, which is not dependent on Arf. Thus, although a mechanism for the proposed activation of Arf in response to v-Src and v-Ras remains unknown, these data suggest that activation of Arf is essential for the elevated PLD levels observed in v-Src- and v-Ras-transformed cells.
Figure 4.
BFA inhibits v-Src- and v-Ras-induced PLD activity. (A) NIH 3T3 cells and NIH 3T3 cells transformed by v-Src and v-Ras were prelabeled with [3H]myristate, and PLD activity was determined by the transphosphatidylation reaction in the presence of butanol (0.8%) by measuring phosphatidylbutanol levels as described previously (32). The data are presented as the percent cpm incorporated into phosphatidylbutanol relative to the total cpm incorporated per culture dish. (B) PLD activity was determined in the presence of BFA (20 μg/ml), which was added 10 min before the addition of butanol. Cells were harvested 20 min after the addition of butanol. The effect of BFA is presented as the PLD activity in the BFA-treated cells as a percentage of untreated cells. The data are the average effect of BFA (± standard error) from at least three independent experiments performed in duplicate where duplicate determinations varied by less than 10%.
DISCUSSION
In this report, we have described an association of Arf with an active RalA–PLD1 complex. The level of Arf protein associated with RalA mutants correlated well with the level of PLD activity in the RalA–PLD1 complexes, and the levels of Arf associated with RalA were substantially increased by a nonhydrolyzable GTP analog that stimulated Arf membrane association. BFA, which blocks the GDP–GTP exchange on Arf, blocked the increased PLD activity in both v-Src- and v-Ras-transformed cells. We demonstrated previously that in cells expressing a temperature-activatable v-Src, PLD activity is elevated within 15 min after temperature shift (7) before substantial protein synthesis can occur or the cells become transformed. Thus, PLD activation is a direct response to tyrosine kinase-induced signaling and not an indirect consequence of transformation or long-term constitutive signaling and therefore likely to be involved in the transduction of signals initiated by v-Src and v-Ras. The data presented in this paper implicate a GTP-dependent association between Arf and a RalA–PLD1 complex that is involved in the activation of PLD1 in response to the oncogenic signals generated by v-Src and v-Ras.
The interaction between RalA and Arf is likely an indirect one. Immobilized RalA was unable to precipitate significant levels of Arf from a partially purified preparation of Arf, suggesting that the association between RalA and Arf is facilitated by another factor. Because the amino terminus of RalA was required for Arf association, the putative factor may bind to this region of RalA. The putative additional factor is not likely to be PLD1 because, as shown in Fig. 2B, Arf was easily detectable in RalA precipitates from GTP[γS]- and cytosol-treated membrane lysates, whereas we are unable to detect the PLD1 that is present in the RalA precipitates due to low levels of PLD1 expression. This factor is also not likely to be the RalA effector domain binding protein Ral-BP1 (39), because the D49N effector domain RalA mutant, which does not bind Ral-BP1 (39), associated with Arf more strongly than with wild-type RalA. Thus, another factor, yet to be identified, likely facilitates the association between RalA and Arf.
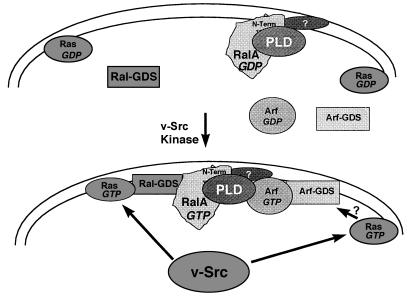
BFA, which inhibits GDP–GTP exchange on Arf, blocked the v-Src- and v-Ras-induced PLD activity. Thus, Arf is apparently activated by v-Src and v-Ras. A mechanism for Arf activation by Src or Ras is not known, but it is not likely mediated by activation of RalA, as an activated form of RalA is unable to elevate PLD activity in cells overexpressing an activated RalA (10). However, because activated Ras does elevate PLD activity, the activation of Arf by activated Ras is likely via a downstream target of Ras other than Ral-guanine nucleotide dissociation stimulator (Ral-GDS) and RalA. Multiple signaling pathways activated by v-Src and mediated by Ras have been reported (40), and multiple downstream targets of Ras are required for transformation (41). Thus, Ras activation by v-Src likely results in both the mobilization of Ral-GDS and RalA and the assembly of another Ras signaling complex that results in the activation of a BFA-sensitive Arf exchange factor. A model for PLD activation via two Ras pathways involving both RalA and a putative Arf exchange factor and Arf is presented in Fig. 5.
Figure 5.
Model for the activation of PLD by v-Src via Ras, RalA, and Arf. It is proposed that in response to the tyrosine kinase activity of v-Src, Ras goes from the inactive GDP form (Ras-GDP) to the active GTP form (Ras-GTP). The activated Ras associates with Ral–GDS (33), resulting in the recruitment of a RalA–PLD1 complex and GDP–GTP exchange on RalA. However, because activated RalA is not sufficient to activate PLD1 (10), whereas activated Ras is sufficient for PLD1 activation, it is proposed that Ras stimulates GDP–GTP exchange on Arf via another mechanism involving an Arf–GDS, leading to an association between RalA and Arf that is dependent upon the RalA amino terminus. An as yet unknown factor is postulated to facilitate the interaction between RalA and activated Arf–GTP as the interaction is apparently indirect.
A role for Arf in vesicle transport has been firmly established (15, 24, 42). Data presented here suggest a role for Arf in the transduction of intracellular signals mediated by Ras, Ral, and PLD1. Arf has also been implicated in the activation of PLD by the m3 muscarinic acetylcholine receptor (43) and insulin (44, 45). Recent data suggest that Arf may promote vesicle budding in the Golgi by activation of PLD and the generation of phosphatidic acid (25–27). A role for Arf in signal transduction raises the question as to whether vesicle transport and membrane trafficking also play a role in the transduction of intracellular signals. It has been proposed recently that intracellular signals may regulate constitutive membrane traffic (46). Alternatively, the transduction of intracellular signals may utilize vesicle movement directly. There are several recent reports suggesting that vesicle movement may be critical for the transduction of intracellular signals. In response to epidermal growth factor (EGF), the EGF receptor is endocytosed (47), and this process is inhibited in cells that have a mutation in dynamin, a GTPase that facilitates release of endocytic vesicles (48). Interestingly, this mutation in dynamin blocked a subset of EGF-induced signals suggesting that EGF-induced signaling involves the internalization of an endocytic vesicle (48). The EGF receptor, like v-Src, has an intrinsic tyrosine kinase and tyrosine phosphorylation, and dephosphorylation has been reported as a requirement for the generation of secretory vesicles in the Golgi (49), further suggesting a functional link between signaling and trafficking. Nerve growth factor and its receptor, TrkA, was recently reported to undergo translocation from distal axons to the cell body via retrograde vesicular movement (50). These data are consistent with a role for vesicle movement in the transduction of intracellular signals.
It may also be relevant that vesicular invaginations along the plasma membrane known as caveolae are enriched with signaling molecules including Src, Ras, and many others (51, 52). In response to platelet-derived growth factor, tyrosine-phosphorylated proteins are restricted to the caveolar membranes (53); in response to epidermal growth factor, increased Ras–Raf interactions are restricted to caveolar membranes (54). Caveolar vesicles, like endocytosed EGF receptors, become internalized in a GTP–dynamin-mediated fission mechanism (55). Interestingly, caveolae also contain the molecular machinery for vesicle budding, docking, and fusion (56). Thus, it would seem that caveolar vesicles with both signaling and trafficking molecules may represent another link between signaling and trafficking. Whether PLD activity is important in the formation of caveolae remains to be established; however, we have found that caveolae are enriched for PLD activity (our unpublished results). Thus, it is provocative to speculate that the formation of “signaling vesicles” may occur in response to PLD1 activation caused by recruitment of Arf into a RalA–PLD1 complex.
Acknowledgments
We are thankful to Paul Sternweis and Alex Brown for providing partially purified Arf protein. We thank Michael Frohman for comments on the manuscript and Dennis Shields for communicating results before publication. The inestimable contribution of Steve Hutkins is gratefully acknowledged. This investigation was supported by National Institutes of Health Grant CA46677, Council for Tobacco Research Grant 3075, American Cancer Society Grant BE-243 (to D.A.F.); a Research Centers in Minority Institutions (RCMI) award from the Division of Research Resources, National Institutes of Health (RR 03037) to Hunter College; National Institutes of Health Grant GM47707 and an American Cancer Society faculty research award (to L.A.F.); and National Institutes of Health Grant GM50388 (to A.J.M.). X.L. was a Howard Hughes Medical Institute Undergraduate Scholar.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BFA, brefeldin A; PC, phosphatidylcholine; PA, phosphatidic acid; PLD, phospholipase D; GST, glutathione S-transferase; GTP[S], guanosine 5′-[γ-thio]triphosphate; PKC, protein kinase C; GDS, guanine nucleotide dissociation stimulator; EFG, epidermal growth factor.
References
- 1.Singer W D, Brown H A, Sternweis P C. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 2.Exton J H. Biochim Biophys Acta. 1994;1212:26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 3.Exton J H. J Biol Chem. 1997;272:15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 4.Foster D A. Cell Signaling. 1993;5:389–399. doi: 10.1016/0898-6568(93)90078-z. [DOI] [PubMed] [Google Scholar]
- 5.Liscivitch M. J Lipid Mediat Cell Signal. 1996;14:215–221. doi: 10.1016/0929-7855(96)00528-7. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel S, Foster D A, Kolesnick R. Curr Opin Cell Biol. 1996;8:159–167. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Pfeffer L M, Foster D A. Mol Cell Biol. 1991;11:4903–4908. doi: 10.1128/mcb.11.10.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Alexandropoulos K, Song J, Foster D A. Mol Cell Biol. 1994;14:3676–3682. doi: 10.1128/mcb.14.6.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, Lu Z, Luo J Q, Wolfman A, Foster D A. J Biol Chem. 1995;270:6006–6009. doi: 10.1074/jbc.270.11.6006. [DOI] [PubMed] [Google Scholar]
- 10.Jiang H, Luo J-Q, Urano T, Frankel P, Lu Z, Foster D A, Feig L. Nature (London) 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 11.Carnero A, Cuadrado A, del Peso L, Lacal J C. Oncogene. 1994;9:1387–1395. [PubMed] [Google Scholar]
- 12.del Paso L, Hernandez R, Esteve P, Lacal J C. J Cell Biochem. 1996;61:599–608. doi: 10.1002/(sici)1097-4644(19960616)61:4<599::aid-jcb14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 13.Cross M J, Roberts S, Ridley A J, Hodgkin M N, Stewart S, Claesson-Welsh L, Wakelam M J O. Curr Biol. 1996;6:588–597. doi: 10.1016/s0960-9822(02)00545-6. [DOI] [PubMed] [Google Scholar]
- 14.Kahn R A, Yucel J K, Malhotra V. Cell. 1993;75:1045–1048. doi: 10.1016/0092-8674(93)90314-g. [DOI] [PubMed] [Google Scholar]
- 15.Boman A L, Kahn R A. Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- 16.Bednarek V A, Orci L, Schekman R. Trends Cell Biol. 1996;6:468–473. doi: 10.1016/0962-8924(96)84943-9. [DOI] [PubMed] [Google Scholar]
- 17.Tsai M-H, Yu C-L, Wei F-S, Stacey D W. Science. 1989;243:522–526. doi: 10.1126/science.2536192. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L. J Biol Chem. 1993;268:10709–10712. [PubMed] [Google Scholar]
- 19.Randazzo P A, Kahn R A. J Biol Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- 20.Moritz A, Graan P N E, Gispen W H, Wirtz K W A. J Biol Chem. 1992;267:7207–7210. [PubMed] [Google Scholar]
- 21.Ghosh S, Strum J C, Sciorra V A, Daniel L, Bell R M. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 22.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 23.Cockroft S, Thomas G M H, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Truong O, Hsuan J J. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 24.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 25.Ktistakis N T, Brown H A, Waters M G, Sternweis P C, Roth M G. J Cell Biol. 1996;134:295–306. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi K, Roth M G, Ktistakis N T. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y-G, Siddhanta A, Austin C D, Hammond S M, Sung T-C, Frohman M A, Morris A J, Shields D. J Cell Biol. 1997;138:495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond S M, Altshuller Y M, Sung T C, Rudge S A, Ross K, Engebrecht J, Morris A J, Frohman M A. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- 29.Luo J-Q, Liu X, Hammond S M, Colley W C, Feig L A, Frohman M A, Morris A J, Foster D A. Biochem Biophys Res Commun. 1997;235:854–859. doi: 10.1006/bbrc.1997.6793. [DOI] [PubMed] [Google Scholar]
- 30.Hammond S M, Jenco J M, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Prestwich G D, Frohman M A, Morris A J. J Biol Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- 31.Colley W C, Sung T-C, Roll R, Jenco J, Hammond S M, Altshuller Y, Bar-Sagi D, Morris A J, Frohman M A. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Foster D A. Biochem J. 1993;294:711–717. doi: 10.1042/bj2940711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urano T, Emkey R, Feig L A. EMBO J. 1996;16:810–816. [PMC free article] [PubMed] [Google Scholar]
- 34.Brown H A, Gutowski S, Kahn R A, Sternweis P C. J Biol Chem. 1995;270:14935–14943. doi: 10.1074/jbc.270.25.14935. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C-J, Rosenwald A G, Willingham M C, Skuntz S, Clark J, Kahn R A. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin A, Brown F D, Hodgkin M N, Bradwell A J, Cook S J, Hart M, Wakelam M J O. J Biol Chem. 1996;271:17397–17403. doi: 10.1074/jbc.271.29.17397. [DOI] [PubMed] [Google Scholar]
- 37.Donaldson J G, Finazzi D, Klausner R D. Nature (London) 1992;360:352–354. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 38.Helms J B, Rothman J E. Nature (London) 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 39.Cantor S B, Urano T, Feig L A. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qureshi S A, Alexandropoulos K, Joseph C K, Rim M, Bruder J, Rapp U R, Foster D A. J Biol Chem. 1992;267:17635–17639. [PubMed] [Google Scholar]
- 41.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 42.Moss J, Vaughn M. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 43.Rumenapp U, Geiszt M, Wahn F, Schmidt M, Jacobs K H. Eur J Biochem. 1995;234:240–244. doi: 10.1111/j.1432-1033.1995.240_c.x. [DOI] [PubMed] [Google Scholar]
- 44.Karnam P, Standaert M L, Galloway L, Farese R V. J Biol Chem. 1997;272:6136–6140. doi: 10.1074/jbc.272.10.6136. [DOI] [PubMed] [Google Scholar]
- 45.Shome K, Vasudevon C, Romero G. Curr Biol. 1997;7:387–396. doi: 10.1016/s0960-9822(06)00186-2. [DOI] [PubMed] [Google Scholar]
- 46.Roth M G, Sternweis P C. Curr Opin Cell Biol. 1997;9:519–526. doi: 10.1016/s0955-0674(97)80028-2. [DOI] [PubMed] [Google Scholar]
- 47.Haigler H T, McKanna J A, Cohen S. J Cell Biol. 1979;81:382–395. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira A V, Lamaze C, Schmid S L. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 49.Austin C D, Shields D. J Cell Biol. 1996;135:1471–1483. doi: 10.1083/jcb.135.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riccio A, Pierchalla B A, Ciarallo C L, Ginty D D. Science. 1992;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- 51.Lisanti M P, Scherer P E, Tang Z, Sargiacomo M. Trends Cell Biol. 1994;4:231–235. doi: 10.1016/0962-8924(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 52.Lisanti M P, Tang Z, Scherer P E, Kubler E, Koleske A J, Sargiacomo M. Mol Membr Biol. 1995;12:121–124. doi: 10.3109/09687689509038506. [DOI] [PubMed] [Google Scholar]
- 53.Liu P, Ying Y, Ko Y-G, Anderson R G W. J Biol Chem. 1996;271:10299–10303. doi: 10.1074/jbc.271.17.10299. [DOI] [PubMed] [Google Scholar]
- 54.Mineo C, James G L, Smart E J, Anderson R G W. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- 55.Schnitzer J E, Oh P, McIntosh D P. Science. 1996;274:239–242. doi: 10.1126/science.274.5285.239. [DOI] [PubMed] [Google Scholar]
- 56.Schnitzer J E, Liu J, Oh P. J Biol Chem. 1995;270:14399–14404. doi: 10.1074/jbc.270.24.14399. [DOI] [PubMed] [Google Scholar]