Abstract
Current knowledge about developmental processes in complex organisms has relied almost exclusively on analyses of fixed specimens. However, organ growth is highly dynamic, and visualization of such dynamic processes, e.g. real-time tracking of cell movement and tissue morphogenesis, is becoming increasingly important. Here, we use live imaging to investigate expansion of the embryonic pancreatic epithelium in mouse. Using time-lapse imaging of tissue explants in culture, fluorescently labeled pancreatic epithelium was found to undergo significant expansion accompanied by branching. Quantification of the real-time imaging data revealed lateral branching as the predominant mode of morphogenesis during epithelial expansion. Live imaging also allowed documentation of dynamic ß-cell formation and migration. During in vitro growth, appearance of newly formed ß-cells was visualized using pancreatic explants from MIP-GFP transgenic animals. Migration and clustering of ß-cells were recorded for the first time using live imaging. Total ß-cell mass and concordant aggregation increased during the time of imaging, demonstrating that cells were clustering to form “pre-islets”. Finally, inhibition of Hedgehog signaling in explant cultures led to a dramatic increase in total ß-cell mass, demonstrating application of the system in investigating roles of critical embryonic signaling pathways in pancreas development including ß-cell expansion. Thus, pancreas growth in vitro can be documented by live imaging, allowing visualization of the developing pancreas in real-time.
Keywords: Real-time imaging, pancreas development, organ culture, MIP-GFP, branching morphogenesis, ß-cell
Introduction
In the developing mouse embryo, the first morphological sign of pancreas formation is a thickening of the dorsal endodermal epithelium caudal to the stomach by embryonic day 9 (e9) (Kim and Hebrok, 2001). Soon thereafter, two ventral buds appear next to the liver diverticulum by e10.5. In the following days, the pancreatic epithelium penetrates into the surrounding mesenchyme by extensive growth and branching. Concomitantly, endocrine, exocrine, and ductal cell fates are specified (Murtaugh and Melton, 2003). These processes are regulated by a cascade of transcription factors that defines pancreatic cell differentiation (Jensen, 2004; Wilson et al., 2003). Pancreatic duodenal homeobox 1 (Pdx1), one of the earliest transcription factors expressed in the specified pancreas, is essential for determination of all cell types in the mature pancreas (Gu et al., 2002; Jonsson et al., 1994). Thus, Pdx1 expression marks a pluripotent population of cells in the epithelium. In addition to transcriptional control, extrinsic factors determine the fates adopted by the pancreatic epithelium. Using pancreatic bud culture, it has been demonstrated that the mesenchyme surrounding the epithelium plays an instructive role in pancreas development (Gittes et al., 1996; Miralles et al., 1998). While essential for exocrine cell development, endocrine differentiation within the pancreatic epithelium is suppressed by close proximity to the mesenchyme (Li et al., 2004).
Branching morphogenesis of the pancreatic epithelium remains poorly understood. Analysis of fixed tissue samples has revealed that the epithelial sheet organizes itself around the lumen to undergo tubulogenesis (Jensen, 2004). However, development of a single layer of epithelium into a complex 3-dimensional aggregate of cells with distinct fates is a highly dynamic process. Although instructive, analysis of fixed specimens only allows visualization of a single time point in development. In contrast, real-time analysis provides a dynamic view of events that are critical to the formation and function of an organ. While real-time imaging in pancreas is still in its infancy, important information has been gained in other tissues as well as other model organisms, including mouse kidney and salivary glands, fruitfly tracheae, and zebrafish vasculature (Costantini, 2006; Davies, 2005; Larsen et al., 2006). Time-lapse imaging was used to document the branching “tree” pattern of the mouse ureteric bud labeled specifically using green fluorescent protein (GFP) (Watanabe and Costantini, 2004). In addition to describing normal development of the ureteric bud, the authors also determined the effect of a MEK inhibitor on branching within the bud. Thus, live imaging allowed the authors to obtain quantitative data about organ formation including the rate of epithelial elongation and the sequence of branching.
Transgenic mouse lines that fluorescently mark pancreatic cells are live-imaging tools for investigating the various aspects of pancreas development, for e.g. describing branching morphogenesis of the epithelium, specification of endocrine cells and their migration to form clusters, and acinar and ductal cell differentiation. The Pdx1-GFP transgenic line labels all the pancreatic progenitors in the epithelium as early as e9.5 (Holland et al., 2006). Tissue explants from Pdx1-GFP embryos thus allow visualization of tissue expansion and morphogenetic movements of the epithelium. During endocrine development in the mouse pancreas, specified cells delaminate from the epithelium while undergoing maturation and clustering to form the classic islet morphology. Although these stages have been identified, the dynamic movement of individual cells into islet clusters is unclear. Transgenic mice with fluorescently labeled ß-cells have been generated (Gunawardhana et al., 2005; Hara et al., 2003; Hara et al., 2006) and these mice provide an opportunity to investigate ß-cell behavior in real-time.
Here, we report the use of real-time imaging to monitor events during pancreatic expansion in culture. The mode of epithelial branching is described using fluorescently marked pancreatic progenitors. In addition, we document the appearance of newly formed ß-cells and migration of these cells to form clusters with other cells. To our knowledge, this is the first study to successfully capture and describe the expansion of the embryonic pancreatic epithelium and ß-cell mass in real-time.
Materials and Methods
Embryonic bud dissection and in vitro culture
Embryonic days were determined by observation of the vaginal plug, which was counted as day 0.5 after mating. Pregnant female mice were sacrificed in accordance with the protocol approved by the Animal Research and Care committee. Embryos of the desired stage were removed and the pancreatic anlage, including pancreatic buds (with the surrounding mesenchyme), caudal stomach, and proximal duodenum, was isolated (denoted as organ rudiments or pancreatic explants). Pancreatic explants were stored in sterile PBS on ice until all dissections were completed. For growth, organ explants were transferred to coverglass-bottomed dishes (MatTek Corp. Ashland, MA), coated with 20μl Matrigel™ (BD Biosciences, San Jose, CA) and placed in a 37°C incubator for 20min to solidify the Matrigel™. The rudiments were then covered in growth medium (DME H-16 50%/F-12 50% with 10% FBS, antibiotics and insulin-transferrin-selenium). The medium was changed every two to three days. The day of dissection was scored as Day 0. For modulation of Hedgehog signaling, cyclopamine (Toronto Research Chemicals, Ontario, Canada) was resuspended in a 1:1 ratio of ethanol and dimethylsulfoxide at a stock concentration of 10mM. Recombinant mouse Sonic Hedgehog (R&D Systems, Minneapolis, MN) was resuspended in PBS with 0.1% bovine serum albumin at a stock concentration of 300μg/ml. Removal from Matrigel™ was carried out using MatriSperse (BD Biosciences, San Jose, CA) as per manufacturer's instructions.
Mouse strains
For histological analysis, wild-type CD1 mice (Jackson Laboratory) were used. The Pdx1-Cre mice were a gift from Dr. Doug Melton (Harvard); the MIP-GFP mice were a gift from Dr. Manami Hara (Univ. of Chicago); the Pdx1-GFP mice were obtained from Dr. Andrew Holland (Monash University), and the Z/EG mice were obtained from the Jackson Laboratory.
RNA preparation, RT-PCR and quantitative PCR analysis
Total RNA was prepared from dissected pancreata using the RNeasy™ Mini Kit (QIAGEN Sciences, Maryland) following manufacturer's instructions. RT-PCR was carried out using random hexamer primers for reverse transcription as per manufacturer's instructions in the SuperScript™ First-strand kit (Invitrogen, Carlsbad). Forward and reverse primers used to amplify the pancreatic genes were as follows: Amylase2, 5' GTG GTC AAT GGT CAG CCT TT 3' and 5' CCA TCA CTG CCA ACA TTC AC 3'; Pancreatic Polypeptide, 5' TAC TGC TGC CTC CCT GTT, and 5' CCA GGA AGT CCA CCT GTG TT 3'; Ngn3, 5' GAG TTG GCA CTC AGC AAA CA 3', and 5' TCT GAG TCA GTG CCC AGA TG 3'; Nkx2.2, 5' GTC GCT GAC CAA CAC AAA GA 3', and 5' GTT GTC GCT GCT GTC GTA GA 3'; Sox9, 5' CGA CTA CGC TGA CCA TCA GA 3', and 5' AGA CTG GTT GTT CCC AGT GC 3'; Sox15, 5' CGG CGT AAG AGC AAA AAC TC 3' and 5' TGG GAT CAC TCT GAG GGA AG 3'; HNF1α, 5' ACT TGC AGC AGC ACA ACA TC 3', and 5' GAA TTG CTG AGC CAC CTC TC 3'; Isl1, 5' TCA TCC GAG TGT GGT TTC AA 3' and 5' CCA TCA TGT CTC TCC GGA CT 3'; Nkx6.1, 5' TCA GGT CAA GGT CTG GTT CC 3' and 5' CGA TTT GTG CTT TTT CAG CA 3'; Actin, 5' AGC CAT GTA CGT AGC CAT CC 3' and 5' CTC TCA GCT GTG GTG GTG AA 3'. Quantitative PCR was carried out as previously described (Sekine et al., 2006), except that RNA was prepared as described above. Cyclophilin A was used as the housekeeping gene. Sequence primers are available on request.
Immunohistochemical analysis
Pancreatic rudiments grown in culture for 6 or 7 days were fixed in 4% (w/v) paraformaldehyde (PFA) for 2 to 3 hours at 4°C. Hematoxilin/Eosin staining was carried out on paraffin sections using previously described conditions (Kim et al., 1997). Immunofluorescence analysis was carried out on tissue whole mounts as follows. Organ rudiments were recovered from the surrounding Matrigel™ using MatriSperse and fixed as above, followed by 3 washes in PBS and incubation in block (5% goat serum in PBS with 0.1% Triton X-100) at 4°C overnight. Primary and secondary antibody incubations were carried out for 10-12 hours at 4°C, with 5 washes with PBS between antibodies. At the end of the secondary incubation, the pancreatic explants were mounted on a glass slide in VectaShield and covered with a cover slip for microscopy. The primary antibodies used for analyses were as follows: guinea pig anti-Pdx1, diluted 1:1000 (a gift from Dr. Mike German); Armenian hamster anti-Mucin 1, diluted 1:200 (NeoMarkers, HM-1630); guinea pig anti-insulin, diluted 1:400 (Linco, 4011-01); rabbit anti-glucagon, diluted 1:400 (Linco 4030-01F); rabbit anti-amylase, diluted 1:500 (Sigma, A8273); and guinea pig anti-pancreatic polypeptide, diluted 1:400 (Linco, 4041-01). The following secondary antibodies were used for immunofluorescence at 1:200 dilutions: Cy3-conjugated anti-guinea pig (Jackson ImmunoResearch), Alexa488-conjugated anti-rabbit (Molecular Probes), Alexa488-conjugated anti-guinea pig (Molecular Probes), Cy3-conjugated anti-Armenian hamster (Jackson ImmunoResearch). Bright-field images of Hematoxilin/Eosin stained samples were acquired using a Zeiss Axio Images D1 scope. The Leica DMIRE2 SP2 Laser Confocal microscope was used to capture fluorescent images.
Real-time imaging and image analysis
Live imaging of the explants was carried out using a Leica DMIRE2 SP2 confocal microscope, with an automated motorized stage, in a humidified heated chamber with CO2. For acquisition, 488nm laser light was used to excite samples under a 10X objective with a 1.5X optical zoom, and digital images were captured through multiple z-steps (8-10μm apart) using a GFP filter set every 20min. The total time of imaging varied from a minimum of 15hrs to a maximum of 60hrs. On average, 15-17 sections were acquired per z-stack for any movie. After imaging, z-stacks were collapsed into maximum intensity projections, and all time points were compiled and exported as a Quicktime (avi) file using the Leica software. For quantification of the MIP-GFP-positive objects, ImageJ (NIH) was used to subtract background followed by setting a threshold to maximum projection grayscale TIFF images. The total area of objects and the average size of each object were quantified. The total area occupied by fluorescent objects is represented as normalized to the first time frame.
Quantification of branching
The first frame of a single time-lapse movie was labeled for the number of protrusions that were clearly discernible as independent outgrowths from the central body of the tissue. These were designated as the “primary” bud, and morphological changes in each such bud were monitored over time. Extensions that underwent splitting i.e., the appearance of another outgrowth either laterally or along the axis of extension, were scored as positive for budding. The mode of budding was noted and expressed as a percent of the total modules that were undergoing branching.
Cell tracking analysis
All distance calculations were accomplished using ImageJ software (NIH). Confocal images acquired at each time point of interest were converted to binary images and a threshold applied to exclude noise. To measure distance between two objects, objects were selected using the wand tool and centroids (center of mass of the object) were calculated. The distance between objects was then measured by manually using the line tool. Mtrack2 plugin was used to track movement of single objects over time. The parameters used for calculating the total path length of migrating ß-cells were: minimum pixel area of 2; maximum pixel area of 80; maximum velocity of 10; number of frames, 10. The maximum area cutoff was 80 pixels; this size corresponded to objects with diameters <15μm. This threshold included single cells and small clusters of ß-cells.
Results
Development of an in vitro culture system to document morphogenesis of mouse embryonic pancreas
To achieve a dynamic representation of pancreas development in the mouse, ex vivo culture of embryonic pancreatic tissue explants was established. While the organ rudiments include distal stomach and proximal duodenum, our analysis focuses exclusively on the development of the pancreas (hereafter, we will denote organ rudiments as pancreatic explants). Explants from wild-type CD1 embryos dissected at day 10.5 (e10.5) were cultured in Matrigel™ for several days (Figs. 1A, B). Significant growth occurs under these culture conditions, marked by the appearance of structures resembling acinar tissue by day 6 (Fig. 1D). Morphologically, the pancreatic epithelium in culture was similar to e15.5 embryonic tissue (Fig. 1C). Immunohistochemical analyses of pancreatic explants in culture for 7 days confirmed the expression of the transcription factor Pdx-1 (Fig. 1E, green), the ductal marker mucin (Fig. 1E, red), the exocrine marker amylase (Fig. 1F), the endocrine markers insulin, glucagon (Fig. 1G, red and green, respectively) and pancreatic polypeptide (Fig. 1H). Thus, overall morphology and protein expression analysis reveals that pancreatic explants express ductal, exocrine and endocrine markers in culture.
Figure 1. In vitro expansion of the mouse pancreatic explant.
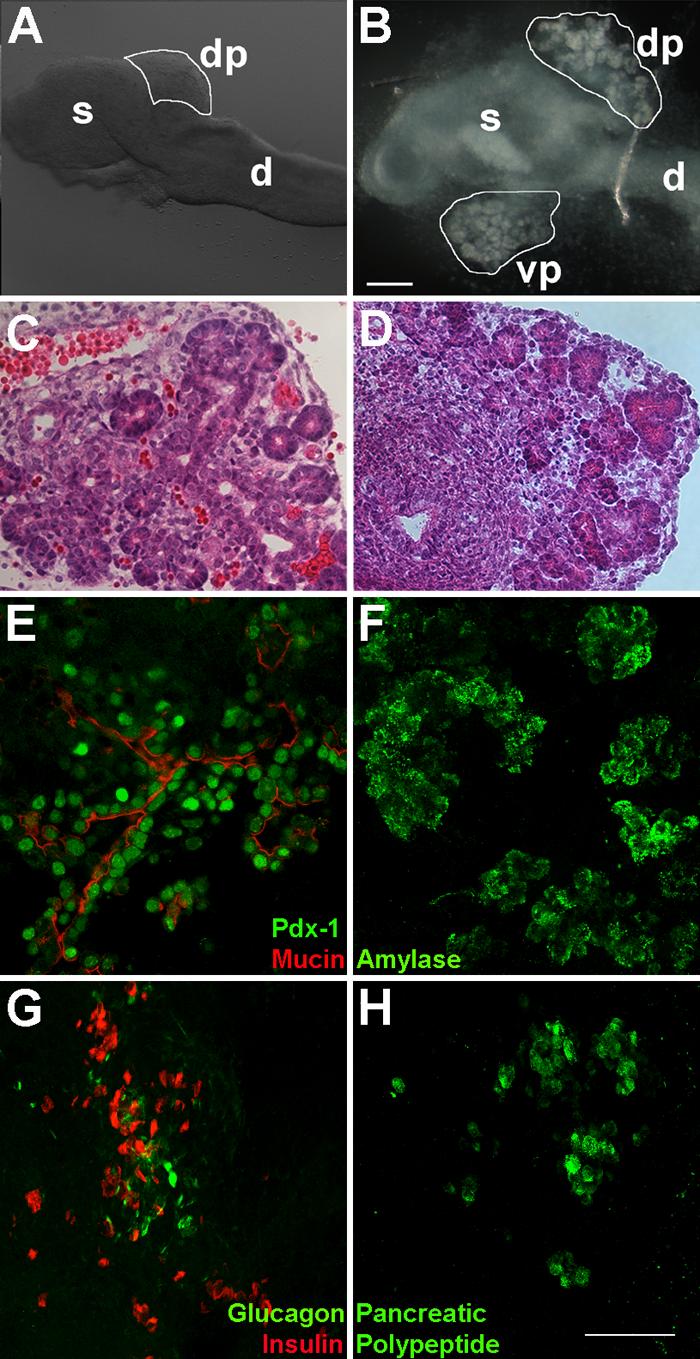
A. e10.5 dorsal pancreatic (dp) bud attached to stomach (s) and the duodenum (d). B. e10.5 explant in culture for 7 days undergoes significant expansion of the dorsal (dp) and ventral buds (vp), resembling acinar structures. Bar, 200μm. To evaluate the morphology of pancreatic growth in vitro, H&E staining was carried out on explants grown in Matrigel™ for 6 days (D). Pancreatic epithelial development closely resembles exocrine morphology at embryonic stage e15.5 (C). Immunohistochemical analysis of pancreatic explants grown in culture for 7 days reveals expression of the pancreatic transcription factor Pdx-1 (green) and the ductal marker mucin (red) (E), exocrine marker amylase (F), and endocrine markers insulin, glucagon (G), and pancreatic polypeptide (H). Bar, 75μm.
Semi-quantitative RT-PCR was used to determine the timeline of mRNA expression of pancreatic markers during culture (Fig. 2). For this, e10.5 pancreatic explants were dissected, embedded in Matrigel™, and cultured in growth media for several days (day 0 being the day of dissection). At various time points (day 1 to day 6), explants were harvested from the Matrigel™ for RNA isolation. RNA was also prepared from uncultured embryonic pancreatic buds harvested at different time points (e10.5-e15.5). As previously reported, the temporal expression of pancreatic marker genes in explants was slightly delayed (approximately 24 hours) (Percival and Slack, 1999). To account for the initial delay, we aligned the gene expression patterns based on the appearance of amylase, a mature pancreatic marker first detected around e13.5 during normal embryonic development. In general, the profile of marker expression in vitro was strikingly similar to the in vivo developmental pattern. Ngn3 expression appeared elevated at day 4 in culture (equivalent to e13.5 in the embryo), marking the onset of the secondary transcription and specification of endocrine progenitors. These data confirm that ex vivo culturing of pancreatic explants occurs in a manner that follows the normal developmental in vivo program.
Figure 2. Pancreatic bud expansion in vitro mimics in vivo marker expression.
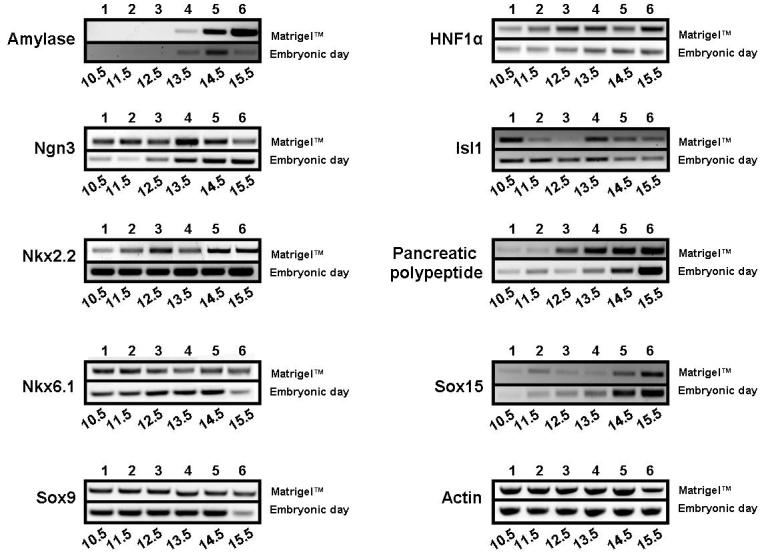
A comparison of the expression profile of pancreas-specific markers during growth in Matrigel™ with in vivo embryonic growth using RT-PCR demonstrates a striking resemblance in expression pattern. Pancreatic explants dissected at e10.5 were grown in Matrigel™ for 6 days, and RNA harvested each day. The top row in each panel represents days in culture. RT-PCR on pancreatic RNA collected from embryos (e10.5 to e15.5) is shown in the lower row in each panel.
Expansion and branching of the pancreatic epithelium in real-time
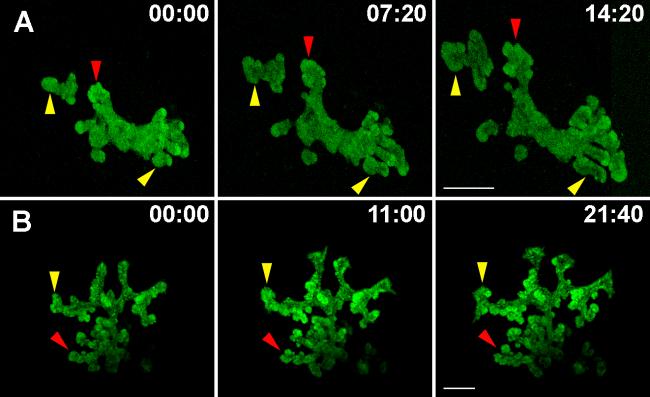
With the goal of documenting dynamic epithelial growth, live imaging was carried out on pancreatic buds in culture. Explants growing in culture dishes were placed in a heated and humidified chamber on top of an inverted laser confocal microscope, and imaged for several hours at regular intervals. To specifically label the pancreatic epithelium during mouse embryonic development, we used the Pdx1-GFP transgenic mouse line, encoding GFP in the Pdx1 locus (Holland et al., 2006). The Pdx1-GFP line is a powerful tool to mark and trace cells that express this key transcription factor. Pancreatic explants were isolated from Pdx1-GFP embryos at e10.5, cultured in Matrigel™ for four days, and imaged in real-time (Fig. 3A and movie 1). There is a clear increase in the overall epithelial mass, as observed in static time frames from the movie (Fig. 3A, yellow arrowheads). In addition, buds appear to pinch off into new buds (Fig. 3A, red arrowheads) indicating tissue re-modeling and re-organization. Pancreatic epithelial expansion and budding can thus be successfully documented using real-time imaging of cultured explants.
Figure 3. Expansion and branching of the embryonic pancreatic epithelium in real-time.
A. Pancreatic explants were dissected at e10.5 from the Pdx1-GFP transgenic embryos and grown in culture for 4 days before imaging. B. Explants expressing Pdx1-Cre-Z/EG were dissected at e11.5 and grown in culture for 4 days followed by imaging. Still images from time-lapse movies show the overall expansion of the pancreatic epithelium (yellow branches) and branching by budding (red arrows). Time of imaging is shown in hours:minutes. Bar, 200μm.
To confirm these observations, the pancreatic epithelium was labeled using the Z/EG transgenic mouse line (Novak et al., 2000). This reporter line encodes the LacZ-stop sequence flanked by LoxP sites, followed by an enhanced GFP coding sequence. Upon Cre recombination, the LacZ-stop is excised and EGFP expression is activated. Crossing the Z/EG strain with a Pdx1-Cre line labels pancreatic progenitors specifically in the epithelium with EGFP. Fluorescently labeled cells are robustly detectable at e10.5-11.5 (Fig. 3B and movie 2). Frames from a time-lapse movie of a Pdx1-Cre;Z/EG explant grown in culture and imaged as described above are shown in figure 3B. Similar to the Pdx1-GFP labeled explant (Fig. 3A), the Pdx1-Cre;Z/EG explant epithelium undergoes expansion and budding during the time of imaging (Fig. 3B, red and yellow arrowheads). Using both labeling techniques, it was observed that the leading edges of the epithelium have greater fluorescence intensity, indicating a higher density of GFP-positive cells. This is in support of previous studies that have observed increased cell division at the “tips” rather than the “dips” of the expanding pancreatic epithelium (Horb and Slack, 2000). Using the assay of growth in culture coupled with live imaging, epithelial expansion was successfully imaged for up to 22 hours (Fig. 3B). An additional advantage of using the Pdx1-Cre line lies in its use for multi-spectral imaging that will enable labeling of the pancreatic epithelium and diverse cell populations with fluorescent markers besides GFP.
Notably, our results indicate that this analysis can be expanded to include early pancreatic tissue. When gut explants from e9.5 Pdx1-GFP embryos were cultured, significant growth was detected as marked by robust GFP expression. As shown in figure 4A (and movie 3), a similar increase in the overall mass of the epithelium and events of budding could be identified (see below). Although the majority of our analysis focuses on explants from e10.5 embryos, this result demonstrates that it is possible to culture the pancreatic epithelium even earlier. This is significant as it provides a tool to observe very early morphogenetic movements within the epithelium.
Figure 4. Description of branching in the pancreatic epithelium during morphogenesis.
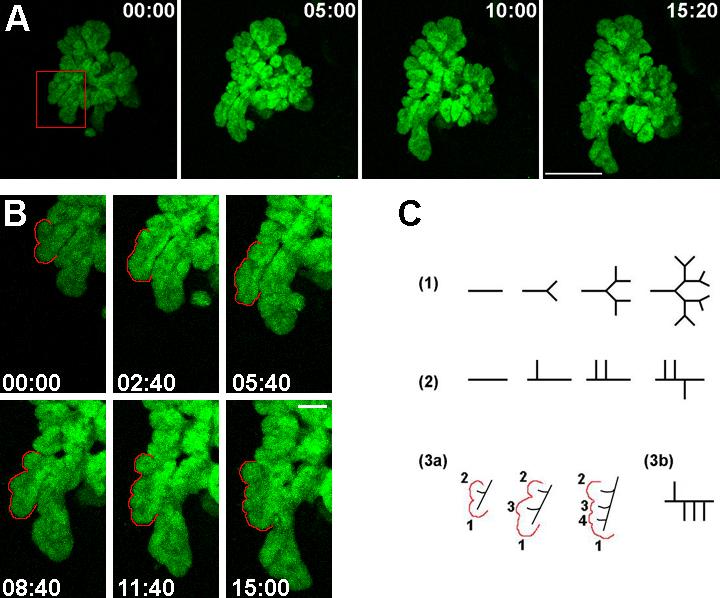
A. Frames from a time-lapse movie of an e9.5 Pdx1-GFP pancreatic explant grown in culture for 4 days before imaging are shown. Bar, 200μm. B. A magnified view of a region of the growing pancreatic epithelium (indicated in the red box in A) depicts branching in the pancreas. Bar, 50μm. Time of imaging is shown in hours:minutes. Lateral buds appear (B, and C, 3a, labeled 2, 3 and 4) as the “primary” bud (labeled as 1) expands. In C, different modes of branching morphogenesis are represented- (1) type “1-2” terminal bifurcation, (2) type “1-3” lateral budding, and (3) lateral branching observed during pancreas growth. The order of appearance of buds exemplifies this lateral mode of expansion in the pancreas.
Lateral branching is the predominant mode of budding in the pancreatic epithelium as observed by real-time imaging
It is clear that branching in the developing pancreas, as in other epithelial organs, is coupled with expansion of the epithelium and fate specification to form the mature structure. Currently, the mode and mechanism of branching in the pancreatic epithelium are not known. Watching the epithelium grow in real-time allows us to address this question. Possible modes of branching characterized by either terminal bifurcation or lateral budding are schematized in figure 4C. The outlined epithelium represented in figure 4B underwent lateral budding with simultaneous elongation of the “primary” bud (labeled as 1 in Fig. 4C, [3a]). Although the pattern of branching represented in the schematic appears restricted to one side of the “primary” bud, protrusions were seen to emerge from either side (data not shown). This mode of branching is similar to that observed in the lung, which predominantly undergoes 1-3 type lateral branching (Fig. 4C, [2]) as opposed to a bifurcation at the tip (Fig. 4C, [1]). However, in contrast to the defined 1-3 lateral branching mode in lungs, a small fraction of the pancreatic epithelium had more than 3 buds emerge laterally from a single “primary” axis during growth (schematized in Fig. 4C, [3b]). Thus, the mode of budding for the pancreas could be identified upon visualization of explants growing in culture.
To quantify what fraction of explants underwent lateral branching as opposed to a bifurcation at the tip (typically seen in the growing ureteric bud), a “branching module” was defined as a single epithelial protrusion that could be discerned as a bud (Fig. 5). Quantification of branching was carried out on movies of explants isolated at different embryonic stages (e9.5, e10.5 and e11.5) and cultured in vitro. Once branching modules were identified, quantification of budding revealed that 86% of modules followed a lateral pattern of branching where a growing epithelium expanded in one direction while buds were formed lateral to it (Figs. 4C, [3], and 5). Only 13% of the modules underwent a bifurcation at the tip (Fig. 4C, [1]). The pancreatic epithelium expands and branches simultaneously in multiple axes, making it a challenge to quantify expansion. Since the analysis is carried out on projected images, the result is a 2-dimensional representation of a 3 dimensional process. This implies an under-representation of the number of lateral budding events, as some might be missed due to the projection perpendicular to the stacked image. Summarily, our analyses reveal that the mode of branching morphogenesis in the pancreatic explants can be visualized, identified, and quantified using live imaging of the expanding epithelium. Furthermore, pancreas branching during growth in culture occurs largely by lateral branching rather than by terminal bifurcation.
Figure 5. Lateral branching is the predominant mode of budding in the pancreatic epithelium during branching morphogenesis.
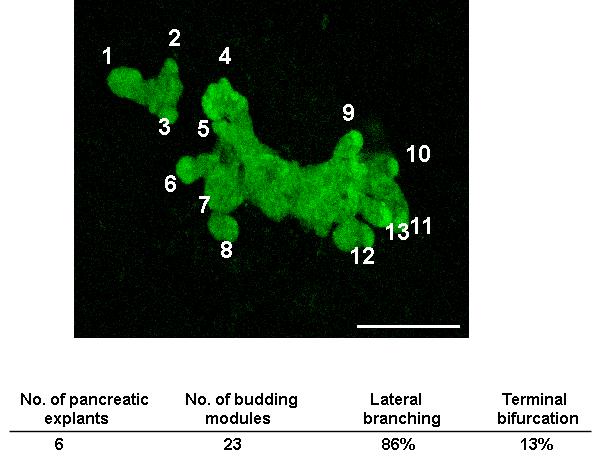
Budding modules were identified and labeled as protrusions in the growing epithelium, and tracked over time for evidence of branching. Quantification of the mode of budding was carried out by counting the modules that underwent lateral budding or terminal bifurcation, and expressed as a percent of total. Bar, 200μm.
Live imaging of insulin-positive ß-cells allows visualization of cellular dynamics in real-time
The branching epithelium gives rise to endocrine progenitor cells that are believed to delaminate and migrate into the surrounding mesenchyme to aggregate into specialized clusters, the islets of Langerhans (Kim and Hebrok, 2001). The MIP-GFP transgenic mouse line expresses GFP under the mouse insulin I promoter, thus allowing for identification and tracking of ß-cells as they are specified through mouse embryonic development (Hara et al., 2003). Using the pancreatic explant culture assay, growth and expansion of ß-cells was visualized using real-time imaging. Approximately 60% of explants in culture expressed GFP-positive cells after 6 days in culture (Fig. 8A, B). As seen in figure 6A and movie 4, numerous ß-cells emerged as GFP-positive objects over 33 hours of imaging. Upon visual analysis, novel green entities appeared over time and the size of these objects increased. Quantification of time-lapse movies was carried out using ImageJ software to calculate the total area occupied by fluorescent objects and the average size of the particles, as described in methods. As seen in figure 6B, there is a significant increase in the total fluorescent area (expressed as a percent of fluorescent area at the start of acquisition). This indicates that over time, ß-cells are specified and accumulate as green fluorescent objects. A similar increase was observed in the average size of all objects, which suggested that ß-cells were clustering together to form aggregates larger than the size of individual cells. Although the classical islet morphology is not formed until the end of gestation in the mouse (Herrera et al., 1991), it is reasonable to suppose that these clusters are precursors to the more mature clusters formed after birth.
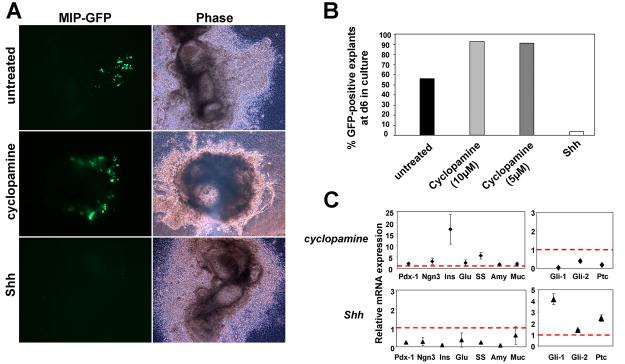
Figure 8. Role of Hedgehog signaling in ß-cell specification in culture.
A. Wide-field images depict the presence of insulin-GFP fluorescence in untreated and cyclopamine treated samples, with negligible GFP signal in the Shh treated sample. Corresponding bright field images show tissue morphology. B. The percentage of pancreatic explants positive for GFP after 6 days in culture were quantified [n= 57 (untreated), 53 (cyclopamine, 10μM), 11 (cyclopamine, 5μM) and 29 (Shh, 300ng/ml)]. C. Real-time quantitative PCR analysis of transcript expression in pancreatic explants in culture for 8 days was carried out (n=2 independent experiments, a total of 12 pancreatic explants for untreated or treated with cyclopamine or Shh). The effects of cyclopamine (5μM) or recombinant Shh (300ng/ml) treatment on pancreas-specific genes as well as components of the Hh signaling pathway are shown. No change in transcript level is indicated by a red dotted line. Treatment with cyclopamine led to a dramatic increase in insulin expression. ins, insulin; glu, glucagon; ss, somatostatin; amy, amylase; muc, mucin; ptc, patched.
Figure 6. Real-time imaging of ß-cells during embryonic pancreatic development.
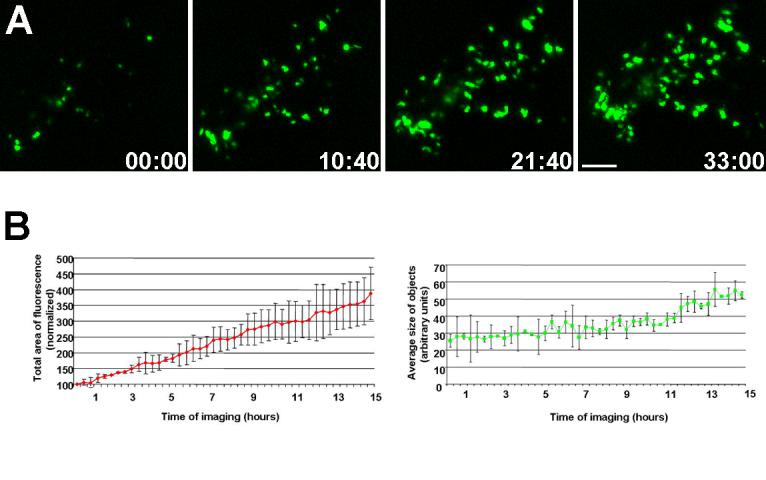
A. Still frames from a time lapse movie depict the increase in the total fluorescence in the field, indicating an increase in ß-cells over time. Time of imaging is shown in hours:minutes. Bar, 100μm. B. Quantification of the fluorescent objects show an increase in the total area of fluorescence and the average size of objects (n=3).
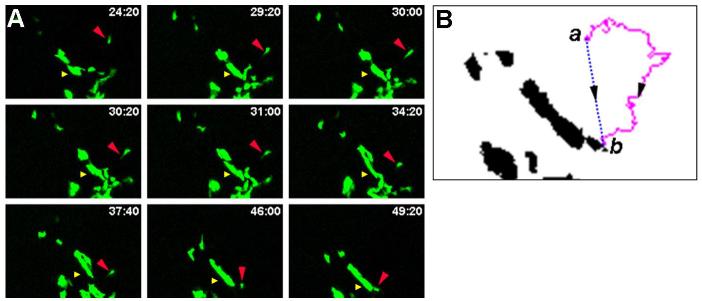
Striking evidence of ß-cell migration was recorded upon live imaging, wherein a subset of cells tracked over significant distances to coalesce with an existing cluster of cells (Fig. 7A and movie 5). A cytoplasmic extension was observed projecting from the individual ß-cell, suggesting communication with the environment. Using the Mtrack2 plugin in ImageJ, the migration of a single cell was measured (Fig. 7A). Over 32hrs, the cell was observed to traverse a path length from point a to point b measuring 365.3μm (average velocity of 11.4μm/hr) (Fig. 7B, magenta tracing). However, the total linear distance it traveled was 102.4μm (Fig. 7B, blue tracing). This demonstrates the necessity of calculating the actual path length as opposed to the total linear distance traveled by a cell to determine the accurate velocity of the cell. This observation further emphasizes the need to visualize cellular dynamics in real-time, as migration of cells through the epithelium is clearly not linear. Significantly, the average velocity of individual ß-cells or ß-cells in small clusters (see materials and methods) was calculated to be 11.27±5.8μm/hr (n=186 objects, 3 independent movies). Time-lapse imaging of GFP-labeled epithelial cells in cultured submandibular salivary gland (SMG) revealed that cells migrated at an average velocity of 16μm/hr (Larsen et al., 2006). Although they do not traverse the basement membrane, epithelial cells in the SMG migrated through meandering paths through the tissue, similar to that observed for ß-cells (Fig. 7B, magenta tracing). These data might suggest a similar role of cell migration for remodeling the epithelium during organ morphogenesis.
Figure 7. Migration of ß-cells towards clusters of cells.
A. Stills from a time-lapse series show migration of a ß-cell towards a larger cluster of cells. The distance between the single cell (red arrowhead) and the cluster of cells (yellow arrowhead) changed from 88.5μm to 49.9μm during imaging. Time of imaging is shown in hours:minutes. B. Migration of a single ß-cell occurred over 365μm (a to b, magenta line) at an average velocity of 11.4μm/hr.
The difference in distance between a cluster of cells (Fig. 7A, yellow arrowhead) and the individual migrating cell (red arrowhead) was also calculated over time. This distance was found to change from 88.5μm (at the point when the cell was specified) to 49.9μm over the time of imaging (Fig. 7A). It should be noted that the distance between objects is calculated from the centroid (or center of mass) of all objects for accurate temporal comparison. Thus, the final distance of ∼50 μm is the distance between the migrating cell and the center of the large cell cluster just prior to coalescence of the two objects. It is important to point out that the cluster of cells is constantly remodeled, and changes in cluster size and shape occur over time as well (movie 5). We conclude that individual ß-cells migrate over significant distances, and clusters of ß-cells undergo morphological changes over time, providing evidence of constant activity during endocrine development. Documenting the normal behavior of these cells enables us to detect modifications in their ability to migrate and/or cluster upon perturbations within the cells or in their environment.
Role of Hedgehog signaling in embryonic pancreas morphogenesis
Hedgehog (Hh) signaling is excluded from the developing embryonic pancreas at early stages, and ectopic expression of this pathway, for e.g. overexpression of the ligand Sonic Hedgehog (Shh) during embryogenesis disrupts organ formation (Apelqvist et al., 1997; Kawahira et al., 2003). In chick embryonic tissue explants, inhibition of Hh signaling leads to ectopic appearance of insulin-positive cells in the stomach and intestine (Kim and Melton, 1998). This suggests that suppression of Hh signaling promotes formation of the endocrine lineage in the Pdx1-expressing domain in the stomach and intestinal tissues. In addition, islet size within the dorsal chick pancreas is larger, although the number of islets is unaffected (Kim and Melton, 1998).
Given our interest in Hh signaling during mammalian embryonic pancreas development, the function of this pathway was tested on pancreatic development in vitro. To block signaling, pancreatic explants were treated with cyclopamine, an inhibitor that blocks the activity of Smoothened, an essential component of the pathway (Chen et al., 2002). Conversely, a recombinant form of Shh was added to pancreatic buds to activate signaling. Treatments were carried out for 8 days after dissection. Explants were imaged between day 4 and day 6 of culture.
Pancreatic explants with fluorescently labeled epithelium (either Pdx1-GFP or Pdx1-Cre;Z/EG) underwent extensive growth and branching when treated with cyclopamine (data not shown), indicating that inhibition of Hh signaling does not block branching within the epithelium. On the contrary, treatment with Shh completely blocked expansion of the pancreatic epithelium, with no expansion or branching (data not shown). These results expand the genetic data from transgenic mice in which ectopic Hh activation disrupts epithelial morphogenesis.
To explore the effects on endocrine development, pancreatic explants from MIP-GFP embryos were similarly treated with either cyclopamine or recombinant Shh for 8 days. Representative wide-field images depict the ß-cell component labeled fluorescently within the explant (Fig. 8A). Bright field images show the overall morphology of the tissue. Visualization of GFP after 6 days in culture revealed a greater percentage of explants with GFP-positive cells upon inhibition of Hh signaling (Fig. 8A, B, >90% with 10μM or 5μM cyclopamine) as compared to untreated samples (56%). A strikingly low percent of explants treated with Shh (thereby activating the pathway in all tissues competent to do so) expressed GFP (3%), which is concordant with the conclusion that activation of Hh signaling adversely affects pancreas development (Fig. 8A, B).
Upon live imaging of explants treated with cyclopamine, an increase in the overall ß-cell mass was observed (supplementary movies 1&2). There was, however, no difference in the ability of these cells to migrate or cluster. Hence, inhibiting Hh signaling within the tissue explants leads to specification of a larger population of ß-cells that migrate and cluster as normal.
Two methods were employed to quantify the increase in insulin-positive cells due to cyclopamine treatment. First, total fluorescent area after several days in culture (with or without cyclopamine) was calculated as described in methods (using ImageJ). The ratio of the total fluorescent area in treated versus untreated samples was calculated to be 7.4 (6 days of treatment, average of 2 controls and 2 explants treated with cyclopamine) and 5.5 (7 days of treatment, one control and one explant treated with cyclopamine). Quantitative analysis therefore provided evidence for an increase in ß-cell mass upon inhibition of Hh signaling. Second, to measure the change in insulin expression level, quantitative PCR was carried out on RNA extracted from pancreatic explants (untreated, or treated with cyclopamine or recombinant Shh) on day 8 of culture. Marker expression level was normalized to the housekeeping gene cyclophilin, and depicted as fold change over untreated samples. The data indicate a prominent increase in insulin expression upon treatment with cyclopamine (Fig. 8C) that correlates well with the increase in ß-cell numbers. Cyclopamine effectively repressed the expression of Gli1 and Patched, known target genes of the Hh signaling pathway, establishing the efficacy of the drug. As expected, treatment with Shh resulted in an upregulation of Hh target genes, providing evidence that signaling was active in tissues competent for it. Furthermore, the expression of pancreas-specific genes, including insulin, was strongly inhibited upon exposure to Shh.
The data presented above demonstrate the application of the current assay for quantifying changes in ß-cell mass during organogenesis. Although we have not documented a migratory phenotype, we believe this assay can be used to screen a host of chemical compounds for a quantitative assessment of their effects on fate specification, migration, or clustering of ß-cells.
Discussion
With the emergence of live cell imaging in recent years, real-time analysis of dynamic developmental processes has become possible. Watching an organ grow, under normal or perturbed conditions, provides greater insight into the physical interplay between the cells that constitute that organ. In vitro culture of pancreatic buds has been accomplished by several labs (Gittes et al., 1996; Huotari et al., 2002; Miralles et al., 1998; Percival and Slack, 1999), and phase contrast images acquired on successive days of bud culture have shown that the epithelial tissue is capable of undergoing branching (Horb and Slack, 2000). Fluorescent tools are fast emerging that can now allow coupling in vitro culture with real-time imaging.
We decided to use an early embryonic stage to culture pancreatic buds in vitro to focus on early morphogenetic events during pancreas growth. While most labs have successfully cultured buds dissected at e11.5 or later, we isolated the pancreatic bud while still attached to the stomach and part of the duodenum at e9.5 or e10.5. We and others have shown that growing embryonic pancreatic buds in culture leads to the expression of numerous mature markers, suggesting that the transcriptional cascade that defines the mature cell types in the pancreas is in place (Figs. 1, 2; Gittes et al., 1996; Huotari et al., 2002). As expected, the epithelium does not grow at the equivalent rate observed in vivo, and stops expanding after several days in culture (>10days). However, our analysis focuses mainly on early organogenesis (4-5 days) during which cells of endocrine, exocrine and ductal lineages are formed and organized as they would in the mature organ.
In the mouse, branching organs such as the pancreas, lung, kidney, and salivary gland appear initially as an epithelial bud, surrounded by dense mesenchymal tissue (Bellusci et al., 1997; Costantini, 2006; Patel et al., 2006; Slack, 1995). The epithelium grows and penetrates into the mesenchyme, with continuous signaling required for accurate development. Organ cultures of mouse lung, kidney and salivary gland have been used extensively to describe morphogenesis of these organs (Bellusci et al., 1997; Costantini, 2006; Patel et al., 2006). In both the developing ureteric bud and the lung bud, in vitro branching corresponds well with the in vivo pattern (Lin et al., 2003). During in vitro growth, approximately 80% of the budding events in the lung occurred by “1-3” mode of lateral branching, characterized by branching occurring lateral to the growth axis of the “primary” bud (Fig. 4C, [2]). In contrast, the dominant mode in the ureteric bud was the 1-2 type of terminal branching, characterized by a bifurcation of the developing tip (Fig. 4C, [1]). Using the culture conditions described here, our analysis reveals that the pancreatic epithelial branching resembles the lung pattern of lateral branching (Fig. 4C, [3]). Epithelial protrusions were numerically labeled at the start of image acquisition, and scored for growth and branching over the duration of imaging (Fig. 5). Although a small fraction of epithelial protrusions bifurcated at the tip, 86% of budding modules exhibited lateral branching (Fig. 5). The lung and the pancreas share several signaling cascades that impact epithelial growth. It is possible that a subset of these pathways regulate morphogenesis similarly between these organs. For example, FGF signaling from the mesenchyme leads to branching of the lung epithelium in culture (Bellusci et al., 1997). Perturbations in FGF signaling in the pancreas block epithelial expansion and branching (Bhushan et al., 2001; Pulkkinen et al., 2003). Using the live imaging assay, we can perturb specific signaling pathways in cultured explants and investigate their effect on branching in the pancreas in future experiments.
In the developing mouse embryo, the secondary transition occurs at e13.5, and is characterized by a dramatic activation of genes that specify the mature exocrine and endocrine compartments. Ngn3, a transcription factor required for the specification of all endocrine cell types, is expressed at the onset of the secondary transition (Gu et al., 2002). Time-lapse microscopy of mouse pancreas explants derived from transgenic mice that express fluorescently tagged neurogenin-3 demonstrate an expansion of Ngn-3-expressing cells over time (Mellitzer et al., 2004). In agreement with in vivo development, activation of Ngn3 expression coincided with the appearance of insulin-GFP positive cells, first observed in culture on day 4 (Fig. 6A). Green objects that appeared de novo represented newly specified ß-cells. After they were marked by GFP fluorescence, ß-cells were seen to migrate actively during the time of imaging. Although we estimate the average velocity of migrating ß-cells, it should be noted that cell migration rates might vary depending on the specific culture conditions used. Cytoplasmic extensions were projected into the environment in some cases (Fig.7A, red arrowhead). Filopodial extensions may occur to sense the presence of other cells in the surrounding milieu. Additionally, a signaling gradient may exist that allows cells to undergo guided migration towards a cluster of ß-cells.
The average size of objects increased over the duration of imaging (Fig. 6B). This indicates the formation of cell aggregates, which may be precursors to islet clusters found in the adult animal. It is unclear how the decision for nucleation of a cluster is made. However, it appears that once a cluster is established, it progressively increases in size. Each cluster appears to be composed of several smaller aggregates that undergo constant remodeling (movie 5). The increase in size of clusters could be due to cell division within the cluster as well as migration of cells from the surrounding epithelium. These questions can now be addressed by inhibiting cell division and migration and testing for the formation of clusters in our culture assay.
In contrast to our results, Gunawardhana et al (2005) report that they do not see the appearance of GFP-positive cells upon culturing pancreatic explants from the MIP-GFP transgenic line in Matrigel™ as compared with collagen. Differences in growth media or the presence of surrounding tissue in explants used in this study might explain the observations. In lung bud cultures, collagen gels do not promote in vitro growth, and Matrigel™, a mixture of ECM molecules, is thought to provide a more conducive environment for endodermal budding (Bellusci et al., 1997).
In the MIP-GFP embryos, GFP expression was detected in the dorsal pancreatic bud at e10.5, which constitutes the first wave of endocrine cells formed early during embryonic development. These cells are lost during the next few days in culture. The cells that are imaged appear to be the true mature ß-cells born during the secondary transition after 4 days in culture (corresponding to the increased Ngn3 expression).
The explant culture and live imaging set up was used to investigate the role of Hh signaling during pancreas development. Inhibition of Hh signaling with cyclopamine led to an increase in ß-cell mass, which could be quantified by measuring the total fluorescent area within the explants, and by real-time PCR to detect for endocrine genes. We are currently investigating the origin of these cells. We did not observe a change in the migratory or clustering behavior of the ß-cells under conditions of blocked Hh signaling. This suggests that the pathway acts upstream during specification of ß-cells from progenitors, and is not involved in migration or communication of ß-cells with each other to form pre-islet clusters in culture.
During embryogenesis, expression of Shh under the Pdx1 promoter in mice leads to pancreas agenesis (Apelqvist et al., 1997). In concordance with the analysis of Pdx-Shh transgenic mice, there was negligible expansion of the pancreatic epithelium with no apparent branching upon treatment with recombinant Shh (Fig. 8A and data not shown). Our attempts to document morphogenesis of cultured Pdx1-GFP labeled explants treated with Shh in real-time proved unsuccessful due to lack of change in the pattern of fluorescence. Thus, ectopic Shh treatment blocks epithelial expansion at the earliest stages of our culture period. By immunohistochemical analysis, Pdx-1 positive cells were still detected at the end of 8 days of Shh treatment, indicating the presence of pancreatic progenitors (data not shown). Ki67 staining of explants treated with Shh revealed the presence of dividing cells within the pancreatic epithelium, indicating that cell division can occur (data not shown). We predict that the presence of excess Shh suppresses the differentiation of these pancreatic progenitors, although further analyses will be required to understand the specific molecular mechanisms through which Hh signaling impacts pancreas growth.
In contrast to our results with recombinant Shh, activation of Hh signaling in the insulinoma cell line Ins-1 leads to an increase in insulin expression (Thomas et al., 2000). Such opposing roles for Hh signaling for ß-cell formation might depend on distinct requirements of the pathway in progenitor fate specification and expansion (during embryogenesis) versus maintenance of adult cells. Thus, the level of activation of the Hh pathway appears critical for normal morphogenesis and maintenance of the endocrine compartment, specifically ß-cells. The results further exemplify the viability of using explant cultures of e10.5 pancreatic buds in exploring the role of small compounds.
A descriptive analysis of pancreas development in culture presents a potentially powerful system for in vitro expansion of specific cell fates. Modulating additional signaling pathways and monitoring effects through live imaging will be highly informative of their role during development. Lineage analysis using multi-fluorescently labeled pancreas is the next step. In sum, we anticipate that evolution of real-time imaging tools in the pancreas will contribute greatly in the studies of highly dynamic processes involved in pancreas development, ß-cell formation and ß-cell maintenance.
Supplementary Movies
Documentation of ß-cell characteristics upon inhibition of Hedgehog signaling. Pancreatic explants from MIP-GFP embryos were imaged after treatment with cyclopamine (10μM) for 5 (movie 1) or 6 (movie 2) days after dissection at e10.5. The movies show an increase in fluorescence over the time of imaging, with motile GFP-labeled ß-cells that undergo clustering.
Pdx1-GFP positive pancreatic explant isolated at e10.5 and cultured for 4 days was imaged for 14hrs20min (Fig. 3A). The movie shows the overall expansion and branching of the epithelium. Leading edges, or “tips”, of the epithelium appear brighter.
Pdx1-Cre-Z/EG positive pancreatic explant isolated at e11.5 and cultured for 4 days was imaged for 22hrs25min (Fig. 3B). The epithelium is highly ramified, and undergoes growth and budding at several branches.
Pdx1-GFP positive pancreatic explant isolated attached to the gut at e9.5 and cultured for 4 days was imaged for 15hrs20min (Fig. 4A). Several branching modules can be identified, accompanied by overall expansion of the epithelium.
MIP-GFP positive pancreatic explant was dissected at e10.5 and cultured for 4 days before imaging for 33hrs20min (Fig. 6A). Cells expressing insulin appear as green entities. Over the time of imaging, several new cells appear and cluster together to form aggregates.
An isolated MIP-GFP positive cell migrates towards a cluster. The pancreatic explant was dissected at e10.5 and cultured for 5 days before imaging for 35hrs. Filopodial extensions protrude from multiple cells in the field that move towards the cluster (Fig. 7A, yellow arrowhead).
Acknowledgements
We would like to thank Drs. Doug Melton, Manami Hara, and Andrew Holland for providing us with mouse lines used in this work, and Dr. Mike German for providing antibody reagents. We also thank Mark White, Regina Burris, Janet Lau, and Dr. Marina Pasca for critical reading of the manuscript. S.P. was supported by a postdoctoral fellowship from the Juvenile Diabetes Research Foundation. Work in M.H.'s laboratory was supported by grants from the NIH (DK60533, CA112537), the Juvenile Diabetes Research Foundation and the Larry L. Hillblom Foundation. Image acquisition was supported by the UCSF Diabetes and Endocrinology Research Center microscopy core (P30 DK63720).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr. Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F. Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation. 2006;74:402–421. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- Davies JA. Watching tubules glow and branch. Curr. Opin. Genet. Dev. 2005;15:364–370. doi: 10.1016/j.gde.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Gittes GK, Galante PE, Hanahan D, Rutter WJ, Debase HT. Lineage-specific morphogenesis in the developing pancreas: role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Gunawardana SC, Hara M, Bell GI, Head WS, Magnuson MA, Piston DW. Imaging Beta cell development in real-time using pancreatic explants from mice with green fluorescent protein-labeled pancreatic Beta cells. In Vitro Cell Dev. Biol. Anim. 2005;41:7–11. doi: 10.1290/0412080.1. [DOI] [PubMed] [Google Scholar]
- Hara M, Wang X, Kawamura T, Bindokas VP, Dizon RF, Alcoser SY, Magnuson MA, Bell GI. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am. J. Physiol. Endocrinol. Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- Hara M, Dizon RF, Glick BS, Lee CS, Kaestner KH, Piston DW, Bindokas VP. Imaging Pancreatic ß-cells in the Pancreas. Am. J. Physiol. Endocrinol. Metab. 2006;290:E1041–1047. doi: 10.1152/ajpendo.00365.2005. [DOI] [PubMed] [Google Scholar]
- Herrera PL, Huarte J, Sanvito F, Meda P, Orci L, Vassalli JD. Embryogenesis of the murine endocrine pancreas; early expression of pancreatic polypeptide gene. Development. 1991;113:1257–1265. doi: 10.1242/dev.113.4.1257. [DOI] [PubMed] [Google Scholar]
- Holland AM, Micallef SJ, Li X, Elefanty AG, Stanley EG. A mouse carrying the green fluorescent protein gene targeted to the Pdx1 locus facilitates the study of pancreas development and function. Genesis. 2006;44:304–307. doi: 10.1002/dvg.20214. [DOI] [PubMed] [Google Scholar]
- Horb LD, Slack JM. Role of cell division in branching morphogenesis and differentiation of the embryonic pancreas. Int. J. Dev. Biol. 2000;44:791–796. [PubMed] [Google Scholar]
- Huotari MA, Miettinen PJ, Palgi J, Koivisto T, Ustinov J, Harari D, Yarden Y, Otonkoski T. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology. 2002;143:4437–4446. doi: 10.1210/en.2002-220382. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev. Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kawahira H, Ma NH, Tzanakakis ES, McMahon AP, Chuang PT, Hebrok M. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development. 2003;130:4871–4879. doi: 10.1242/dev.00653. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M. Intercellular signals regulating pancreas development and function. Genes Dev. 2001;15:111–127. doi: 10.1101/gad.859401. [DOI] [PubMed] [Google Scholar]
- Kim SK, Melton D. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc. Natl. Acad. Sci. U S A. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J. Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- Li Z, Manna P, Kobayashi H, Spilde T, Bhatia A, Preuett B, Prasadan K, Hembree M, Gittes GK. Multifaceted pancreatic mesenchymal control of epithelial lineage selection. Dev. Biol. 2004;269:252–263. doi: 10.1016/j.ydbio.2004.01.043. [DOI] [PubMed] [Google Scholar]
- Lin Y, Zhang S, Tuukkanen J, Peltoketo H, Pihlajaniemi T, Vainio S. Patterning parameters associated with the branching of the ureteric bud regulated by epithelial-mesenchymal interactions. Int. J. Dev. Biol. 2003;47:3–13. [PubMed] [Google Scholar]
- Mellitzer G, Martin M, Sidhoum-Jenny M, Orvain C, Barths J, Seymour PA, Sander M, Gradwohl G. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Mol. Endocrinol. 2004;18:2765–2776. doi: 10.1210/me.2004-0243. [DOI] [PubMed] [Google Scholar]
- Miralles F, Czernichow P, Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development. 1998;125:1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Melton DA. Genes, signals, and lineages in pancreas development. Annu. Rev. Cell Dev. Biol. 2003;19:71–89. doi: 10.1146/annurev.cellbio.19.111301.144752. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation. 2006;74:349–364. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Percival AC, Slack JM. Analysis of pancreatic development using a cell lineage label. Exp. Cell Res. 1999;247:123–132. doi: 10.1006/excr.1998.4322. [DOI] [PubMed] [Google Scholar]
- Pulkkinen MA, Spencer-Dene B, Dickson C, Otonkoski T. The IIIb isoform of fibroblast growth factor receptor 2 is required for proper growth and branching of pancreatic ductal epithelium but not for differentiation of exocrine or endocrine cells. Mech. Dev. 2003;120:167–175. doi: 10.1016/s0925-4773(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Sekine S, Lan BY, Bedolli M, Feng S, Hebrok M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- Thomas MK, Rastalsky N, Lee JH, Habener JF. Hedgehog signaling regulation of insulin production by pancreatic beta-cells. Diabetes. 2000;49:2039–2047. doi: 10.2337/diabetes.49.12.2039. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Costantini F. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev. Biol. 2004;271:98–108. doi: 10.1016/j.ydbio.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mech. Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pdx1-GFP positive pancreatic explant isolated at e10.5 and cultured for 4 days was imaged for 14hrs20min (Fig. 3A). The movie shows the overall expansion and branching of the epithelium. Leading edges, or “tips”, of the epithelium appear brighter.
Pdx1-Cre-Z/EG positive pancreatic explant isolated at e11.5 and cultured for 4 days was imaged for 22hrs25min (Fig. 3B). The epithelium is highly ramified, and undergoes growth and budding at several branches.
Pdx1-GFP positive pancreatic explant isolated attached to the gut at e9.5 and cultured for 4 days was imaged for 15hrs20min (Fig. 4A). Several branching modules can be identified, accompanied by overall expansion of the epithelium.
MIP-GFP positive pancreatic explant was dissected at e10.5 and cultured for 4 days before imaging for 33hrs20min (Fig. 6A). Cells expressing insulin appear as green entities. Over the time of imaging, several new cells appear and cluster together to form aggregates.
An isolated MIP-GFP positive cell migrates towards a cluster. The pancreatic explant was dissected at e10.5 and cultured for 5 days before imaging for 35hrs. Filopodial extensions protrude from multiple cells in the field that move towards the cluster (Fig. 7A, yellow arrowhead).