Abstract
The past decade has seen the rapid evolution of small-molecule gene-silencing strategies, driven largely by enhanced understanding of gene function in the pathogenesis of disease. Over this time, many genes have been targeted by specifically engineered agents from different classes of nucleic acid-based drugs in experimental models of disease to probe, dissect, and characterize further the complex processes that underpin molecular signaling. Arising from this, a number of molecules have been examined in the setting of clinical trials, and several have recently made the successful transition from the bench to the clinic, heralding an exciting era of gene-specific treatments. This is particularly important because clear inadequacies in present therapies account for significant morbidity, mortality, and cost. The broad umbrella of gene-silencing therapeutics encompasses a range of agents that include DNA enzymes, short interfering RNA, antisense oligonucleotides, decoys, ribozymes, and aptamers. This review tracks current movements in these technologies, focusing mainly on DNA enzymes and short interfering RNA, because these are poised to play an integral role in antigene therapies in the future.
Over the past century, our appreciation of the pathogenesis of human disease has continued to evolve with corresponding therapeutic developments. In particular, more recent significant advances in genomics have led to a substantial shift away from conventional perceptions and dogma to focus on intricate molecular and cellular pathways regulated by an array of key genes. It is at this interface that nucleic acid molecules are emerging as a potent force in further characterizing important molecular pathways and in defining themselves as a sustainable therapeutic class of agent. The ability to selectively attenuate the expression of specifically targeted genes represents an appealing method of therapy and a means of dissecting molecular function. As such, strategies to specifically knockdown gene expression have received considerable attention.
Paterson et al1 demonstrated the utility of nucleic acids in modulating gene expression approximately 30 years ago. Zamecnik and Stephenson2 soon after showed the capacity of antisense molecules to inhibit viral replication. The field of nucleic acid therapeutics has since evolved considerably with numerous gene targets and methods comprising both naturally occurring and synthetic molecules that have been applied in vitro and in vivo in a variety of contexts with varying degrees of success. Although target selection is clearly vital, the method used in achieving this is of equal importance. Strategies have included DNA enzymes (DNAzymes), siRNA, antisense oligonucleotides, decoys, ribozymes, and aptamers, all of which attenuate gene expression by interfering with cytosolic mRNA or translated protein. Currently, a number of these approaches are being evaluated in human and higher animal trials and are poised to offer considerable inroads and additions to our current therapeutic armamentarium where an unmet clinical need exists. Issues that underpin the clinical feasibility of “antigene” nucleic acid strategies are many and include i) that the agent is gene specific and functionally active with temporal relevance to the particular disease process; ii) that the target gene should play a key role in the disease process and that its role should not readily be compensatable by other genes; iii) that target gene inhibition should not adversely influence normal physiological processes; iv) local versus systemic delivery routes; v) the choice of endogenous (eg, gene/viral transcription) or exogenous (eg, synthetic nucleic acid) nucleic acid delivery; vi) the choice of delivery agent (eg, naked DNA, polymer, cationic lipid, PEGylated liposome, protein/nucleic acid chimera, or complex); and vii) that nucleic acid modification/stabilization made to avoid degradation of the agent may contribute to nonspecific (off-target) effects. This review will focus mainly on DNAzyme and siRNA strategies and briefly cover recent developments in antisense oligonucleotides, decoys, ribozymes, and aptamers.
DNAzymes
DNA enzymes (DNAzymes or deoxyribozymes), like ribozymes, may be perceived as gene-specific molecular scissors. Catalytic DNA has not been observed in nature, and all existing molecules have been derived by in vitro selection processes similar to those used to identify aptamers (see below). The most well-characterized DNAzyme is the “10-23” subtype comprising a cation-dependent catalytic core of 15 deoxyribonucleotides3 that binds to and cleaves its target RNA (Figure 1) between an unpaired purine and paired pyrimidine through a de-esterification reaction, producing a 2′,3′-cyclic phosphate terminus and a 5′-hydroxyl terminus. Sequence conservation in the border regions of the catalytic core is important for the maintenance of catalytic activity.4 This core is flanked by complementary binding arms of 6 to 12 nucleotides in length that confer target mRNA specificity.
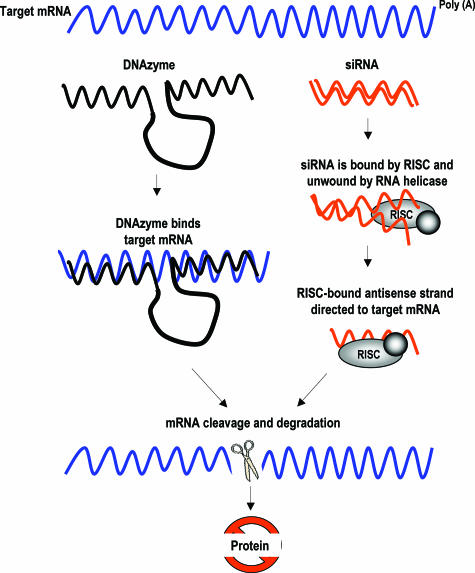
Figure 1.
Schematic representation of DNAzyme- and siRNA-mediated degradation of target mRNA. Left: DNAzymes recognize the complementary mRNA sequence of its hybridizing arms via Watson-Crick base pairing and catalyze degradation of the target mRNA, producing two products, one containing a 2′,3′-cyclic phosphate terminus and the other a 5′-hydroxyl terminus. Right: siRNA is recognized by RNA-induced silencing complexes (RISC). RNA helicases unwind the double-stranded siRNA, and the antisense strand guides RISC to the complementary mRNA. Targeted mRNA is cleaved by RISC and rapidly degraded.
The 10-23 DNAzyme, named by virtue of its selection process in vitro, catalyzes sequence-specific RNA cleavage in a manner akin to the hammerhead ribozyme and hence has substantial utility as a gene-silencing agent. In vitro cleavage experiments have shown that the 10-23 DNAzyme is highly specific and sensitive to small changes in target sequence.5,6 DNAzyme activity is dependent on the prevailing secondary structure of long-target RNA at the cleavage site.6 For this reason, it is important to test a range of molecules to identify those that display a high level of activity against biologically relevant target molecules. In terms of biological specificity, an important control in the assessment of DNAzyme antigene efficacy and specificity is the “scrambled DNAzyme,” wherein the sequence of nucleotides in the binding arms of the DNAzyme is jumbled while the catalytic core is preserved. This produces a molecule of identical size, the same percentage composition of nucleic acids, and the same net charge with a binding sequence that is not matched to the target gene. DNAzymes with nonsense or mismatch sequences in the binding arms or with point mutations in the catalytic core that render the DNAzyme enzymatically inactive can serve as additional controls. In vitro gene and cell inhibitory applications of the 10-23 DNAzyme are summarized in Table 1.
Table 1.
In Vitro Applications of 10-23 DNAzymes
| Gene | Cell type | Assay conditions | Proliferation assay | Cell death assay | References |
|---|---|---|---|---|---|
| c-myc | Rat aortic SMCs | SS | Yes | No | 7 |
| Transforming growth factor-β1 | Rat mesangial cells | SS | No | No | 8 |
| Protein kinase C-ε | Human pulmonary artery SMCs | SS | No | No | 9 |
| bcr-abl | K562 cells | SFC | Yes | Yes | 10 |
| survivin | PANC-1 cells | SCC | Yes | Yes | 11 |
| ftsZ | DH5alphapro cells | SFC | Yes | No | 12 |
| PML/RARa | NB4 cells | SS | Yes | Yes | 13 |
| K562 cells | SS | Yes | Yes | 13 | |
| PB2 | MDBK cells | SD | Yes | No | 14 |
| β1 and β3 integrin subunits | EA.hy 926 | SCC | No | Yes | 15 |
| K562 | SCC | No | Yes | 16 | |
| HIV-1 TATRev | HeLa | SS | No | Yes | 17 |
| Cos-1 | SS | No | Yes | 17 | |
| THP-1 | SS | No | Yes | 17 | |
| VEGF receptor-2 | BAEC | SCC | Yes | Yes | 18 |
| HUVEC | SCC | Yes | Yes | 18 | |
| MDA-MB-435 | SCC | Yes | Yes | 18 | |
| Urokinase-type plasminogen activator receptor | Saos-2 | SS | No | Yes | 19 |
| Egr-1 | MCF-7 cells | SS | Yes | No | 20,21 |
| HMEC-1 | SS | Yes | No | 20 | |
| Human vascular SMCs | SS | Yes | No | 22 | |
| Porcine vascular SMCs | SS | Yes | No | 23 | |
| Rat aortic SMCs | SS | Yes | Yes | 23 | |
| c-Jun | Human vascular SMCs | SS | Yes | No | 24 |
| Porcine vascular SMCs | SS | Yes | No | 24 | |
| HMEC-1 | |||||
| T79-Squamous CC | SS | Yes | Yes | 25 | |
| LK2-Squamous CC | SS | Yes | No | 26 | |
| SS | Yes | No | 26 | ||
| VDUP1 | H9C2 | SCC | No | Yes | 27 |
SS, serum stimulation; SD, serum deprivation; SFC, serum-free conditions; SCC, serum-constant conditions.
A number of structural modifications have been used to enhance the stability and to improve the potency of DNAzymes. An important, commonly used modification is the incorporation of a 3′-3′ inverted nucleotide at the 3′ end of the DNAzyme to prevent exonuclease degradation. This can dramatically increase stability of the molecule, extending the half-life from ∼70 minutes to >21 hours in human serum.28 In addition, DNAzymes with this modification can remain functionally intact for at least 24 to 48 hours after exposure to serum compared with its unmodified counterpart23,28 with little change in the kinetics.7 Phosphorothioate (PS) linkages, which enhance stability by rendering the oligonucleotide more resistant to endogenous nucleases, have been used with DNAzymes.29 The introduction of PS modifications may affect cleavage efficiency28,30 and has been associated with toxicity,31 immunological responsiveness,32 and increased affinity for cellular proteins, resulting in sequence-independent effects.33,34
Locked nucleic acids (LNAs), more recently, have been attractive monomers for modifying oligonucleotides35 and DNAzymes,30,36,37 in an attempt to increase binding affinity. LNA bases comprise a 2′-O 4-C methylene bridge that locks in a C3′-endo conformation,35 which places constraint on the ribose ring, increasing affinity for complementary sequences.38,39 The advantages of LNAs include increased thermal stability of duplexes toward complementary DNA or RNA, stability toward 3′-exonucleolytic degradation, solubility due to structural similarities to nucleic acids, easy automated synthesis with complete modified LNA or chimeric (LNA/DNA or LNA/RNA) oligonucleotides,40,41 and straightforward cellular delivery using standard transfection reagents.31,36,42,43 LNA incorporation into DNAzymes may influence catalytic activity under single-turnover conditions30,36,37,43 and biological potency.36,43 DNAzymes with an inverted nucleotide at the 3′ end are catalytically more efficient compared with their LNA-modified counterparts because of a slower product release rate.30,44
Accumulating evidence indicates the utility, efficacy, and potency of DNAzymes in a variety of animal models of disease, allowing characterization of key molecular pathways underlying pathogenesis and use as a therapeutic agent. For instance, DNAzymes targeting the “master-regulator” zinc finger transcription factor Egr-145,46 have shown promise in experimental models of restenosis via inhibition of smooth muscle cell hyperplasia. We have demonstrated inhibition of neointima formation in the rat carotid artery after both balloon injury (first demonstration of DNAzyme efficacy in an animal model) and carotid artery ligation.23,47 Furthermore, intracoronary administration of DNAzymes targeting human Egr-1 reduced neointima formation in porcine coronary arteries after stent implantation.22 Likewise, Egr-1 DNAzymes attenuated neointima formation in human internal mammary arteries ex vivo.48 These and other in vivo applications of DNAzymes are summarized in Table 2.
Table 2.
In Vivo Applications of DNAzymes
| Gene | Model | Applications | Reference |
|---|---|---|---|
| c-Jun | Rabbits, rats, mice | Restenosis, neovascularization, inflammation, tumor growth | 24,25,26,49,50 |
| Egr-1 | Pigs, rats, mice | Restenosis, tumor growth, neovascularization, ureteral obstruction | 20,21,22,23,36,47,51,52 |
| Xylosyltransferase-1 | Mice | Spinal regeneration | 53 |
| Transforming growth factor-β1 | Rats | Glomerulonephritis | 8 |
| mPer1 | Mice | Morphine addiction | 54 |
| PAI-1 | Rats | Myocardial infarction | 55 |
| 56 | |||
| Vitamin D3 up-regulated protein 1 | Rats | Myocardial infarction | 27 |
| Tumor necrosis factor-α | Rats | Congestive cardiac failure | 57 |
| VEGF-2 | Mice | Tumor growth | 18 |
We have also recently evaluated Egr-1 DNAzymes in the setting of myocardial infarction and demonstrated that intramyocardial delivery of Egr-1 DNAzymes in rats undergoing myocardial ischemia-reperfusion resulted in a 50% reduction in infarct size, myocardial neutrophil infiltration, and intercellular cell adhesion molecule-1 (ICAM-1) expression.51 Four other studies have used DNAzymes to target the myocardium. Itescu and colleagues55,56 conducted two separate studies in which intramyocardial delivery of DNAzymes targeting PAI-1 in a rodent model of myocardial infarction resulted in a reduction in apoptosis, improved functional recovery, and enhanced myocardial capillary density. The same group also used intramyocardial administration of DNAzymes targeting the vitamin D3 up-regulated protein 1, which promotes cellular oxidative stress, and demonstrated a reduction in cardiomyocyte apoptosis when delivered at the time of myocardial infarction in rats.27 Iversen et al57 delivered tumor necrosis factor-α DNAzymes via peritoneal osmotic minipumps in rats with myocardial infarction-induced heart failure and demonstrated improved hemodynamic performance compared with controls.
DNAzymes targeting a second immediate-early gene, the leucine zipper transcription factor c-Jun, also show promise. We have demonstrated that c-Jun DNAzymes (Dz13) play a modulatory role in the inflammatory process by disrupting the expression of key downstream molecules, including the cell adhesion molecules ICAM-1 and VCAM-1, and E-selectin and VE-cadherin.49 Dz13 rapidly abolished the processes of leukocyte rolling, adhesion, and extravasation in response to interleukin-1β stimulation in a rat microcirculation model. Intranasal administration of Dz13 abolished pulmonary inflammation in a murine lung sepsis model and joint inflammation in a murine arthritis model after intra-articular delivery. This represents a key area for future therapeutic exploitation because inflammation is integral in the pathogenesis of many diseases and current strategies are still far from optimal.
By exploiting the reliance of cancers on angiogenesis for growth, various transcription factors mediating this process have been targeted successfully to inhibit tumor growth. Zhang et al18 first applied this technology in vivo against tumors with DNAzymes targeting vascular endothelial growth factor (VEGF) receptor 2 attenuating tumor growth in rats. Intratumorally administered DNAzymes targeting Egr-1 also demonstrated potent reduction in tumor growth with an associated reduction in tumor angiogenesis.20 A direct antitumor effect was achieved with a humanized Egr-1 DNAzyme.21 Furthermore, the Egr-1 DNAzyme inhibited de novo VEGF-induced neovascularization of the rat cornea.25 Dz13, targeting c-Jun, attenuated solid melanoma and squamous cell carcinoma growth in mice in part via its suppression of tumor angiogenesis.25,26 Further evidence of the antiangiogenic properties of Dz13 is demonstrated by its inhibition of corneal neovascularization in rats25 and retinal neovascularization in mice induced by exposure to hyperoxia-normoxia.49 Although in vitro assessment of DNAzyme efficacy helps establish gene and sequence specificity and facilitates high-throughput screening, the clinical utility of these agents, like any other potential therapeutic, can only be gauged in animal models, where important issues such as delivery, biodistribution, pharmacokinetics, metabolism, toxicity, and pharmacodynamics can be explored. Dz13, for example, has the capacity to inhibit restenosis,24 angiogenesis,25 tumor growth,25,26 and as already discussed, inflammation49 in animal models consistent with its activity in a variety of in vitro systems.
DNAzymes have been used in a variety of other animal models. For example, DNAzymes targeting transforming growth factor-β1, important in extracellular matrix accumulation, delivered by injection into the renal artery followed by electroporation in a rat anti-Thy-1 model of glomerulonephritis led to reduced extracellular matrix accumulation.8 DNAzymes have also proved efficacious in the CNS when delivered via the intracerebroventricular route in a study in mice examining the role of the circadian “clock gene” mPer-1 in morphine addiction. Investigators found less morphine dependence in mice whose mPer-1 expression had been attenuated with mPer-1-targeting DNAzymes compared with those that did not.54 The potential role of DNAzymes in augmenting spinal regeneration was explored in a study in which DNAzymes were designed to disrupt the enzyme xylosyltransferase-1, which is important in glycosylating the protein backbone of proteoglycans. The investigators found enhanced axonal regeneration in newborn rats with DNAzyme treatment and reduced glycosaminoglycan chains on proteoglycans.53 These studies, taken together, demonstrate the potential of DNAzymes as gene-specific molecular tools. That DNAzymes possess a number of advantages over other gene-silencing techniques, including lower production cost and relative serum stability, makes these attractive therapeutic candidates en route to the clinic.
A recent in vitro study58 has related DNAzyme catalysis using a short synthetic substrate with cell death as a measure of biological activity, making comparisons with published data on Dz13 and several other DNAzymes using different cleavage conditions, biological systems, and methodologies. Although oligonucleotides with some motifs, particularly those containing runs of multiple guanines, can induce nonspecific cellular effects59,60 such as by interacting with particularly abundant cellular proteins, conclusions cannot be drawn in the absence of experiments appropriate for the targeted gene (eg, inducible immediate-early genes versus constitutively expressed genes) and without proper characterization of phenotypic effects (eg, problematic correlation of cell-free cleavage experiments with short substrates and cytotoxicity, synonymity of proliferation with survival and toxicity, lack of stimulating conditions, or translation in vivo) as was the case in that study.58 Dz13 activity nonetheless was found to be sequence- and dose-dependent, and the oligonucleotide lacked quadruplex structure.58 Unexpected toxic side effects have not been observed in vivo, in the numerous animal models of disease treated with this DNAzyme to date (Table 2).
Although singular targets have been used in biological systems thus far, it is possible that greater biological efficacy may be achieved using combinations of DNAzymes and/or other small-molecule nucleic acid strategies targeting the same factor or multiple factors. In cancer for instance, there is increasing realization that for effective tumor stasis, a combinatorial approach may be preferable in efforts to prevent neoplastic cells evolving mechanisms to avoid single agent-based therapy.61 These strategies might also be used as adjuncts with conventional therapies such as thrombolytic agents (eg, urokinase-type plasminogen activator and warfarin) or antiproliferatives (eg, taxol and rapamycin).
siRNA
The advent of RNA interference (RNAi) as a gene-silencing strategy represents an exciting development in the field of small-molecule nucleic acid-based therapeutics. RNA interference was first described in 1998 by the 2006 Nobel Laureates Andrew Fire and Craig Mello, who demonstrated double-stranded RNA-mediated degradation of target mRNA in Caenorhabditis elegans and has subsequently been demonstrated in diverse eukaryotes. Short interfering RNA (siRNA) of 21 to 23 nucleotides processed by the RNase III family member Dicer are incorporated into an RNA-induced silencing complex (RISC). The sense strand of the double-stranded siRNA is cleaved during the formation of the RISC complex.62 RNA helicases unwind the double-stranded siRNA, and the antisense strand guides RISC to the complementary target mRNA, which is cleaved by RISC (Figure 1).63,64,65 siRNA avoids the problem of long double-stranded RNA-mediated activation of the interferon pathway, which can shutdown general protein synthesis and cause nonspecific mRNA degradation in mammalian cells,66 although siRNAs synthesized from the T7 RNA polymerase system have been found to trigger interferon responses in a variety of cell lines.67 Short-hairpin RNAs transcribed from RNA polymerase II or III promoters from plasmid- and virus-based vectors provide alternative strategies for RNA-mediated gene silencing.68 Vector-based short-hairpin RNA are processed by Dicer into siRNA duplexes. These strategies have been recently applied in mammalian cancer genetics69 and models of neurodegeneration,70 and vector libraries are now commercially available for high-throughput screens.71 Mechanisms of RNAi-mediated gene silencing in mammalian systems have been reviewed elsewhere.71,72
Some of the major concerns arising with siRNA applications in vivo, as with all small-molecule nucleic acid agents, are tissue specificity and the ability to withstand degradation by nucleases. The latter is of particular significance because the molecule has to “survive” within the host if it intends to provide lasting effects in the biological milieu. Tissue-specific delivery continues to present a key challenge for small-molecule nucleic acid therapeutics.73 Although in vivo application of siRNA has attracted attention particularly in cancer therapeutics, systemic delivery would provide more clinical appeal than local intratumoral administration. It is becoming more common to formulate (polyplex or nanoplex) siRNA to incorporate compounds, ligands, or peptides to achieve tissue specificity and nuclease resistance, thus eliminating, as much as possible, non-tissue-specific uptake of siRNA. The targeted tissue would include the tumor itself, inhibiting cell proliferation or the neovasculature, inhibiting angiogenesis, and starving the tumor of a blood supply. Kim et al74 have nanoplexed an siRNA to the polymer TargeTran, comprising a branched polyethylenimine, polyethylene glycol, and arginine-glycine-aspartate peptide sequence.75 A nanoimmunodelivery system devised by Pirollo et al76 demonstrated specific uptake of 6-FAM-labeled nanoplexed siRNA in primary tumors 20 minutes after systemic delivery. Furthermore, the nanoplexed siRNA, composed of an anti-transferrin receptor antibody, specifically penetrated deep into the tumors. Specificity for the tumor tissue was conferred by a lack of fluorescence (6-FAM) by blood vessels surrounding the tumor, suggesting uptake of siRNA by tumor cells and not endothelial cells.
The issue of whether siRNA diminishes over time because of degradation or dilution due to rapidly dividing cells was addressed by Bartlett and Davis.77 siRNA polyplexed to transferrin, targeting the luciferase gene, specifically inhibited luciferase activity in nondividing hepatocytes in mice. Inhibition of luciferase activity after a bolus intravenous injection lasted 4 weeks, suggesting that stability of siRNA was not a limiting factor in gene silencing. Schiffelers et al78 systemically delivered PEGylated siRNA to an arginine-glycine-aspartate peptide ligand specific to the VEGF receptor-2 transcript. They demonstrated a reduction in tumor progression of pre-established tumors xenografted in mice, and tumor regression was paralleled by a reduction in blood vessel formation surrounding the tumor.78 Atelocollagen, a highly purified type I collagen, complexed with siRNA confers increased resistance to nucleases, efficiency in transducing cells and prolonged gene silencing.79 Intratumoral injections of siRNA complexed with atelocollagen targeting VEGF inhibited tumor growth and tumor angiogenesis over 40 days after four repeated injections every 10th day.80 The effectiveness of locally delivered VEGF siRNA in tumors was further complemented by Takeshita et al,81 who systemically delivered atelocollagen siRNA. They demonstrated that atelocollagen complexed to siRNA improved cellular uptake in tumor tissue sixfold compared with naked siRNA with efficient inhibition of metastatic tumor growth in bone tissue.81 More importantly, systemic delivery of atelocollagen siRNA failed to elicit an interferon or interleukin-12 response in vivo.
Local and systemic delivery of siRNA directed against target genes responsible for the progression of disease has been successful in animal models.82 Furthermore, siRNA-mediated specific cleavage of targeted mRNA, in vitro and in vivo, has also been successfully demonstrated.83,84 Local delivery of siRNA has been used as a preventative for ocular neovascularization for the treatment of age-related macular degeneration and diabetic retinopathy.74,85,86 VEGF or its receptors have been prime targets of siRNA for exerting antiangiogenic effects. Local, conjunctival, or periocular administration of siRNA targeting murine VEGF receptor-1 has been compared with systemic delivery.74,84 A reduction in ocular neovascularization was observed by both groups. Shen et al84 used a mouse model of laser-induced choroidal neovascularization (CNV), whereas Kim et al74 used herpes simplex virus DNA containing bioactive CpG motifs to stimulate VEGF and subsequent neovascularization. More potent inhibition of ocular vascularization was evident through local administration rather than the systemic route. Furthermore, Kim et al74 delivered a cocktail of siRNAs targeting VEGF, VEGF receptor-1, and VEGF receptor-2 at a 1:1:1 ratio and observed synergistic effects. More potent inhibition of ocular neovascularization and corneal VEGF protein and mRNA was achieved with this cocktail compared with siRNA targeting each transcript separately. Subretinal delivery of siRNA targeted to murine VEGF transcript inhibited choroidal neovascularization after laser photocoagulation.85 Tolentino et al86 locally delivered siRNA targeting VEGF to inhibit laser-induced choroidal neovascularization in nonhuman primates. Single intravitreal injection of Cand5 (VEGF siRNA) inhibited choroidal neovascularization in a dose-dependent manner and was sustained for 36 days with no evidence of inflammation, cataract formation, retinal detachment, or vitreous hemorrhage. Lipid-based systems may be useful for the systemic intracellular delivery of siRNA. Zimmerman et al,87 for example, delivered apolipoprotein B siRNA encapsulated in stable nucleic acid lipid particles into nonhuman primates by intravenous injection, reducing levels of apolipoprotein B mRNA and protein, serum cholesterol, and low-density lipoprotein. The clinical interrogation of siRNA has commenced. There are at least two siRNA molecules in clinical trials. For example, Cand5 is in Phase II trials for age-related macular degeneration (Acuity Pharmaceuticals, Philadelphia, PA), and siRNA targeting the VEGF-receptor-1 (SiRNA-027) has successfully completed phase I (Merck & Co., Inc., Whitehouse Station, NJ) for the same condition.84,86
Antisense Oligonucleotides, Decoys, Ribozymes, and Aptamers
The wave of small-molecule nucleic acid-based gene-silencing strategies includes a mix of old and new players, such as antisense oligonucleotides, oligonucleotide decoys, ribozymes, and aptamers. These molecules differ in their mechanisms of action, and many are under clinical development for a wide range of disorders.
Antisense Oligonucleotides
Antisense oligonucleotides (ASOs) are single-stranded segments of DNA or RNA generally 15 to 25 bp in length. Although their precise mechanism of action is not fully understood, their function is mediated by interaction with target mRNA via hydrogen bonding, blocking translation into protein by steric hindrance of ribosomal movement along the transcript, or by activation of endogenous RNase H for targeted destruction of the DNA/RNA heteroduplex, resulting in mRNA degradation.88 Unmodified ASO molecules are prone to degradation, and their negative charge makes cellular membrane penetration inefficient. As such, these molecules have evolved with a variety of modifications that enhance stability and efficacy. These have included the PS backbone modification, which increases oligonucleotide half-life. However, the introduction of PS into the backbone of ASOs increases the propensity of nonspecific interaction with other proteins, resulting in sequence-independent phenotypic effects or cytotoxicity.33,34 High concentrations of PS ASOs can also bind and inhibit DNA polymerases and RNase H.89,90 To eliminate off-target effects spurred by the introduction of PS ASOs, other substitutes have since been made. LNAs, described above, have been incorporated into the backbone of ASOs as LNA/DNA gapmers, increasing both target binding affinity and, more importantly, stability.40,41,91,92 An alkyl modification, such as an 2′-O-alkyl modification to the ribose ring (2′-O-methyl or 2′-O-methoxyethoxy) averts the need for PS modifications, providing stability and efficacy.93 3′-3′-inverted T modifications have also remarkably increased oligonucleotide stability.23,24,94 Takei et al94 demonstrated greater stability of ASOs with 5′- and 3′-inverted T additions compared with PS-ASOs. In addition, 5′- and 3′-inverted T-modified ASOs inhibited tumor growth more effectively compared with PS ASOs after intratumoral injection. Comparison of siRNA with ASOs, each targeting green fluorescent protein, revealed more efficient inhibition by the siRNA in both cell culture and in mice.95
Around 50 clinical studies have used antisense strategies spanning a variety of disease processes, including cancer, cardiovascular disease, inflammation, and infection.96 Fomivirsen, or Vitravene, which targets the immediate-early RNA encoded by human CMV DNA, has been approved by the United States Food and Drug Administration for use in humans in treatment of CMV retinitis via intravitreal administration.97 Other antisense approaches that are currently entering Phase III trials include the ICAM-1 antisense Alicaforsen, which has shown promise in the treatment of inflammatory bowel disease when administered as a retention enema.98,99 Recently, in a Phase III trial, the addition of an antisense oligonucleotide targeting protein kinase C-α, Aprinocarsen, to a standard chemotherapeutic regimen for advanced non-small-cell lung carcinoma failed to improve outcomes.100 The bcl-2 oligonucleotide oblimersen, or Genasense, is currently in phase II/III for a variety of cancers, including chronic lymphocytic leukemia, acute myelocytic leukemia, melanoma, and multiple myeloma, and has been administered via intravenous and subcutaneous routes.101
Decoys
In contrast to antisense approaches that target mRNA, oligonucleotide decoys are short, double-stranded DNA molecules that contain binding elements for a variety of protein targets that competitively inhibit promoter binding and gene expression. Several types of decoys have been developed, including unmodified oligonucleotide duplexes, α-β-anomeric oligonucleotides, duplexes with methylphosphonate- and phosphorothioate-modified bonds, and circular dumbbell double-stranded oligodeoxynucleotides.102 Morishita et al103 demonstrated suppression of neointima formation with a decoy oligonucleotide to E2F in a rat model of carotid injury. This was extended further in a rabbit model of vein-conduit arterial bypass grafting in cholesterol-fed rabbits with a reduction in the incidence of neointima formation and atherosclerosis at 6 months in animals treated ex vivo with the E2F decoys.104 Following from the PREVENT trial, which established feasibility and safety of the E2F decoy Edifoligide in infra-inguinal vascular bypass surgery,105 the recently reported Phase III PREVENT IV study evaluated the efficacy of Edifoligide in preventing vein graft failure in patients undergoing coronary artery bypass grafting. Although safe and well tolerated, no significant improvements in graft failure rate or angiographic appearances of vein grafts at 12 months was achieved.106 Whether the established effects of Edifoligide on smooth muscle cell proliferation translates into longer-term benefits in this context remains to be seen. Other issues affecting the potential clinical use of molecules include susceptibility to nuclease degradation, propensity to induce a host immunological response, and cell transfection difficulties necessitating higher concentration requirements.
Ribozymes
Ribozymes are catalytically active RNA molecules capable of site-specific cleavage of target mRNA and, unlike DNAzymes, can occur naturally. Like DNAzymes and ASOs, ribozymes need access to their binding sites in the target RNA. Several subtypes have been described; those most commonly studied are hammerhead and hairpin ribozymes,44 which differ in their catalytic response to changes in solvent pH rather than their capacity to bind and ligate cleavage products or reliance on metal ions.107 Ribozyme catalytic activity and stability can be improved by substituting deoxyribonucleotides for ribonucleotides at noncatalytic bases.108 Chimeric DNA-RNA hammerhead ribozymes targeting platelet-derived growth factor A-chain mRNA have been shown to inhibit intimal thickening in balloon-injured rat carotid arteries after local delivery,109 whereas those targeting transforming growth factor-β protect against renal injury in hypertensive rats after systemic (intraperitoneal) delivery.110 Clinically, ribozymes have been explored therapeutically in several small trials. Hammerhead anti-HIV ribozymes have been used in T-lymphocyte expansion strategies ex vivo followed by infusion into patients.111,112,113,114 Hammerhead ribozymes targeting a highly conserved portion of 5′-untranslated region of hepatitis C virus HEPTAZYME115 showed promise in phase I and II trials. However, because of toxicological concerns, the study was suspended.116 Ribozymes have also been evaluated as potential adjuncts in cancer therapy. These include the synthetic antiangiogenic ANGIOZYME, which targets the VEGF receptor VEGF R1 (Flt-1) in a variety of solid tumors,117 and HERzyme, which targets human epidermal growth factor-2 overexpressed in breast and ovarian cell carcinoma.118
Aptamers
Finally, aptamers (from the Latin aptus, “to fit”) are synthetic oligonucleotide ligands that have been derived by in vitro selection from a combinatorial library of nucleic acid sequences that like decoys (but unlike antisense approaches) bind their target protein with high affinity and specificity, inhibiting function. The clinical appeal of aptamers has been enhanced by the introduction of chemical modifications, such as substitutions of the 2′-OH groups of the ribose backbone to provide resistance against enzymatic degradation.119 Pegaptanib, an RNA aptamer targeting VEGF165, has been evaluated in patients with neovascular age-related macular degeneration. Intravitreal delivery of this agent results in less visual loss and other clinically relevant improvements as early as 6 weeks, and this agent has been approved by the United States Food and Drug Administration for use against age-related macular degeneration.120 RNA and DNA aptamers have also been developed that inhibit HIV-1 function by directly interfering with key proteins at critical stages in the viral replication cycle.121 Other antiviral aptamers under development include those targeting hepatitis C virus and influenza virus.122 More recently, DNA or RNA molecules have been selected based on their capacity to bind targets with high affinity and specificity using the systematic evolution of ligands by exponential enrichment combinatorial oligonucleotide library-based in vitro selection approach.123
Parting Remarks
Gene targeting using nucleic acid strategies has now entered a new era with the evolution of stable, potent, and effective molecules. In particular, DNAzymes, siRNA, and antisense oligonucleotides by virtue of their relative specificity and stability have enabled precise targeting of genes regulating pivotal processes in the pathogenesis of disease, providing an exciting class of potential therapeutic tool and a means of understanding complex transcriptional and molecular pathways. Current studies have demonstrated their versatility and potency in disrupting pathophysiologically important pathways, via a variety of different delivery routes with relative specificity of action and in vivo stability.
With the ongoing identification of new genes and an appreciation of their regulatory pathways and pathological roles, small-molecule antigene strategies have not only emerged as an important molecular approach to delineate the functions of these genes but also are now a clinical reality inching closer to mainstream therapeutics. Progress over the next few years will determine the feasibility of small-molecule nucleic acids to silence disease-causing genes in man specifically and with minimal toxicity.
Acknowledgments
We thank Dr. Gerald F. Joyce (Departments of Chemistry and Molecular Biology, Scripps Research Institute, La Jolla, CA) for critical review of this manuscript.
Footnotes
Address reprint requests to Levon M. Khachigian, Ph.D., D.Sc., Centre for Vascular Research, Department of Pathology, The University of New South Wales, Sydney, NSW 2052, Australia. E-mail: L.Khachigian@unsw.edu.au.
References
- Paterson BM, Roberts BE, Kuff EL. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc Natl Acad Sci USA. 1977;74:4370–4374. doi: 10.1073/pnas.74.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska Z, Furste JP, Erdmann VA, Kurreck J. Sequence requirements in the catalytic core of the “10-23” DNA enzyme. J Biol Chem. 2002;277:40617–40622. doi: 10.1074/jbc.M207094200. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- Cairns MJ, Hopkins TM, Witherington C, Wang L, Sun L-Q. Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol. 1999;17:480–486. doi: 10.1038/8658. [DOI] [PubMed] [Google Scholar]
- Sun L-Q, Cairns MJ, Gerlach WL, Witherington C, Wang L, King A. Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J Biol Chem. 1999;274:17236–17241. doi: 10.1074/jbc.274.24.17236. [DOI] [PubMed] [Google Scholar]
- Isaka Y, Nakamura H, Mizui M, Takabatake Y, Horio M, Kawachi H, Shimizu F, Imai E, Hori M. DNAzyme for TGF-β suppressed extracellular matrix accumulation in experimental glomerulonephritis. Kidney Int. 2004;66:586–590. doi: 10.1111/j.1523-1755.2004.00777.x. [DOI] [PubMed] [Google Scholar]
- Nunamaker EA, Zhang HY, Shirasawa Y, Benoit JN, Dean DA. Electroporation-mediated delivery of catalytic oligodeoxynucleotides for manipulation of vascular gene expression. Am J Physiol. 2003;285:H2240–H2247. doi: 10.1152/ajpheart.00350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, McMahon R, Rossi JJ, Forman SJ, Snyder DS. Inhibition of bcr-abl oncogene by novel deoxyribozymes (DNAzymes). Hum Gene Ther. 1999;10:2847–2857. doi: 10.1089/10430349950016573. [DOI] [PubMed] [Google Scholar]
- Liang Z, Wei S, Guan J, Luo Y, Gao J, Zhu H, Wu S, Liu T. DNAzyme-mediated cleavage of survivin mRNA and inhibition of the growth of PANC-1 cells. J Gastroenterol Hepatol. 2005;20:1595–1602. doi: 10.1111/j.1440-1746.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Tan XX, Rose K, Margolin W, Chen Y. DNA enzyme generated by a novel single-stranded DNA expression vector inhibits expression of the essential bacterial cell division gene ftsZ. Biochemistry. 2004;43:1111–1117. doi: 10.1021/bi035164h. [DOI] [PubMed] [Google Scholar]
- Kabuli M, Yin JA, Tobal K. Targeting PML/RARα transcript with DNAzymes results in reduction of proliferation and induction of apoptosis in APL cells. Hematol J. 2004;5:426–433. doi: 10.1038/sj.thj.6200535. [DOI] [PubMed] [Google Scholar]
- Toyoda T, Imamura Y, Takaku H, Kashiwagi T, Hara K, Iwahashi J, Ohtsu Y, Tsumura N, Kato H, Hamada N. Inhibition of influenza virus replication in cultured cells by RNA-cleaving DNA enzyme. FEBS Lett. 2000;481:113–116. doi: 10.1016/s0014-5793(00)01974-8. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Niewiarowska J, Nawrot M, Koziolkiewicz M, Stec WJ, Cierniewski CS. DNAzymes to β1 and β3 mRNA down-regulate expression of the targeted integrins and inhibit endothelial cell capillary tube formation in fibrin and matrigel. J Biol Chem. 2002;277:6779–6787. doi: 10.1074/jbc.M102325200. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Szymanski J, Adamiak RW, Cierniewski CS. Structural rearrangements of the 10-23 DNAzyme to beta 3 integrin subunit mRNA induced by cations and their relations to the catalytic activity. J Biol Chem. 2003;278:47987–47996. doi: 10.1074/jbc.M300504200. [DOI] [PubMed] [Google Scholar]
- Unwalla H, Banerjea AC. Novel mono- and di-DNA-enzymes targeted to cleave TAT or TAT-REV RNA inhibit HIV-1 gene expression. Antiviral Res. 2001;51:127–139. doi: 10.1016/s0166-3542(01)00144-9. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gasper WJ, Stass SA, Ioffe OB, Davis MA, Mixson AJ. Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res. 2002;62:5463–5469. [PubMed] [Google Scholar]
- de Bock CE, Lin Z, Itoh T, Morris D, Murrell G, Wang Y. Inhibition of urokinase receptor gene expression and cell invasion by anti-uPAR DNAzymes in osteosarcoma cells. FEBS J. 2005;272:3572–3582. doi: 10.1111/j.1742-4658.2005.04778.x. [DOI] [PubMed] [Google Scholar]
- Fahmy RG, Dass CR, Sun LQ, Chesterman CN, Khachigian LM. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- Mitchell A, Dass CR, Sun L-Q, Khachigian LM. Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumor growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic Acids Res. 2004;32:3065–3069. doi: 10.1093/nar/gkh626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe HC, Fahmy RG, Kavurma MM, Baker A, Chesterman CN, Khachigian LM. Catalytic oligodeoxynucleotides define a key regulatory role for early growth response factor-1 in the porcine model of coronary in-stent restenosis. Circ Res. 2001;89:670–677. doi: 10.1161/hh2001.097867. [DOI] [PubMed] [Google Scholar]
- Santiago FS, Lowe HC, Kavurma MM, Chesterman CN, Baker A, Atkins DG, Khachigian LM. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth factor injury. Nat Med. 1999;5:1264–1269. doi: 10.1038/15215. [DOI] [PubMed] [Google Scholar]
- Khachigian LM, Fahmy RG, Zhang G, Bobryshev YV, Kaniaros A. c-Jun regulates vascular smooth muscle cell growth and neointima formation after arterial injury: inhibition by a novel DNAzyme targeting c-Jun. J Biol Chem. 2002;277:22985–22991. doi: 10.1074/jbc.M200977200. [DOI] [PubMed] [Google Scholar]
- Zhang G, Dass CR, Sumithran E, Di Girolimo NR, Sun L-Q, Khachigian LM. Effect of deoxyribozymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J Natl Cancer Inst. 2004;96:683–696. doi: 10.1093/jnci/djh120. [DOI] [PubMed] [Google Scholar]
- Zhang G, Luo X, Sumithran E, Pua VSC, Barnetson RS, Halliday GM, Khachigian LM. Squamous cell carcinoma growth in mice and in culture is regulated by c-Jun and its control of matrix metalloproteinase-2 and -9 expression. Oncogene. 2006;25:7260–7266. doi: 10.1038/sj.onc.1209726. [DOI] [PubMed] [Google Scholar]
- Xiang G, Seki T, Schuster MD, Witkowski P, Boyle AJ, See F, Martens TP, Kocher A, Sondermeijer H, Krum H, Itescu S. Catalytic degradation of vitamin D up-regulated protein 1 mRNA enhances cardiomyocyte survival and prevents left ventricular remodeling after myocardial ischemia. J Biol Chem. 2005;280:39394–39402. doi: 10.1074/jbc.M502966200. [DOI] [PubMed] [Google Scholar]
- Dass CR, Saravolac EG, Li Y, Sun LQ. Cellular uptake, distribution, and stability of 10-23 deoxyribozymes. Antisense Nucleic Acid Drug Dev. 2002;12:289–299. doi: 10.1089/108729002761381276. [DOI] [PubMed] [Google Scholar]
- Lu ZX, Ye M, Yan GR, Li Q, Tang M, Lee LM, Sun LQ, Cao Y. Effect of EBV LMP1 targeted DNAzymes on cell proliferation and apoptosis. Cancer Gene Ther. 2005;12:647–654. doi: 10.1038/sj.cgt.7700833. [DOI] [PubMed] [Google Scholar]
- Schubert S, Gul DC, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J. RNA cleaving ‘10-23’ DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003;31:5982–5992. doi: 10.1093/nar/gkg791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluiter K, ten Asbroek AL, de Wissel MB, Jakobs ME, Wissenbach M, Olsson H, Olsen O, Oerum H, Baas F. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003;31:953–962. doi: 10.1093/nar/gkg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell P, O’Connor WJ, King K, Goldstein NI, Zhang LM, Stein CA. Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc Natl Acad Sci USA. 1997;94:6523–6528. doi: 10.1073/pnas.94.12.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvakova MA, Yakubov LA, Vlodavsky I, Tonkinson JL, Stein CA. Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem. 1995;270:2620–2627. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Wengel J. Synthesis of novel 2′,3′-linked bicyclic thymine ribonucleosides. J Org Chem. 1998;63:2778–2781. doi: 10.1021/jo972239c. [DOI] [PubMed] [Google Scholar]
- Fahmy RG, Khachigian LM. Locked nucleic acid-modified DNA enzymes targeting early growth response-1 inhibit vascular smooth muscle cell growth. Nucleic Acids Res. 2004;32:2281–2285. doi: 10.1093/nar/gkh543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Lundberg LB, Sorensen MD, Babu BR, Douthwaite S, Wengel J. LNAzymes: incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J Am Chem Soc. 2002;124:13682–13683. doi: 10.1021/ja0276220. [DOI] [PubMed] [Google Scholar]
- Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Wengel J. Synthesis of 3′-C- and 4′-C-branched oligonucleotides and the development of locked nucleic acid (LNA). Acc Chem Res. 1999;32:301–310. [Google Scholar]
- Petersen M, Nielsen CB, Nielsen KE, Jensen GA, Bondensgaard K, Singh SK, Rajwanshi VK, Koshkin AA, Dahl BM, Wengel J, Jacobsen JP. The conformations of locked nucleic acids (LNA). J Mol Recognit. 2000;13:44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2′-thio-LNA. Bioorg Med Chem Lett. 1998;8:2219–2222. doi: 10.1016/s0960-894x(98)00366-7. [DOI] [PubMed] [Google Scholar]
- Arzumanov A, Walsh AP, Rajwanshi VK, Kumar R, Wengel J, Gait MJ. Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–14654. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]
- Fluiter K, Frieden M, Vreijling J, Koch T, Baas F. Evaluation of LNA-modified DNAzymes targeting a single nucleotide polymorphism in the large subunit of RNA polymerase II. Oligonucleotides. 2005;15:246–254. doi: 10.1089/oli.2005.15.246. [DOI] [PubMed] [Google Scholar]
- Schubert S, Kurreck J. Ribozyme- and deoxyribozyme-strategies for medical applications. Curr Drug Targets. 2004;5:667–681. doi: 10.2174/1389450043345092. [DOI] [PubMed] [Google Scholar]
- Khachigian LM. Catalytic DNA as therapeutic agents and molecular tools to dissect biological function. J Clin Invest. 2000;106:1189–1195. doi: 10.1172/JCI11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- Lowe HC, Chesterman CN, Khachigian LM. Catalytic antisense DNA molecules targeting Egr-1 inhibit neointima formation following permanent ligation of rat common carotid arteries. Thromb Haemost. 2002;87:134–140. [PubMed] [Google Scholar]
- Lowe HC, Khachigian LM. Coating stents with antirestenotic drugs: the blunderbuss or the magic bullet? Circulation. 2002;105:E29. [PubMed] [Google Scholar]
- Fahmy R, Waldman A, Zhang G, Mitchell A, Tedla N, Cai H, Chesterman CN, Geczy CR, Perry MA, Khachigian LM. Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat Biotechnol. 2006;24:856–863. doi: 10.1038/nbt1225. [DOI] [PubMed] [Google Scholar]
- Murrell M, Khachigian L, Ward MR: The role of c-jun in PDTC-sensitive flow-dependent restenosis after angioplasty and stenting. Atherosclerosis 2006, [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bhindi R, Khachigian LM, Lowe HC. DNAzymes targeting the transcription factor Egr-1 reduce myocardial infarct size following ischemia-reperfusion in rats. J Thromb Haemost. 2006;4:1479–1483. doi: 10.1111/j.1538-7836.2006.02022.x. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Isaka Y, Tsujie M, Rupprecht HD, Akagi Y, Ueda N, Imai E, Hori M. Introduction of DNA enzyme for Egr-1 into tubulointerstitial fibroblasts by electroporation reduced interstitial alpha-smooth muscle actin expression and fibrosis in unilateral ureteral obstruction (UUO) rats. Gene Ther. 2002;9:495–502. doi: 10.1038/sj.gt.3301681. [DOI] [PubMed] [Google Scholar]
- Grimpe B, Silver J. A novel DNA enzyme reduces glycosaminoglycan chains in the glial scar and allows microtransplanted dorsal root ganglia axons to regenerate beyond lesions in the spinal cord. J Neurosci. 2004;24:1393–1397. doi: 10.1523/JNEUROSCI.4986-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Wan C, Zhou W, Peng T, Wang Z, Li G, Cornelisson G, Halberg F. The role of mPer1 in morphine dependence in mice. Neuroscience. 2005;130:383–388. doi: 10.1016/j.neuroscience.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Schuster MD, Seki T, Kocher AA, Eshghi S, Boyle A, Itescu S. Down-regulation of plasminogen activator inhibitor 1 expression promotes myocardial neovascularization by bone marrow progenitors. J Exp Med. 2004;200:1657–1666. doi: 10.1084/jem.20040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Schuster MD, Seki T, Witkowski P, Eshghi S, Itescu S. Downregulated expression of plasminogen activator inhibitor-1 augments myocardial neovascularization and reduces cardiomyocyte apoptosis after acute myocardial infarction. J Am Coll Cardiol. 2005;46:536–541. doi: 10.1016/j.jacc.2005.04.047. [DOI] [PubMed] [Google Scholar]
- Iversen PO, Nicolaysen G, Sioud M. DNA enzyme targeting TNF-alpha mRNA improves hemodynamic performance in rats with postinfarction heart failure. Am J Physiol. 2001;281:H2211–H2217. doi: 10.1152/ajpheart.2001.281.5.H2211. [DOI] [PubMed] [Google Scholar]
- Rivory L, Tucker C, King A, Lai A, Goodchild A, Witherington C, Gozar MM, Birkett DJ. The DNAzymes Rs6, Dz13, and DzF have potent biologic effects independent of catalytic activity. Oligonucleotides. 2006;16:297–312. doi: 10.1089/oli.2006.16.297. [DOI] [PubMed] [Google Scholar]
- Burgess TI, Fisher EF, Ross SL, Bready JV, Qian Y-X, Bayewitch IA, Cohen AM, Herrera CJ, Hu SS-F, Framer TB, Lott FD, Martin FH, Pierce GF, Simonet L, Farrell CL. The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc Natl Acad Sci USA. 1995;92:4051–4055. doi: 10.1073/pnas.92.9.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Baldwin AS. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awada A, Mano M, Hendlisz A, Piccart M. New anticancer agents and therapeutic strategies in development for solid cancers: a clinical perspective. Expert Rev Anticancer Ther. 2004;4:53–60. doi: 10.1586/14737140.4.1.53. [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- Diaz-Hernandez JI, Almeida A, Delgado-Esteban M, Fernandez E, Bolanos JP. Knockdown of glutamate-cysteine ligase by small hairpin RNA reveals that both catalytic and modulatory subunits are essential for the survival of primary neurons. J Biol Chem. 2005;280:38992–39001. doi: 10.1074/jbc.M507065200. [DOI] [PubMed] [Google Scholar]
- Sandy P, Ventura A, Jacks T. Mammalian RNAi: a practical guide. Biotechniques. 2005;39:215–224. doi: 10.2144/05392RV01. [DOI] [PubMed] [Google Scholar]
- Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–449. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- Sørensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- Kim B, Tang Q, Biswas PS, Xu J, Schiffelers RM, Xie FY, Ansari AM, Scaria PV, Woodle MC, Lu P, Rouse BT. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165:2177–2185. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers RM, Storm G. ICS-283: a system for targeted intravenous delivery of siRNA. Expert Opin Drug Deliv. 2006;3:445–454. doi: 10.1517/17425247.3.3.445. [DOI] [PubMed] [Google Scholar]
- Pirollo KF, Zon G, Rait A, Zhou Q, Yu W, Hogrefe R, Chang EH. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17:117–124. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi Y, Takeshita F, Kosaka N, Sasaki H, Yamamoto Y, Kouno M, Honma K, Nagahara S, Hanai K, Sano A, Kato T, Terada M, Ochiya T. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y, Hanai K, Kato T, Sano A, Ochiya T. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11:67–73. doi: 10.1016/S1359-6446(05)03668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Shen J, Samul R, Silva RL, Akiyama H, Liu H, Saishin Y, Hackett SF, Zinnen S, Kossen K, Fosnaugh K, Vargeese C, Gomez A, Bouhana K, Aitchison R, Pavco P, Campochiaro PA. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, Bennett J, Tolentino MJ. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- Tolentino MJ, Brucker AJ, Fosnot J, Ying GS, Wu IH, Malik G, Wan S, Reich SJ. Intravitreal injection of vascular endothelial growth factor small interfering RNA inhibits growth and leakage in a nonhuman primate, laser-induced model of choroidal neovascularization. Retina. 2004;24:660. doi: 10.1097/00006982-200408000-00039. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Schwartz SM. Antisense therapy for angioplasty restenosis: some critical considerations. Circulation. 1995;92:1981–1993. doi: 10.1161/01.cir.92.7.1981. [DOI] [PubMed] [Google Scholar]
- Gao WY, Han FS, Storm C, Egan W, Cheng YC. Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications forantisense technology. Mol Pharmacol. 1992;41:223–229. [PubMed] [Google Scholar]
- Gao WY, Stein CA, Cohen JS, Dutschman GE, Cheng YC. Effect of phosphorothioate homo-oligodeoxynucleotides on herpes simplex virus type 2-induced DNA polymerase. J Biol Chem. 1989;264:11521–11526. [PubMed] [Google Scholar]
- Nielsen CB, Singh SK, Wengel J, Jacobsen JP. The solution structure of a locked nucleic acid (LNA) hybridized to DNA. J Biomol Struct Dyn. 1999;17:175–191. doi: 10.1080/07391102.1999.10508352. [DOI] [PubMed] [Google Scholar]
- Nielsen KE, Singh SK, Wengel J, Jacobsen JP. Solution structure of an LNA hybridized to DNA: nMR study of the d(CT(L)GCT(L)T(L)CT(L)GC):d(GCAGAAGCAG) duplex containing four locked nucleotides. Bioconjug Chem. 2000;11:228–238. doi: 10.1021/bc990121s. [DOI] [PubMed] [Google Scholar]
- Knight DA, Briggs BR, Bennett CF, Harindranath N, Waldman WJ, Sedmak DD. Attenuation of cytomegalovirus-induced endothelial intercellular adhesion molecule-1 mRNA/protein expression and T lymphocyte adhesion by a 2′-O-methoxyethyl antisense oligonucleotide. Transplantation. 2000;69:417–426. doi: 10.1097/00007890-200002150-00019. [DOI] [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Itoh H, Sato W, Nakazawa K, Kubota S, Muramatsu T. 5′-,3′-Inverted thymidine-modified antisense oligodeoxynucleotide targeting midkine: its design and application for cancer therapy. J Biol Chem. 2002;277:23800–23806. doi: 10.1074/jbc.M112100200. [DOI] [PubMed] [Google Scholar]
- Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- Persidis A. Antisense therapeutics. Nat Biotechnol. 1999;17:403–404. doi: 10.1038/7973. [DOI] [PubMed] [Google Scholar]
- Yacyshyn BR, Chey WY, Goff J, Salzberg B, Baerg R, Buchman AL, Tami J, Yu R, Gibiansky E, Shanahan WR. Double blind, placebo controlled trial of the remission inducing and steroid sparing properties of an ICAM-1 antisense oligodeoxynucleotide, alicaforsen (ISIS 2302), in active steroid dependent Crohn’s disease. Gut. 2002;51:30–36. doi: 10.1136/gut.51.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer SJ, Tami JA, Wedel MK. A randomised, controlled, double blind, escalating dose study of alicaforsen enema in active ulcerative colitis. Gut. 2004;53:1646–1651. doi: 10.1136/gut.2003.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares L, Douillard JY, Koralewski P, Manegold C, Smit EF, Reyes JM, Chang GC, John WJ, Peterson PM, Obasaju CK, Lahn M, Gandara DR. Phase III study of gemcitabine and cisplatin with or without aprinocarsen, a protein kinase C-α antisense oligonucleotide, in patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2006;24:1428–1434. doi: 10.1200/JCO.2005.04.3299. [DOI] [PubMed] [Google Scholar]
- Pirollo KF, Rait A, Sleer LS, Chang EH. Antisense therapeutics: from theory to clinical practice. Pharmacol Ther. 2003;99:55–77. doi: 10.1016/s0163-7258(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Lee IK, Ahn JD, Kim HS, Park JY, Lee KU. Advantages of the circular dumbbell decoy in gene therapy and studies of gene regulation. Curr Drug Targets. 2003;4:619–623. doi: 10.2174/1389450033490821. [DOI] [PubMed] [Google Scholar]
- Morishita R, Gibbons GH, Horiuchi M, Ellison KE, Nakama M, Zhang L, Kaneda Y, Ogihara T, Dzau VJ. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc Natl Acad Sci USA. 1995;92:5855–5859. doi: 10.1073/pnas.92.13.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan A, Mann MJ, Dell’Acqua G, Dzau VJ. Long-term stabilization of vein graft wall architecture and prolonged resistance to experimental atherosclerosis after E2F decoy oligonucleotide gene therapy. J Thorac Cardiovasc Surg. 2001;121:714–722. doi: 10.1067/mtc.2001.111204. [DOI] [PubMed] [Google Scholar]
- Mann MJ, Whittemore AD, Donaldson MC, Belkin M, Conte MS, Polak JF, Orav EJ, Ehsan A, Dell’Acqua G, Dzau VJ. Ex-vivo gene therapy of human vascular bypass grafts with E2F decoy: the PREVENT single-centre, randomised, controlled trial. Lancet. 1999;354:1493–1498. doi: 10.1016/S0140-6736(99)09405-2. [DOI] [PubMed] [Google Scholar]
- Alexander JH, Ferguson TB, Jr, Joseph DM, Mack MJ, Wolf RK, Gibson CM, Gennevois D, Lorenz TJ, Harrington RA, Peterson ED, Lee KL, Califf RM, Kouchoukos NT. The project of ex-vivo vein graft engineering via transfection IV (PREVENT IV) trial: study rationale, design, and baseline patient characteristics. Am Heart J. 2005;150:643–649. doi: 10.1016/j.ahj.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Burke JM. Hairpin and hammerhead ribozymes: how different are they? Biochem Soc Trans. 2002;30:1115–1118. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Taylor NR, Kaplan BE, Swiderski P, Li H, Rossi JJ. Chimeric DNA-RNA hammerhead ribozymes have enhanced in vitro catalytic efficiency and increased stability in vivo. Nucleic Acids Res. 1992;20:4559–4565. doi: 10.1093/nar/20.17.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani M, Fukuda N, Ando H, Hu WY, Kunimoto S, Saito S, Kanmatsuse K. Chimeric DNA-RNA hammerhead ribozyme targeting PDGF A-chain mRNA specifically inhibits neointima formation in rat carotid artery after balloon injury. Cardiovasc Res. 2003;57:265–276. doi: 10.1016/s0008-6363(02)00607-7. [DOI] [PubMed] [Google Scholar]
- Tahira Y, Fukuda N, Endo M, Ueno T, Matsuda H, Saito S, Matsumoto K, Mugishima H. Chimeric DNA-RNA hammerhead ribozyme targeting transforming growth factor-beta1 mRNA ameliorates renal injury in hypertensive rats. J Hypertens. 2007;25:671–678. doi: 10.1097/HJH.0b013e3280122f22. [DOI] [PubMed] [Google Scholar]
- Brower V, Chahine K, Dorey E, Dove A, Francisco M, Hodgson J, Michael A, Marshall A. All clear for HIV-targeting ribozyme in phase II. Nat Biotechnol. 1998;16:123. doi: 10.1038/nbt0298-123. [DOI] [PubMed] [Google Scholar]
- Rowe PM. Ribozymes enter clinical trials for HIV-1 treatment. Lancet. 1996;348:1302. doi: 10.1016/S0140-6736(05)65767-4. [DOI] [PubMed] [Google Scholar]
- Macpherson JL, Boyd MP, Arndt AJ, Todd AV, Fanning GC, Ely JA, Elliott F, Knop A, Raponi M, Murray J, Gerlach W, Sun LQ, Penny R, Symonds GP, Carr A, Cooper DA. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J Gene Med. 2005;7:552–564. doi: 10.1002/jgm.705. [DOI] [PubMed] [Google Scholar]
- Wong-Staal F, Poeschla EM, Looney DJ. A controlled, phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum Gene Ther. 1998;9:2407–2425. doi: 10.1089/hum.1998.9.16-2407. [DOI] [PubMed] [Google Scholar]
- Usman N, Blatt LM. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J Clin Invest. 2000;106:1197–1202. doi: 10.1172/JCI11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi A. Prospects for antiviral ribozymes and deoxyribozymes. Rev Med Virol. 2004;14:47–64. doi: 10.1002/rmv.415. [DOI] [PubMed] [Google Scholar]
- Weng DE, Masci PA, Radka SF, Jackson TE, Weiss PA, Ganapathi R, Elson PJ, Capra WB, Parker VP, Lockridge JA, Cowens JW, Usman N, Borden EC. A phase I clinical trial of a ribozyme-based angiogenesis inhibitor targeting vascular endothelial growth factor receptor-1 for patients with refractory solid tumors. Mol Cancer Ther. 2005;4:948–955. doi: 10.1158/1535-7163.MCT-04-0210. [DOI] [PubMed] [Google Scholar]
- Zinnen SP, Domenico K, Wilson M, Dickinson BA, Beaudry A, Mokler V, Daniher AT, Burgin A, Beigelman L. Selection, design, and characterization of a new potentially therapeutic ribozyme. RNA. 2002;8:214–228. doi: 10.1017/s1355838202014723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. RNA aptamers: from basic science towards therapy. Handb Exp Pharmacol. 2006;173:305–326. doi: 10.1007/3-540-27262-3_15. [DOI] [PubMed] [Google Scholar]
- Gragoudas ES, Adamis AP, Cunningham ET, Jr, Feinsod M, Guyer DR, Group VISiONCT Pegaptanib for neovascular age-related macular degeneration. N Engl J Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- Bunka DH, Stockley PG. Aptamers come of age: at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Ulrich H, Trujillo CA, Nery AA, Alves JM, Majumder P, Resende RR, Martins AH. DNA and RNA aptamers: from tools for basic research towards therapeutic applications. Comb Chem High Throughput Screen. 2006;9:619–632. doi: 10.2174/138620706778249695. [DOI] [PubMed] [Google Scholar]