Abstract
Sphingomyelin is an abundant constituent of the plasma membranes of mammalian cells. Ceramide, its primary catabolic intermediate, is released by either acid sphingomyelinase or neutral sphingomyelinase (nSMase) and has emerged as a potential lipid signaling molecule. nSMase is regarded as a key enzyme in the regulated activation of the “sphingomyelin cycle” and cell signaling. We report here the cloning, identification, and functional characterization of murine and human nSMase, a ubiquitously expressed integral membrane protein, which displays all established properties of the Mg2+-dependent nSMase of the plasma membrane. Stably nSMase-overexpressing U937 and human embryonic kidney cell lines have been generated for the study of the role of nSMase in signal transduction pathways. Their stimulation by tumor necrosis factor α leads only to a moderately elevated ceramide concentration. Activation of Jun kinase and NFκB and poly(ADP-ribose) polymerase cleavage are identical in mock- and nSMase-transfected cells. Tumor necrosis factor α triggers the ERK1 pathway in none of the cell lines. The cloned nSMase will facilitate further controlled experiments aiming at the definition of a possible role of ceramide as signal transduction molecule.
Sphingomyelin is an abundant constituent in the outer leaflet of the lipid bilayer of plasma membranes of all mammalian cells (1). In addition, the catabolic metabolism of sphingomyelin generates several lipid intermediates with potential second messenger functions, e.g., ceramides (2), sphingosine (4trans-sphingenine) (3), sphingosine 1-phosphate (4). Only intermediates with the desaturated sphingenine long chain base, the double bond of which is introduced at the ceramide level during biosynthesis, are potential lipid signal molecules. Two sphingomyelinases (SMase; sphingomyelin phosphodiesterase, EC 3.1.4.12), the lysosomal acid sphingomyelinase (aSMase) (5, 6) and the plasma membrane-bound neutral sphingomyelinase (nSMase) (7), determine the major route of sphingomyelin degradation in a phospholipase C-like hydrolysis reaction, yielding ceramides and phosphocholine. Activation of the “sphingomyelin pathway” by acidic and/or neutral SMase in several normal and myeloid cell lines has been described to increase the production of ceramides, which subsequently trigger signaling pathways leading to either cell proliferation and differentiation or to apoptosis (2, 8). Whereas the aSMase is a well characterized enzyme, nSMase has remained elusive despite several purification attempts (9–11). The molecular characterization of this enzyme is a prerequisite for understanding the role of ceramide in cellular signal transduction processes mediating the response to agonists like tumor necrosis factor α (TNF-α), interleukin 1β, interferon-γ, and 1α,25-dihydroxyvitamin D3. Previous studies addressing this question had to either rely on the external administration of membrane-permeable ceramide analogues with their potential membrane-toxic side effects or had to use bacterial SMases, which lack the regulatory features of their mammalian counterparts. Moreover, the bacterial SMases are soluble enzymes and are likely to generate ceramide in other subcellular compartments than the one accessed by the plasma membrane-bound mammalian nSMase.
Here, we describe the cloning, identification, and functional characterization of mouse and human nSMase. Myeloid leukemia cell line U937 was transfected with the nSMases. Stably overexpressing clones were triggered with TNF-α for the study of the activation of the kinases JNK2 and ERK1 and the transcription factor NF-κB as well as apoptosis. No significantly different responses of mock- and nSMase-transfected cells were observed.
MATERIALS AND METHODS
Mouse and Human nSMase cDNA.
Complete mouse and human cDNAs were assembled from mouse (accession numbers AA028477 and AA013912) and human EST clones (accession numbers W32352 and AA056024), respectively. cDNAs were cloned into the HindIII/NotI sites of the multiple cloning site of the eukaryotic expression vector pRc/CMV (Stratagene). Their nucleotide sequences were verified by DNA sequencing with a Perkin-Elmer automatic DNA sequencer model 377A.
Preparation of Poly(A)+ RNA.
Total RNA was isolated from different organs of eight 3-week-old CD1 mice, and poly(A)+ RNA was isolated by affinity purification on oligo(dT)-cellulose according to the manufacturer’s protocol (Boehringer Mannheim).
Cell Culture and Stable Transfection.
Human embryonic kidney 293 (HEK293) cells were grown in DMEM medium supplemented with 10% FCS, 100 μg/ml penicillin/streptomycin, and 1 mM sodium pyruvate. U937 cells were grown in RPMI 1640 medium supplemented with 10% FCS, 100 μg/ml penicillin/streptomycin, and 0.03% glutamine at 37°C under 5% CO2. 5 × 106 cells were transfected with 1 μg of linearized plasmid DNA by electroporation in a gene pulser (Bio-Rad). Stable clones were selected under 1 mg/ml Geneticin (G418, Life Technologies, Gaithersburg, MD).
Measurement of nSMase Activity.
Enzymatic activity was determined in cells and murine tissues. Cells were washed twice with ice-cold PBS and sedimented at 1,000 × g. The cell sediment was resuspended in lysis buffer (50 mM Tris⋅HCl, pH 7.4/5 mM DTT/1 × Complete without EDTA (protease inhibitor set, Boehringer Mannheim)/5 mM EGTA/2 mM EDTA) and disrupted by repeated freezing and thawing. The lysate was centrifuged for 2 min at 2,500 × g. Sediments were extracted with lysis buffer (0.2% Triton X-100) and centrifuged for 15 min at 100,000 × g. Variable amounts of protein or homogenized tissues were incubated with 10 nM (80,000 dpm) [N-14CH3]sphingomyelin for 30 min at 37°C in a total volume of 200 μl. Then 100 μl of double distilled H2O was added, and unreacted substrate was removed by extraction with CHCl3/CH3OH (2:1; vol/vol). The radioactivity of the aqueous upper phase containing the enzymatically released phosphocholine was measured in a liquid scintillation counter.
Tissues of 3-week-old mice were homogenized in cold lysis buffer (0.32 M sucrose/5 mM DTT/50 mM Tris⋅HCl, pH 7.4/1 × Complete/5 mM EDTA) and directly subjected to nSMase activity measurement.
Generation of Polyclonal Antibodies.
Rabbit anti-nSMase antibodies raised against the synthetic peptide NH2-CDPHSDKPFSDHE-COOH resembling residues 261–273 of murine nSMase coupled to keyhole limpet hemocyanin were purified by chromatography on ceramic hydroxyapatite (Bio-Rad) and affinity chromatography on ethyl aminohexyl-Sepharose (Pharmacia) covalently linked to the synthetic peptide antigen. Protein concentration was determined by the Bradford procedure with BSA as standard.
Metabolic Labeling.
Mock-transfected and nSMase-overexpressing U937 cells (5 × 106) were metabolically labeled with 2 μCi/ml 1-[14C]acetate (specific activity, 54 mCi/mmol, Amersham Buchler, Braunschweig, Germany) for 48 h in medium containing 10% fetal bovine serum. After stimulation with TNF-α cells were sedimented by centrifugation and washed once with PBS, and total lipids were extracted according to the established method (12). Samples containing equal amounts of radioactivity were spotted on two different 20-cm silica gel thin layer chromatography plates. One of the plates was developed with chloroform/methanol (15:1; vol/vol) for the separation of ceramides and the other one with chloroform/methanol/water (65:25:4; vol/vol/vol) for the separation of total lipid extract. The plates were imaged simultaneously and quantified on a PhosphorImager 445 SI (Molecular Dynamics) by using ImageQuant software with phosphatidylcholine as internal standard.
Assays for JNK2 and ERK1.
2 × 107 cells were stimulated with TNF-α (6.6 × 106 units/mg, kindly provided by BASF-Knoll, Germany) for the indicated time intervals, and the ERK1 (ERK1-CT, Upstate Biotechnology) and JNK2 (JNK2-FL, Santa Cruz) assays were carried out as described by the manufacturer.
Poly(ADP-ribose) Polymerase (PARP) Cleavage.
107 cells were exposed to TNF-α/cycloheximide (CHX), and assays for PARP proteolysis (Boehringer Mannheim) were carried out following the manufacturer’s instructions. Goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma) was used to visualize protein bands with SuperSignal substrate (Pierce).
RESULTS
Cloning of the Murine and Human nSMase cDNAs.
Because the biochemical purification of mammalian nSMase proved difficult (9–11), we used a different approach for the identification of this enigmatic enzyme. Several bacteria, among them Bacillus cereus, Clostridium perfringens, and Leptospira interrogans, are known to contain SMases with pH optima and ion requirements similar to the plasma membrane-associated Mg2+-dependent mammalian nSMase (reviewed in ref. 13). We constructed generalized profiles (14) based on multiple sequence alignments of these bacterial SMases, either by using the complete sequences or by restricting the profile to the most highly conserved regions. In both cases, profile searches in the protein sequence databases identified a single significant match in the ORF Yer019w from the yeast Saccharomyces cerevisiae. By searching the dbEST database of expressed sequence tags we identified several mammalian sequences with high homology to the yeast sequence. We confirmed this homology by cloning and sequencing of the full-length cDNAs assembled from overlapping mouse and human EST sequences, respectively. Subsequent inclusion of the yeast, human, and murine sequences into iterative cycles of profile construction and database searches resulted in significant matches with a large family of Mg2+-dependent phosphodiesterases including exonuclease III and deoxyribonuclease I from the bacterium Escherichia coli. The crystallographic structure of these two nucleases allows the delineation of residues important for Mg2+ binding and for catalysis (15). As indicated in Fig. 1, these residues are totally conserved in the bacterial SMases, the yeast ORF, and the mammalian nSMase candidates, lending further credibility to this newly established superfamily. Comprehensive phylogenetic distance analysis revealed that the bacterial SMases, the yeast ORF Yer019w, and the mammalian sequences form a cluster within the superfamily, thus suggesting the mammalian clones as good candidates for Mg2+-dependent nSMases.
Figure 1.
Alignment of two bacterial SMases Bacillus cerus and Leptospira interrogans, with ORF Yer019w from S. cerevisiae and neutral SMases from mouse and man. The first block contains the catalytic domain present in both prokaryotic and eukaryotic nSMases, and the second block contains the C-terminal extension of the eukaryotic enzymes with the two transmembrane regions boxed. Residues identical or similar in more than 50% of the sequences are shown on black or gray background, respectively. The numbering refers to the mature sequences for the secreted bacterial enzymes and to complete ORFs for the eukaryotic enzymes devoid of signal sequences. The Mg2+-complexing glutamic acid (triangle), asparagine involved in substrate binding (arrow), and the general base histidine (filled circle) are indicated above the sequence.
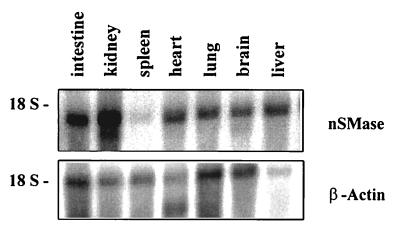
The murine and human nSMase candidates show a high degree of sequence relatedness (Fig. 1). Their ORFs encode proteins of 419 residues (mouse, 47.5 kDa) and 423 residues (human, 47.6 kDa), respectively. In contrast to the bacterial SMases the mammalian cDNA-derived amino acid sequences contain no signal sequence at their N terminus. The hydropathy plot suggests two adjacent hydrophobic membrane-spanning domains at the C terminus separated by eight amino acid residues. The nSMase candidates thus appear to be integral membrane proteins with their catalytic domain facing the cytosol and only a minor portion of the protein facing the extracellular environment. This, too, is in contrast to the bacterial SMases that are secreted soluble proteins but in accordance with the reported properties of mammalian Mg2+-dependent nSMase. The 1.7-kb mRNA of the murine nSMase candidate appeared to be ubiquitously expressed as visualized by Northern blot hybridization analysis (16). RNA of intestine, kidney, brain, liver, heart, and lung showed a strong signal, whereas the expression in spleen appeared to be low (Fig. 2). The signal intensities of the nSMase mRNAs in the multitissue Northern blot did not parallel the observed enzymatic activity in the corresponding tissues (Table 1). This indicates a posttranscriptional regulation of nSMase by a so far unknown mechanism. No change in expression level was observed in RNA and enzymatic activity from aSMase-deficient mice (17).
Figure 2.
Expression of nSMase in mouse tissues. Northern blot analysis of poly(A)+ mRNA derived from different murine tissues. A 770-bp sequence at the 5′ end of murine cDNA was used as a hybridization probe. The membrane was rehybridized with a β-actin probe for standardization.
Table 1.
Specific activity of nSMase in different tissues from aSMase-deficient mice (aSMase−/−)
| Tissue | Specific activity, nmol sphingomyelin/mg protein/h |
|---|---|
| Intestine | 113 ± 27 |
| Kidney | 1.6 ± 0.1 |
| Spleen | 1.5 ± 0.2 |
| Heart | 4.8 ± 0.1 |
| Lung | 1.2 ± 0.1 |
| Brain | 582 ± 98 |
| Liver | 2.0 ± 0.5 |
aSMase−/− mice were used to avoid contamination of the enzyme assay by aSMase activity. Results represent mean values (±SD) derived from three independent measurements.
The Cloned Phosphodiesterase Shows All Characteristics of Mammalian nSMase.
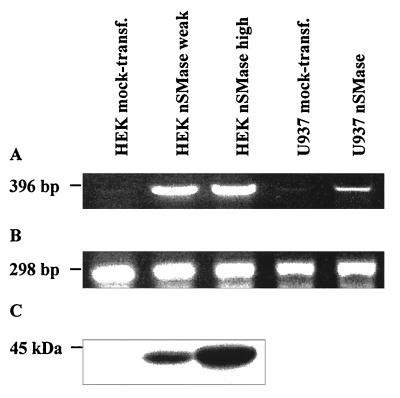
For the comprehensive characterization of the functions of the nSMase candidates, we generated HEK293 cell lines stably expressing the murine nSMase candidate protein. Enzymatic assays of membrane extracts showed a strong increase of the nSMase activity compared with mock-transfected cells. Various cell lines exhibited specific nSMase activities between 0.3 and 10 μmol/mg of protein/h. The expression level of the construct and the specific nSMase activity measured were closely correlated as visualized by reverse transcription–PCR (RT-PCR) (Fig. 3 A and B and Table 2). Western blot analysis, with the peptide-derived anti-mouse-nSMase antibody, revealed a similar correlation with the amount of immunoreactive material (Fig. 3C).
Figure 3.
Analysis of nSMase expression by Northern and Western blot hybridization in different stably transfected human cell lines. (A) RT-PCR from total cell RNA with primer pairs hybridizing to both human and murine nSMase cDNA. (B) RT-PCR from total cell RNA with primer pairs fitting to human β-actin cDNA. (C) Western blot analysis of plasma membrane protein extracts from different HEK293 cell lines, separated by 1% SDS-polyacrylamide electrophoresis, with peptide-derived polyclonal anti-nSMase antibody. The sensitivity was insufficient to detect nSMase protein in extracts from U937 cells and murine tissues.
Table 2.
Specific activity of murine nSMase in stably transfected cell lines
| Cell line | Specific activity, nmol sphingomyelin/mg protein/h |
|---|---|
| HEK mock-transfected | 10.0 ± 0.5 |
| HEK weakly expressed nSMase | 685 ± 33 |
| HEK highly expressed nSMase | 9,960 ± 308 |
| U937 mock-transfected | 28.2 ± 0.8 |
| U937 expressed nSMase | 93.1 ± 7.7 |
Results represent mean values (±SD) derived from three independent measurements.
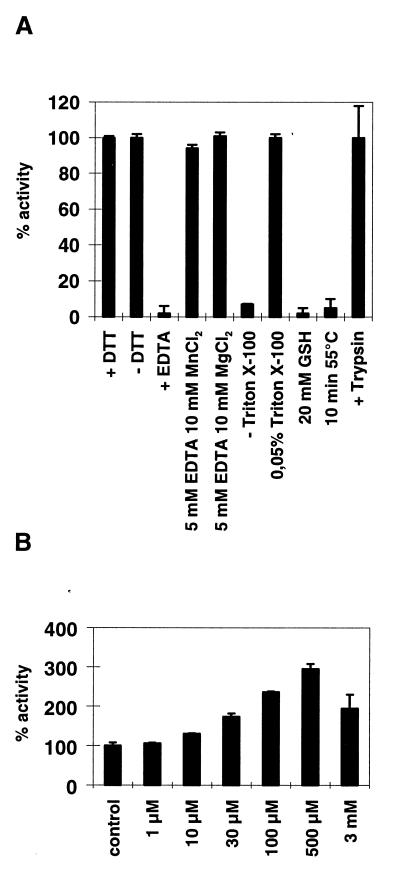
Cell fractionation experiments showed the SMase activity to be present in the plasma membrane-enriched fraction (2,500 × g sediment). Extracts of this fraction were used for the further characterization of the enzyme. The pH optimum was found between 6.5 and 7.5; the Km value for long chain (C18) sphingomyelin amounts to 1.0–1.5 × 10−5 M. Mg2+ ions are required. EDTA addition led to a complete inhibition of SMase activity, which could be reversed by the addition of Mn2+ and Mg2+ ions. Triton X-100 concentration required for optimal enzymatic activity was between 0.03 and 0.05%. Whereas nSMase activity was unaffected by treatment with DTT or 2-mercaptoethanol, addition of 20 mM glutathione led to a complete inhibition (Fig. 4A). A 3-fold activation of nSMase activity in the HEK cell overexpression system was observed on treatment of membrane extracts with 0.5 mM arachidonic acid (Fig. 4B). A similar activation of Mg2+-dependent nSMase activity had previously been observed in HL60 cells and brain homogenate, respectively (18, 19). The hydrolysis capacity of the cloned nSMase is not strictly limited to sphingomyelin; the structurally related phosphatidylcholine was found to be cleaved with approximately 30% efficiency. The enzymatic degradation of sphingomyelin follows a phosphodiesterase reaction. The cloned enzyme does not catalyze a phosphocholine or choline transfer from sphingomyelin to diacylglycerol or phosphatidic acid and vice versa from phosphatidylcholine to ceramide (data not shown). The mammalian nSMases contain a C-terminal extension relative to their bacterial homologues, including the two membrane-spanning regions. This extension, or at least a part of it, is required for enzymatic activity. A truncated murine nSMase (residues 1–282) in which the two putative transmembrane domains were deleted did not show increased SMase activity when expressed in HEK293 cells.
Figure 4.
Enzymatic properties of murine nSMase. (A) Influence of reducing agents on murine nSMase activity, ion requirement, dependence on the nonionic detergent Triton X-100, heat inactivation, and trypsin sensitivity. Standard assay mixtures contain 5 mM MgCl2, 0.05% Triton X-100, 5 mM DTT, 50 mM Tris⋅Cl, pH 7.4, and 1 × complete without EDTA. Trypsin treatment of nSMase-overexpressing cells was carried out for 5 min at 37°C following protease inactivation by the addition of culture medium containing 10% FCS. (B) Stimulation of nSMase activity by arachidonic acid was essentially carried out as described (18, 19). Briefly, different amounts of arachidonic acid were added directly to the assay mixture containing membrane extracts of mock-transfected and nSMases overexpressing HEK293 cells. Results represent mean values (±SD) derived from three independent measurements.
The localization of nSMase in the plasma membrane and all the enzymatic properties described above correspond well to those reported for the neutral SMase in tissues, notably in brain. We therefore conclude that the cloned phosphodiesterase is the long sought Mg2+-dependent neutral SMase.
The Role of nSMase in Signal Transduction.
In some cell systems, ceramide is produced by SMase in response to cytokine challenge, e.g., TNF-α (20, 21), interleukin 1β (22, 23), and CD95/Fas (24–26). For the activation of the plasma membrane nSMase by TNF-α the protein FAN (factor associated with nSMase) has been proposed to link the nSMase activation domain of the p55 TNF receptor and nSMase and to promote proinflammatory cellular responses (27, 28). We studied the influence of nSMase overexpression on the response of HEK293 and U937 cells to a challenge with TNF-α. Four essential parameters of the postulated nSMase-mediated signaling pathways were analyzed as described in the following paragraphs: (i) the kinetics of ceramide production; (ii) activation of NF-κB; (iii) activation of mitogen-activated protein kinases; and (iv) PARP proteolysis, a hallmark of apoptosis.
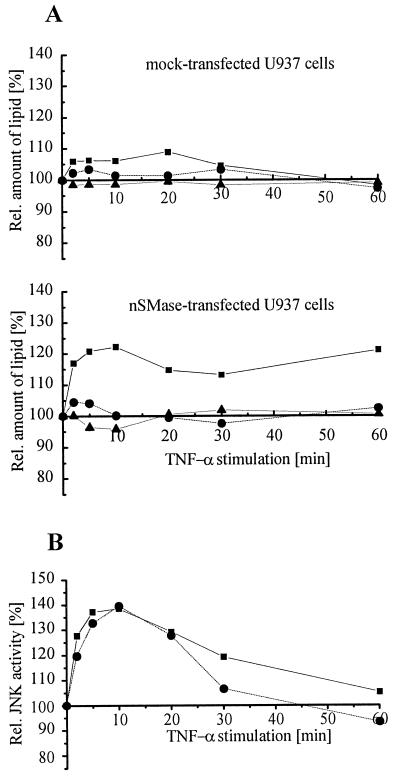
(i) In HEK293 cells prelabeled with 1-[14C]palmitate, the amount of ceramide formation on TNF-α stimulation was found to be independent of the degree of nSMase expression and identical to the level observed in mock-transfected cells. Moreover, no increase in NF-κB activation and no apoptotic features (PARP cleavage, nuclear chromatin fragmentation) were observed (data not shown). In contrast, in U937 cells moderately overexpressing the nSMase construct the TNF-α-triggered ceramide formation was increased by 25% within the first 10 min and remained 15–20% above the level at zero time or in mock-transfected cells (Fig. 5A) (12). In these experiments cellular ceramide concentrations were not determined by the commonly used diacylglycerol kinase assay (29) because its reliability appears questionable in the light of a recent study (30). Instead, we used the radiolabeling technique as described previously (31). Minor changes in the sphingomyelin concentration may escape the sensitivity of the assay because of the high concentration of sphingomyelin in plasma membrane.
Figure 5.
Cellular response of nSMase-overexpressing U937 cells challenged with TNF-α. (A) Kinetics of ceramide release after TNF-α stimulation of mock-transfected and nSMase-overexpressing U937 cells. Cells were stimulated with 100 ng/ml human TNF-α for the times indicated in the figure, and lipids were extracted as described (12). Ceramide (▪), sphingomyelin (•), and phosphatidylethanolamine (▴) were normalized to phosphatidylcholine. (B) Kinetics of JNK2 activation in mock-transfected (▪) and nSMase-overexpressing U937 cells (•) after treatment with 100 ng/ml human TNF-α.
(ii) Following the protocol of our previous study on the effect of TNF-α on aSMase-deficient mouse fibroblasts (31) we studied the kinetics of TNF-α-induced NF-κB activation in nSMase-transfected U937 cells. The electrophoretic mobility shift analysis revealed kinetics identical with that of mock-transfected cells (data not shown) making a role of nSMase in NF-κB activation in this cell type unlikely.
(iii) Ceramide generated by the nSMase has been proposed to trigger an anti-apoptotic pathway via a ceramide-activated protein kinase in response to TNF-α (32). This eventually leads to the activation of the extracellular signal-related kinases. Significant ERK1 activation could not be measured in mock-transfected or in nSMase-overexpressing U937 cells after stimulation with TNF-α (not shown). Additionally, no difference in the activation of Jun kinase was observed in mock-transfected and nSMase-transfected U937 cells (Fig. 5B).
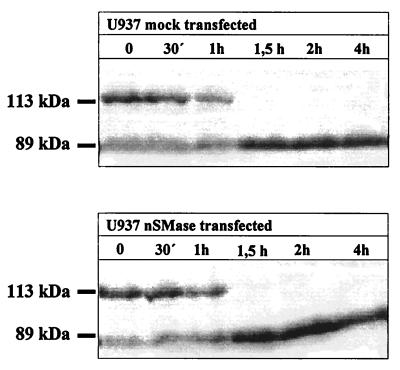
(iv) Apoptosis in U937 cells transfected with nSMase was demonstrated by the determination of PARP cleavage (33). This method clearly indicated that nSMase overexpressing U937 cells and mock-transfected U937 cells showed no difference in the kinetics of the occurring PARP cleavage after TNF-α/CHX stimulation, which was complete within 120 min (Fig. 6). Treatment of the cells with TNF-α alone led only to partial PARP cleavage, which remained incomplete for at least 18 h. CHX alone had no effect (data not shown). Furthermore, overexpression of nSMase even in the highly effective HEK293 cell system had no obvious effect on cell growth and morphology.
Figure 6.
Kinetics of PARP cleavage after treatment of transfected U937 cells with 200 ng/ml human TNF-α and 10 μg/ml CHX.
DISCUSSION
We report here the cloning and molecular characterization of the mouse and human neutral SMase, a phosphodiesterase that belongs to a large family of Mg2+-dependent phosphodiesterases with neutral pH optimum. The mouse and human nSMases are 419 and 423 amino acid residues long, respectively. In the hydropathy plot the primary structure reveals two putative hydrophobic transmembrane domains at the C-terminal end by which the enzyme is bound as a bitopic protein to the plasma membrane. The membrane-bound nSMase overexpressed in transfected HEK293 cells is insensitive to proteolytic enzymes. This suggests that the short loop of 6–10 amino acid residues connecting the two transmembrane helices (boxed sequences in Fig. 1) at the outer cell surface is resistant to proteolysis and the catalytic one is located intracellularly.
Although nSMase is expressed in all organs except spleen with comparable intensities on the mRNA level a high enzymatic activity occurs only in brain. This suggests a posttranscriptional regulation in the other organs, the mechanism of which has to be unraveled.
Lysosomal acid (aSMase) and plasma membrane-integrated Mg2+-dependent nSMase have been suggested to be the final target of several agonists (e.g., TNF-α, interleukin 1β, NGF, 1α,25-dihydroxyvitamin D3, interferon γ) leading to complex cellular responses such as growth arrest, cell differentiation, and apoptosis. This implicates that ceramide, the reaction product of the SMase reaction, is an important second messenger (33).
The postulated aSMase activation by TNF receptor 1 or Fas with the release of ceramide as second messenger followed by NF-κB activation or the induction of apoptosis has been questioned (30, 31). Ceramide released by the induction of a plasma membrane-bound Mg2+-dependent nSMase has been postulated as intracellular signal for apoptosis. nSMase activation has also been linked to extracellular signal-regulated ERK1 cascade and proinflammatory response (32). In contrast to these reports our nSMase-overexpressing U937 cells release only moderately elevated concentrations of ceramide on TNF-α stimulation. We observed no activation of ERK1.
In summary we were not able to detect any biological effects influenced by the overexpressing of nSMase in contrast to the effects expected from studies applying bacterial nSMase or membrane-permeable ceramides. It cannot be excluded that some of the more dramatic effects in the former reports are because of the nonphysiological setup of the experiments. On the other hand, it cannot be ruled out that the mammalian nSMase gives rise to more pronounced effects in very specific cell types and/or requires particular activating factors in vivo that were not present in sufficient amounts in the experiments described here. This hypothesis is supported by the observation that even HEK cells highly overexpressing nSMase show no obviously altered rate of sphingomyelin hydrolysis compared with mock-transfected cells probably because of an effective inhibition (regulation) of the enzyme activity in the plasma membrane. By far the highest enzymatic activity of nSMase of all tissues is measured in the central nervous system. This selectivity is not reflected in the ubiquitous distribution of the nSMase message suggesting a brain-specific activation process of nSMase. The nerve growth factor receptor (p75NTR) belongs to the TNF receptor family and regulates cell death of retinal neurons in early development (34). Later in development the binding of its ligand NGF to p75NTR selectively activates NF-κB (35) and ERK1 and -2 (36). NF-κB activity is constitutively present in nuclei of neurons and inducible in the cytoplasm and the synapse (for review see ref. 37). Deficiency of NGF leads to neuronal death. Binding of NGF to p75NTR activates SMase with the release of ceramide (38, 39). Ceramide produced by exogenously added bacterial SMase has been described to overcome NGF deprivation (40). The high specific activity of nSMase in neuronal tissue might thus be essential for neuronal survival.
The major question to identify and unravel the mechanisms of the activation of SMases involved in agonist-induced degradation of sphingomyelin remains to be answered. However, the characterization of the molecular properties of nSMase described here will facilitate studies addressing these questions.
The regulated activity of nSMase also might play an important role in maintaining the equilibrium between sphingomyelin and cholesterol in the plasma membrane. Treatment of CHO-7 (Chinese hamster ovary) cells with bacterial SMase led to the mobilization and intracellular accumulation of cholesterol followed by an inhibition of sterol regulatory element binding protein proteolysis and of transcription of sterol regulatory element-containing genes (41). The characterization of the molecular properties of nSMase described here will facilitate also studies addressing the role of plasma membrane fluidity in signal transduction, which might be regulated by the stoichiometry of sphingomyelin and cholesterol.
In addition, the molecular characterization of nSMase will essentially contribute to our understanding of signaling pathways mediated by sphingolipid metabolites. A “loss of structure and loss of function” mouse model will reveal insight into the complex regulation of the enzymatic activity of nSMase in the plasma membrane and its functions in cellular signaling pathways.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Project Sto32/36-1 (to W.S.) and by the Federal Ministry of Education, Science, Research and Technology (BMBF), Project 01 KS 9502 in the Interdisciplinary Center of Molecular Medicine, Cologne (ZMMK), Project 24. cDNA clones were obtained from the Resource Center/Primary Database of the German Human Genome Project, Berlin, Germany. S.T. is a fellow of the Graduiertenkolleg “Molekularbiologische Grundlagen pathophysiologischer Vorgänge.”
ABBREVIATIONS
- SMase
sphingomyelinase
- aSMase
acid sphingomyelinase
- nSMase
neutral sphingomyelinase
- TNF
tumor necrosis factor
- PARP
poly(ADP-ribose) polymerase
- CHX
cycloheximide
- RT-PCR
reverse transcription–PCR
- NGF
nerve growth factor
Footnotes
References
- 1.Slotte J P, Härmälä A-S, Jansson C, Pörn M I. Biochim Biophys Acta. 1990;1030:251–257. doi: 10.1016/0005-2736(90)90301-4. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki T, Bell R M, Hannun Y A. J Biol Chem. 1989;264:19076–19080. [PubMed] [Google Scholar]
- 3.Zhang H, Desai N, Olivera A, Seki T, Brooker G, Spiegel S. J Cell Biol. 1991;114:155–167. doi: 10.1083/jcb.114.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivera A, Spiegel S. Nature (London) 1993;365:557–560. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 5.Brady R, Kanfer J N, Mock M B, Fredickson D S. Proc Natl Acad Sci USA. 1965;55:366–369. doi: 10.1073/pnas.55.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barenholz Y, Roitman A, Gatt S. J Biol Chem. 1966;241:3731–3737. [PubMed] [Google Scholar]
- 7.Hostetler K Y, Yazaki P. J Lipid Res. 1979;20:456–515. [PubMed] [Google Scholar]
- 8.Pena L A, Fuks Z, Kolesnick R. Biochem Pharmacol. 1997;53:615–621. doi: 10.1016/s0006-2952(96)00834-9. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson A. Biochim Biophys Acta. 1968;164:575–584. doi: 10.1016/0005-2760(68)90187-2. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee S, Ghosh N. J Biol Chem. 1989;264:12554–12561. [PubMed] [Google Scholar]
- 11.Maruyama E N, Arima M. J Neurochem. 1989;52:611–618. doi: 10.1111/j.1471-4159.1989.tb09163.x. [DOI] [PubMed] [Google Scholar]
- 12.Bligh E, Dyer W. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Spence M W. Adv Lipid Res. 1993;26:3–23. [PubMed] [Google Scholar]
- 14.Bucher P, Karplus K, Moeri N, Hofmann K. Comput Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 15.Mol C, Kuo C, Thayer M, Cunningham R, Tainer J. Nature (London) 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Otterbach B, Stoffel W. Cell. 1995;81:1053–1061. doi: 10.1016/s0092-8674(05)80010-8. [DOI] [PubMed] [Google Scholar]
- 18.Jayadev S, Linardic C M, Hannun Y A. J Biol Chem. 1994;269:5757–5763. [PubMed] [Google Scholar]
- 19.Larrick J W, Wright S C. FASEB J. 1990;4:3215–3223. doi: 10.1096/fasebj.4.14.2172061. [DOI] [PubMed] [Google Scholar]
- 20.Yanaga F, Watson S P. FEBS Lett. 1992;314:297–300. doi: 10.1016/0014-5793(92)81493-6. [DOI] [PubMed] [Google Scholar]
- 21.Dressler K A, Mathias S, Kolesnick R N. Science. 1992;255:1715–1718. doi: 10.1126/science.1313189. [DOI] [PubMed] [Google Scholar]
- 22.Ballou L R, Chao C P, Holness M A, Barker S C, Rhagow R. J Biol Chem. 1992;267:20040–20050. [PubMed] [Google Scholar]
- 23.Mathias S, Younes A, Kan C C, Otlow I, Joseph C, Kolesnick R N. Science. 1993;259:519–522. doi: 10.1126/science.8424175. [DOI] [PubMed] [Google Scholar]
- 24.Cifone M G, De Maria R, Roncaioli P, Rippo M R, Azuma M, Lanier L L, Santoni A, Testi R. J Exp Med. 1994;180:1547–1552. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cifone M G, Roncaioli P, De Maria R, Camarda G, Santoni A, Ruberci G, Testi R. EMBO J. 1995;14:5859–5868. doi: 10.1002/j.1460-2075.1995.tb00274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamen S, Marzo I, Anel A, Pineiro A, Naval J. FEBS Lett. 1996;390:232–237. doi: 10.1016/0014-5793(96)00666-7. [DOI] [PubMed] [Google Scholar]
- 27.Adam-Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider-Mergener J, Krönke M. Cell. 1996;86:937–947. doi: 10.1016/s0092-8674(00)80169-5. [DOI] [PubMed] [Google Scholar]
- 28.Adam D, Wiegmann K, Adam-Klages S, Ruff A, Krönke M. J Biol Chem. 1996;271:14617–14622. doi: 10.1074/jbc.271.24.14617. [DOI] [PubMed] [Google Scholar]
- 29.Schütze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Krönke M. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 30.Watts J D, Gu M, Polverino A J, Patterson S D, Aebersold R. Proc Natl Acad Sci USA. 1997;94:7292–7296. doi: 10.1073/pnas.94.14.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zumbansen M, Stoffel W. J Biol Chem. 1996;272:10904–10909. doi: 10.1074/jbc.272.16.10904. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin X H, Basu S, McGinley M, Chan-Hui P Y, Lichenstein H, Kolesnick R. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 33.Hannun Y A, Obeid L M. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 34.Frade J, Rodriguez-Tebar A, Barde Y. Nature (London) 1996;383:166–168. doi: 10.1038/383166a0. [DOI] [PubMed] [Google Scholar]
- 35.Carter B, Kaltschmidt C, Kaltschmidt B, Offenhauser N, Bohm-Matthaei R, Baeuerle P, Barde Y. Science. 1996;272:542–545. doi: 10.1126/science.272.5261.542. [DOI] [PubMed] [Google Scholar]
- 36.Volonte C, Loeb D, Greene L. J Neurochem. 1993;61:664–672. doi: 10.1111/j.1471-4159.1993.tb02171.x. [DOI] [PubMed] [Google Scholar]
- 37.O’Neill L, Kaltschmidt C. Trends Neurosci. 1997;20:252–258. doi: 10.1016/s0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- 38.Dobrowsky R, Werner M, Castellino A, Chao M, Hannun Y. Science. 1994;265:1596–1599. doi: 10.1126/science.8079174. [DOI] [PubMed] [Google Scholar]
- 39.Dobrowsky R, Jenkins G, Hannun Y. J Biol Chem. 1995;270:22135–22142. doi: 10.1074/jbc.270.38.22135. [DOI] [PubMed] [Google Scholar]
- 40.Ito A, Horigome K. J Neurochem. 1995;65:463–466. doi: 10.1046/j.1471-4159.1995.65010463.x. [DOI] [PubMed] [Google Scholar]
- 41.Scheek S, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1997;94:11179–11183. doi: 10.1073/pnas.94.21.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]