Abstract
Etk/Bmx is the newest member of Btk tyrosine kinase family that contains a pleckstrin homology domain, an src homology 3 domain, an src homology 2 domain, and a catalytic domain. Unlike other members of the Btk family kinases, which are mostly hemopoietic cell-specific, Etk/Bmx is preferentially expressed in epithelial and endothelial cells. We first identified this kinase in prostate cancer [Robinson, D., He, F., Pretlow, T. & Kung, H. J. (1996) Proc. Natl. Acad. Sci. USA 93, 5958–5962). Here we report that Etk is engaged in phosphatidylinositol 3-kinase (PI3-kinase) pathway and plays a pivotal role in interleukin 6 (IL-6) signaling in a prostate cancer cell line, LNCaP. Our evidence that PI3-kinase is involved in Etk activation includes: (i) Wortmannin, a specific inhibitor of PI3-kinase, abolished the activation of Etk by IL-6; (ii) a constitutively active p110 subunit of PI3-kinase was able to activate Etk in the absence of IL-6; and (iii) a dominant negative p85 subunit of PI3-kinase mutant blocked the activation of Etk by IL-6. Interestingly, IL-6 treatment of LNCaP induced a remarkable neuroendocrine-like differentiation phenotype, with neurite extension and enhanced expression of neuronal markers. This phenotype could be abrogated by the overexpression of a dominant-negative Etk, indicating Etk is required for this differentiation process.
Keywords: cytokine, signal transduction, phosphatidylinositides, LNCaP
Prostate cancer (PCA) represents the most frequently diagnosed malignancy in men. The growth of PCA cells can often be modulated by peptide growth factors such as type α transforming growth factor and cytokines such as interleukin 6 (IL-6). The signals that regulate the growth and apoptosis of PCA, however, remain largely unexplored. In an effort to define the pathways involved, we previously established a tyrosine kinase expression profile of PCA and uncovered several novel tyrosine kinases (1). Etk (epithelial and endothelial tyrosine kinase) is one of them. This kinase was independently cloned and isolated by Tamagnone et al. (2), and referred to as Bmx. We shall thus use Etk/Bmx and Etk interchangeably in this report. Etk is preferentially expressed in epithelial and endothelial cells including all PCA samples thus far surveyed (ref. 2, Y.Q. and D.R., unpublished data). Etk has a structure resembling the Btk family of kinases with a pleckstrin homology domain (PH), an src homology 3 (SH3) domain, an SH2 domain, and a catalytic (kinase) domain. The other three members of this family are Btk (3, 4), Itk (5, 6), and Tec (7). Btk (also known as BPK or ATK), the prototype of this family of kinases, plays a pivotal role in B cell activation and development (8). Likewise, Itk/Tsk, the T cell analog of Btk, is crucial in T cell development (6) and Tec is involved in IL-3 signaling in hematopoietic stem cells (7). These kinases are found to be important in a variety of signal transduction processes such as radiation-induced apoptosis, IL-induced growth, and others (6, 7, 9–11). Most of the previously described Btk family kinases are highly expressed in hematopoietic cells but not in epithelial cells. Our knowledge of the role of Btk family kinases in signal transduction is therefore very much limited to cells of hematopoietic origin. Etk, on the other hand, has the reverse expression profile and is the only member of this family expressed in epithelial cells (1, 2). Here, we report the initial characterization of Etk and its potential role in modulating prostate cell growth.
Recent structural analysis of Itk reveals that the regulation of the Btk family of kinases is through the internal folding of the molecule (12). In its native “closed” form, the SH3 domain interacts with an adjacent proline-rich domain and the PH domain with the kinase domain. Signaling molecules that bear phosphotyrosine- and proline-rich domains can disrupt the intramolecular interactions, resulting in the unfolding of the kinase domain and leaving the potential phosphorylation sites exposed to Src family kinases (13–15). Through intermolecular transphosphorylation, the “pseudosubstrate” tyrosine within the kinase domain is phosphorylated, and the subsequent autophosphorylation leads to complete activation. Analogous to the activation of the PH-containing serine/threonine kinase Akt (16–18), this family of kinases may also be activated by phosphotidylinositol-3,4,5 triphosphate, a product of phosphotidylinositol 3-kinase (PI3-kinase), through binding to the PH domain (19). In this paper, we show that Etk is an effector of PI3-kinase, which in turn can be activated by the cytokine IL-6.
IL-6 was initially identified as a regulator of immune and inflammatory response. Recent studies suggest that IL-6 may also play a role in regulation of PCA cell growth (20, 21). The IL-6/IL-6 receptor autocrine loop seems to be present in all PCA cell lines tested (20, Qiu et al. unpublished results). The signals involved in PCA are largely unknown. In hematopoietic cells, IL-6 induces the activation of the tyrosine kinase Jak/Tyk2 (22), and consequently transcription factors STAT1 and STAT3 (23). Additional pathways such as mitogen-activating kinase (24–26) and PI3-kinase (27) have been also implicated. Whether similar pathways are activated in prostate epithelial cells has yet to be determined.
We report here that Etk plays an important role in IL-6 signaling in PCA cells. We demonstrate that IL-6 induces the activation of Etk and this activation is mediated by PI3-kinase. In addition, we show that IL-6 induces profound morphological changes resembling neuroendocrine differentiation in the PCA cell line LNCaP. This differentiation process is accompanied by the heightened level of neuronal markers such as neuron-specific enolase (NSE), and is dependent upon the activity of Etk.
MATERIALS AND METHODS
Cell Culture, Transfection, and Selection.
LNCaP cells (American Type Culture Collection, Rockville, MD) were maintained in RPMI1640 with 10% fetal bovine serum and CV-1 ATCC) were maintained in DMEM with 10% fetal bovine serum. Transfections were performed by using LipofectAmine (GIBCO/BRL) according to the manufacturer’s instruction. At 24hrs posttransfection, the cells were serum starved for 24 hr followed by treatment with IL-6 (Upstate Biotechnology, Lake Placid, NY) as indicated in the figure legends. The stable cell lines LN-EDN that overexpress the dominant-negative Etk were obtained by selection for G418 resistance (600 μg/ml) from the pool of LNCaP cells transfected with pcDNA3-T7-EtkDN, and their identity was confirmed by Western blot analysis of total cell lysates by using anti-T7 antibody. The morphology of the cells was recorded by using a phase-contrast microscope under conditions described in the figure legends.
Cloning, Plasmids, and Mutagenesis.
The full length Etk cDNA was cloned by using Marathone rapid amplification of cDNA ends (RACE) kit (CLONTECH). Briefly, the polyA RNA was isolated from human prostate xenograft CWR22 as described (1). The cDNA synthesis and the ligation of the adaptor followed the manufacturer’s protocol. The sequences of the primers used for 5′ RACE and 3′ RACE are: 5′-CATACGTCTGACTTGCTGC-3′ and 5′-GACAAGGTATGTTCTTGATG-3′, respectively. The full-length Etk cDNA was obtained by fusing the 5′ and 3′ RACE products together by using the High Fidelity PCR system (Boehringer Mannheim). The full-length cDNA was digested with NotI and cloned into pBSII, KS(+) (Stratgene). Three individual clones were sequenced by using the sequencing kit provided by United States Biochemical. Subsequently, full-length Etk was subcloned into the NotI site of mammalian expression vector pcDNA3 (Invitrogen) The T7-tag sequence or hemagglutinin (HA)-tag sequence was added to the 5′ of the coding sequence of Etk by PCR using the primers: 5′-GCGGTACCATGGCTAGCATGACTGGTGGACAGCAAATGGGTATGAAA3′ or 5′-GCGGTACCATGGCTTACCCTTATGATGTGCCAGATTATGCCTCCCGGGATACAAAATCTATTCTAG-3′. The tagged Etk sequences were then cloned into pcDNA3. All the mutants were generated by using PCR of the mutated region followed by relegation back to the wild-type construct lacking the corresponding region. The mutations were confirmed by sequencing.
Antibodies.
Antiserum against Etk was raised by immunizing the rabbits with purified glutathione S-transferase fusion protein containing the amino acids 53–279 of Etk and developed by Cocalico Biological, Reamstown, PA. Anti-T7 antibody (Novagen), anti-HA antibody (Babco, Richmond, CA), anti-NSE antibody, anti-pY antibody (Upstate Biotechnology), and anti-p85 (Upstate Biotechnology) antibody were purchased from the respective companies.
Immunoprecipitation, Western Blot Analysis, in Vitro Kinase Assays.
Immunoprecipitation and Western blot analysis were followed the procedures described by Grasso et al. (28). The in vitro kinase assays of Etk were performed as described by T. Hunter (29). Briefly, the immunoprecipitates were washed twice with Pipes and Nacl buffer (20 mM Pipes, pH 7.0/100 mM NaCl) and resuspended in 20 μl of the kinase reaction buffer [30 mM Pipes, pH 7.0/10 mM MnCl2/10 μM ATP/1 mM sodium orthovanadate/10 μCi γ-32P-ATP (1 Ci= 37 GBq)] containing 5 μg enolase (Sigma). The reactions were incubated at room temperature for 10 min and stopped by adding equal volume of 2× SDS loading buffer.
RESULTS
The Cloning and Structure of Etk/Bmx.
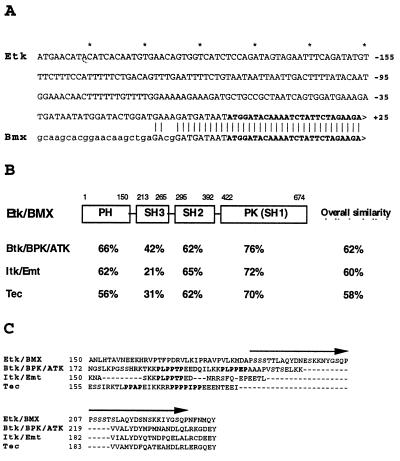
We initially identified Etk as a kinase-domain reverse transcription–PCR fragment derived from prostate carcinoma cells (1). Subsequently, we cloned the full-length cDNA sequence by the RACE method (30, 31). This cDNA has a 674-amino acid ORF. The coding sequence of our clone is identical to that reported by Tamagnone et al. (2); interestingly, these two clones differ substantially in the 5′ untranslated region (Fig. 1A), possibly due to alternative splicing in different tissues. A schematic diagram of the structure of Etk/Bmx is shown in Fig. 1B. The homology of the PH, SH2, SH3, and kinase domains to those of the Btk family is indicated (Fig. 1B). It is noteworthy, however, that Etk lacks the proline-rich region present in all other members of this family, which has been implicated in an intramolecular interaction with its adjacent SH3 domain (12). Instead, Etk has a sequence that is composed of two 22-amino acids repeats, which share about 70% homology to each other (the sequence marked by arrows in Fig. 1C).
Figure 1.
The structure of Etk/Bmx and its homology with the Btk family kinases. (A) The 5′ untranslated region of Etk is different from that of Bmx (plain text). The coding regions of Etk and Bmx are shown in boldface. (B) The domain-similarity of Etk/Bmx with other Btk family kinases are shown (based on the analysis using the GCG program). The overall similarity is shown in the last column. PK(SH1): protein kinase SH1. (C) Etk lacks the proline-rich region commonly present in other Btk family kinases. The proline-rich sequences PXXPXP are highlighted. The two 22-amino-acid repeats of Etk are marked by arrows.
The Activation of Etk by IL-6.
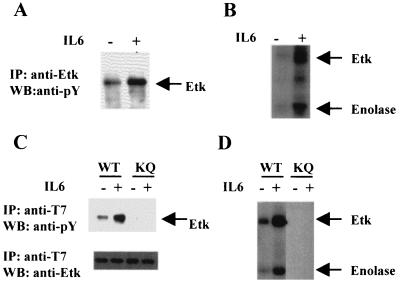
In hematopoietic cells, the Btk family kinases are often activated by cytokine receptors, antigen receptors or lymphoid-specific cell-surface receptors (6, 7, 24, 26). Most of these receptors are absent in epithelial cells. IL-6 and IL-6 receptors, however, are expressed in prostate epithelial cells, and IL-6 has been shown to have a profound effect on the growth of PCA cells. We therefore explored the possibility that Etk could be activated by IL-6. Treatment of LNCaP with IL-6 resulted in the activation of Etk, as evidenced by the increased tyrosine-phosphorylation of Etk in vivo (Fig. 2A), as well as in immune-complex kinase assay (Fig. 2B). Furthermore, the in vitro kinase activity of Etk toward an exogenous substrate such as enolase is significantly increased (Fig. 2B).
Figure 2.
Activation of Etk by IL-6 in LNCaP and CV-1 cells. (A) IL-6 induces the tyrosine phosphorylation of Etk in LNCaP. The cell extracts from the IL-6-treated(+) or untreated (−) cells were subjected to immunoprecipitation by the antiserum against Etk respectively, followed by Western blot analysis with anti-phosphotyrosine antibody. (B) IL-6 stimulates the kinase activity of Etk in LNCaP. The Etk immunoprecipitates from A were subjected to in vitro kinase assay with enolase as a substrate. The phosphorylated Etk and exogenous substrate enolase are indicated. (C) IL-6 induces tyrosine phosphorylation of exogenous T7-tagged wild-type Etk (WT), but not EtkK444Q mutant (KQ) in CV-1 cells. The CV-1 cells were transfected by pcDNA3-T7-Etk (wild type) or pcDNA3-T7-EtkKQ, respectively. The cell extracts from the IL-6-treated (+) or untreated (−) cells were subjected to immunoprecipitation by anti-T7 antibody respectively, followed by Western blot analysis with antiphosphotyrosine antibody. (D) IL-6 stimulates the in vitro kinase activity of Etk in CV-1 cells. The T7 immunoprecipitates from C were subjected to in vitro kinase assay. The phosphorylated Etk and exogenous substrate Enolase are indicated.
To corroborate the above finding, we overexpressed a T7 epitope-tagged Etk cDNA in the CV-1 cell line. This cell line is of epithelial origin and expresses endogenous IL-6 receptor. As shown in Fig. 2C, immunoprecipitation with anti-T7-tag antibody demonstrated increased Etk tyrosine phosphorylation following IL-6 treatment, both in vivo and in vitro. The exogenous activity of Etk toward enolase was likewise increased (Fig. 2D). To demonstrate that the observed kinase activity was derived from Etk itself and not from other associated kinases, lysine444 in the ATP binding pocket of Etk was mutated to glutamine. As shown in Fig. 2 C and D, the tyrosine phosphorylation of EtkK444Q (lanes KQ) is undetectable, indicating that the tyrosine phosphorylation of Etk induced by IL-6 is largely due to autophosphorylation.
The Activation of Etk by IL-6 via PI3-Kinase Pathway.
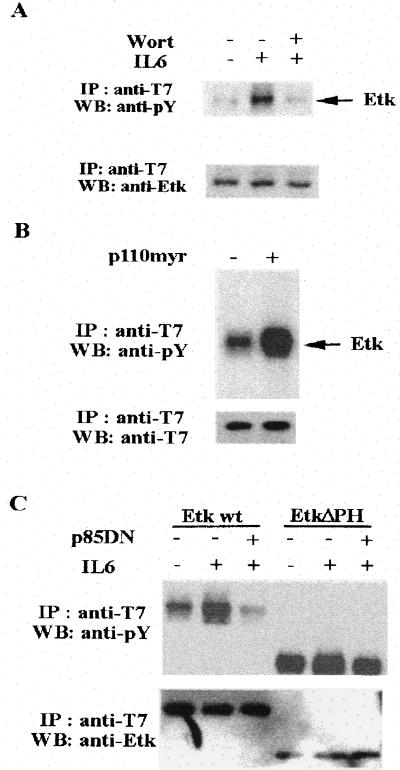
Having demonstrated that Etk can be activated by IL-6, we wished to study that pathways were involved in the activation of Etk. We initially noticed that IL-6 treatment of LNCaP led to a significant increase in tyrosine phosphorylation of the p85 subunit of PI3-kinase and its kinase activity (data not shown). Based on a recent study of Btk (19), its PH domain represents a binding site for the PI3-kinase product, phosphoinositol-triphosphate. We therefore tested whether IL-6 induced Etk activation requires PI3-kinase activity. We did this in three ways. First, the cells were pretreated with wortmannin, a specific inhibitor of PI3-kinase (32). Then the effect of IL-6 on tyrosine phosphorylation of Etk was examined. As shown in Fig. 3A, in the presence of wortmannin, the Etk activity was diminished to a level comparable to that of cells untreated with IL-6. Second, a constitutively active PI3-kinase, p110 myr, was developed by attaching a src myristylation site to the N terminus of the catalytic subunit p110 (33). Coexpression of p110 myr with Etk elevated the Etk activity (Fig. 3B). Finally, a dominant negative p85-PI3-kinase, p85DN, was constructed in which the domain that interacts with p110 was deleted (34). Coexpression of Etk with p85DN in cells reduced the Etk activity to basal level (Fig. 3C). These experiments taken together strongly suggest that IL-6 activates Etk via the PI3-kinase pathway, and Etk is downstream from PI3-kinase. The PH domain of Etk appears to be required for respond to PI3-kinase, because deletion of PH domain leads to a constitutively active Etk, which was no longer inhibitable by p85DN coexpression (see Fig. 3C).
Figure 3.
IL-6 activates Etk through PI3-kinase. (A) Wortmannin (Wort) blocks the activation of Etk by IL-6. The CV-1 cells were transfected with pcDNA3-T7-Etk. At 24 hr posttransfection, the cells were serum starved for 24 hr. The cells were then pretreated with 100 nM wortmannin (+) or dimethyl sulfoxide Mock (−) for 30 min followed by IL-6 treatment for 30 min. The cell extracts were subjected to immunoprecipitation by anti-T7 antibody respectively, followed by Western blot analysis with antiphosphotyrosine antibody (Upper) and anti-T7 antibody (Lower). The tyrosine phosphorylation of the T7-tagged Etk is indicated. (B) A myrislated p110 activates Etk. The CV-1 cells were transfected by pcDNA3-T7-Etk in the presence (+) or absence (−) of pcDNA3-p110 myr. At 24 hr posttransfection, the cells were serum starved for 24 hr. The cell extracts were subjected to immunoprecipitation by anti-T7 antibody respectively, followed by Western blot analysis with anti-phosphotyrosine antibody (Upper) and anti-Etk antibody (Lower). (C) A dominant negative p85 blocks the activation of Etk by IL-6. The CV-1 cells were transfected with pcDNA3-T7-Etk (wild type) or pcDNA3-T7-EtkΔPH(deletion of PH domain) in the presence (+) or absence (−) of pcDNA3-p85DN. At 24 hr posttransfection, the cells were serum starved for 24 hr followed by IL-6 treatment for 30 min. The cell extracts from each treatment were subjected to immunoprecipitation by anti-T7 antibody, followed by Western blot analysis with antiphosphotyrosine antibody (Upper) and anti-Etk antibody (Lower).
Modulation of Etk Kinase Activity by a Point Mutation in Its PH Domain.
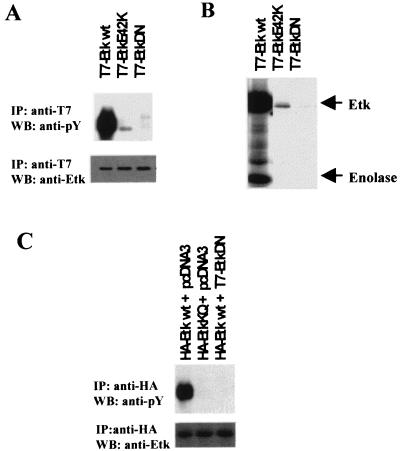
The experiments described in the previous section suggest that the PH domain regulates Etk kinase activity presumably by binding phosphotidylinositol polyphosphates. In the case of Btk, several PH domain mutations have been shown to associate with a hereditary immune disease, X-linked agammaglobulinaemia (8, 35). A glutamate residue (E41) in the Btk PH domain lies close to the predicted lipid binding site (36) and substitution of this residue with lysine (E41K) has a profound effect on the kinase activity of Btk (37). This glutamate is also present in Etk. Substitution of this glutamate residue of Etk (E42) with lysine significantly diminished the Etk kinase activity both in vivo and in vitro (Fig. 4 A and B). This finding reaffirms the regulatory role of the PH domain on kinase activity. Coupling this point mutation with one in the ATP binding loop of kinase domain (K444Q), we have developed a kinase-dead, dominant-negative mutant EtkDN. As shown in Fig. 4 A and B, the in vivo and in vitro kinase activity of EtkDaN mutant (lanes T7-EtkDaN) is completely absent. Coexpression of EtkDaN with wild-type Etk completely abolished Etk activity (Fig. 4C). This mutant is particularly useful in studying the biological function of Etk (see below).
Figure 4.
Modulation of Etk kinase activity by point mutations in PH and kinase domains. (A) Substitution of glutamate 42 of Etk with lysine diminishes the Etk activity in vivo. The CV-1 cells were transfected with pcDNA3-T7-Etk (wild type), pcDNA3-T7-EtkE42K or pcDNA3-T7-EtkDN (E42K+ K444Q). At 48 hr posttransfection, the cells were lysed and cell extracts from each treatment were subjected to immunoprecipitation by anti-T7 antibody, respectively, followed by Western blot analysis with antiphosphotyrosine antibody (Upper) and anti-Etk antibody (Lower). (B) In vitro kinase assays of the immunoprecipitates from A reveals diminished kinase activity of EtkE42K and abolished activity of EtkDN. (C) EtkDN is a dominant negative mutant. The CV-1 cells were transfected with the plasmids indicated. The ratio between the HA-tagged Etk plasmids and T7-tagged Etk plasmids or vector (pcDNA3) used in the transfections is 1:4. At 48 hr posttransfection, the cells were lysed and cell extracts were subjected to immunoprecipitation by anti-HA antibody respectively, followed by Western blot analysis with antiphosphotyrosine antibody (Upper) and anti-HA antibody (Lower).
IL-6 Induces Neuroendocrine Differentiation of LNCaP.
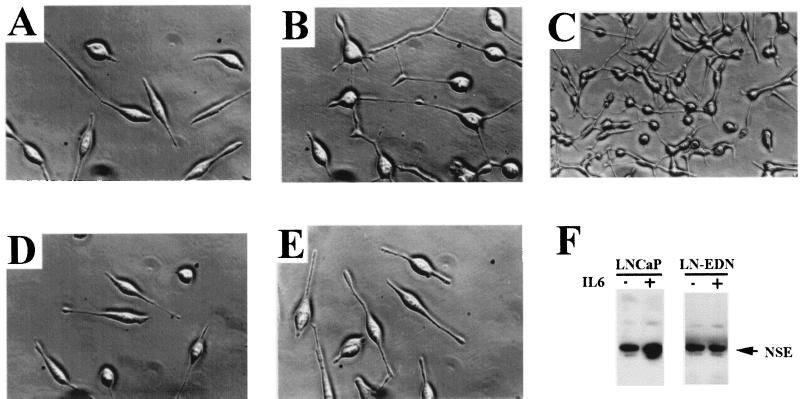
We observed the striking morphological changes associated with IL-6 treatment of LNCaP. The treated cells became rounded and sent out long, often branched, processes. This effect was noted about 24 hr after treatment with 50 ng/ml IL-6 (cf Fig. 5 A and B), and became more pronounced after 72 hr (Fig. 5C). This phenomenon is reminiscent of the neuroendocrine (NE) differentiation phenotype induced by dibutyryl-cAMP as reported (38). To confirm that the IL-6-treated LNCaP cells possess neuronal properties, we looked for the expression of neuron-specific makers. The expression of NSE, as determined by Western blot analysis, is significantly elevated after IL-6 treatment (Fig. 5F). Other neuronal markers tested included neurofilaments and chromogranin A, all of which showed strong immunostaining in LNCaP, reflecting the neuroendocrine origin of this cell line (data not shown).
Figure 5.
Etk is involved in the IL-6-induced neuroendocrine differentiation of LNCaP. (A–C) IL-6 induces morphological changes of LNCaP. (A) LNCaP cells in the serum-free medium (×400 magnification); (B) LNCaP cells treated with 50 ng/l IL-6 for 24 hr (×400 magnification); (C) LNCaP cells treated with 50 ng/ml IL-6 for 72 hr (×100 magnification). (D, E) Overexpression of the dominant negative Etk (EtkDN) in LNCaP blocks the IL-6-induced morphological changes. LNCaP cells were transfected with pcDNA3-T7-EtkDN and selected in the presence of G418, resulting in LNCaP cells overexpressing EtkDN (LN-EDN). (D) LN-EDN in serum-free medium; (E) LN-EDN treated with 50 ng/ml IL-6 for 24 hr. (F) IL-6 induces the enhanced expression of NSE in LNCaP, but not in LN-EDN. 50 μg of the cell extracts from the IL-6-treated (+) or untreated (−) LNCaP or LN-EDN were resolved on an SDS/7.5% polyacrylamide gel and followed by Western blot analysis using antibody specific for NSE.
Etk Activity Is Required for IL-6-Induced Differentiation.
To test whether Etk plays a role in IL-6-induced NE differentiation of LNCaP, we introduced the dominant negative mutant of Etk (EtkDN) into LNCaP. In stark contrast with the parental LNCaP, LN-EDN, the cells stably expressing EtkDaN, are resistant to IL-6 induced differentiation; >90% of LN-EDN cells remained elongated, with scant process formation (see Fig. 5 D and E). In addition, the elevated expression of NSE induced by IL-6 was blocked (Fig. 5F, LN-EDN panel). The overexpression of EtkDN in LNCaP was confirmed by Western blot analysis with anti-T7 antibody and its lack of activity by immunoprecipitation with anti-Etk antibody, followed by antiphospho-tyrosine blot analysis (data not shown). These results point to the involvement of Etk in the neuroendocrine differentiation process.
DISCUSSION
This report provides several new insights into the regulation of PCA cell growth. The first and foremost is the identification of a new Btk family kinase, Etk/Bmx, which is expressed in PCA cells. In a previous study (2), Etk was detected in prostate tissues; however it was not clear whether the relatively weak signal was derived from prostate epithelial cells or from the contaminating stromal cells. Here, we used an established PCA cell line LNCaP that is androgen-sensitive and releases prostate-specific antigen. This together with our previous finding that Etk is expressed in a human prostate xenograft provide conclusive evidence to the prostate origin of Etk. This is significant, because no other Btk family kinases were detected in prostate epithelial cells and Etk may therefore be considered the epithelial analog of Btk. However, there are several important structural differences between Etk and other Btk family kinases, such as the presence of the 22-amino acid repeats and the lack of the proline-rich region. Recent structural analysis of Itk implicates the interaction between the proline-rich motif and the adjacent SH3 domain in the tight regulation of the kinase activity (12). It is unclear whether Etk would be regulated in the same way. Although the PH domain of Etk shares the high degree of homology (66%) with Btk, their affinity toward inositol phosphates are quite different (39). The equivalent point mutation (E42K in Etk, E41K in Btk) resulted in opposite effects on the kinase activity (ref. 37, this report), suggestive of different modes of regulation. We are presently testing the effects of other Etk PH domain mutations on the kinase activity.
IL-6 is a regulator of prostate growth and as shown here, an effective agonist for Etk. Among the different signal pathways known to be engaged by the IL-6 receptor, we found PI3-kinase is capable of activating Etk. This was demonstrated by using constitutively active p110-PI3-kinase and dominant negative p85-PI3-kinase as well as specific PI3-kinase inhibitor wortmannin. Given the similarity between the structure of Etk and Btk, it is likely that Etk is also activated by binding of phosphoinositides to the PH domain. Such a binding presumably could translocate Etk to the membrane and facilitate the formation of an active dimer, resulting in transphophorylation and kinase activation.
Although the downstream pathways of Etk have yet to be identified, one of the consequences of activation of this pathway seems to be neuroendocrine differentiation. The neuroendocrine components of certain prostate carcinomas has long been recognized; neuroendocrine cells are present in a high percentage of prostate carcinomas and tumor cells that coexpress neuroendocrine markers and prostate-specific antigen have been described (40–42). Perhaps most germane to our finding is the interesting report by Bang et al., where they showed that cAMP induces neuroendocrine differentiation of LNCaP cells (38). These authors demonstrated in dibutyryl-cAMP treated cells, extensive neurite-extension and expression of neuronal markers similar to this study. Recent work further reveals the involvement of PKA in this induction process (M. Cox and S. Parsons, personal communication). Whether cAMP and IL-6 induction share the same signaling pathway is presently unclear, although it is known that cAMP is able to induce the transcription of IL-6 due to the presence of four cAMP responsive element sites in the promoter region of IL-6 (43). In this report, we further demonstrate that Etk is a critical element in the differentiation process. This was evidenced by introduction of a dominant negative Etk rendering LNCaP resistant to IL-6-induced differentiation. Elucidation of the Etk pathway thus may also contribute to our understanding of neuronal differentiation of prostate cells. Our finding that the PI3-kinase/Etk pathway is required for the differentiation of LNCaP does not necessarily mean that it is sufficient. It is likely that other pathways activated by IL-6 such as Jak/STATs and mitogen-activating protein kinase are also involved. We note that not all agonists that activate PI3-kinase pathway induce neuronal differentiation of LNCaP (28). Conversely, not all prostate cells that harbor Etk are induced to differentiate by IL-6. This underscores the importance of the cellular context. A complete understanding of the neuronal differentiation pathway of PCA cells would require the knowledge of the downstream substrates of Etk as well as the signal molecules in parallel pathways to which Etk cross-talks.
Acknowledgments
We thank A. W. Grasso, K Everiss, and J. Yustein for critical readings of the manuscript. This work was supported by National Institutes of Health Grants CA39207, CA57179, and CA60171. Y.Q. is a recipient of a National Cancer Institute Research Oncology Training Fellowship (CA59366).
ABBREVIATIONS
- PI3-kinase
phosphatidylinositol 3-kinase
- PCA
prostate cancer
- IL-6
interleukin 6
- PH
pleckstrin homology
- SH
src homology
- NSE
neuron-specific enolase
- HA
hemagglutinin
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AP045459).
References
- 1.Robinson D, He F, Pretlow T, Kung H J. Proc Natl Acad Sci USA. 1996;93:5958–5962. doi: 10.1073/pnas.93.12.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamagnone L, Lahtinen I, Mustonen T, Virtaneva K, Francis F, Muscatelli F, Alitalo R, Smith C I, Larsson C, Alitalo K. Oncogene. 1994;9:3683–3688. [PubMed] [Google Scholar]
- 3.Tsukada S, Saffaran D, Rawlings D, Witte O. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 4.Vetrie D, Vorechovsky I, Sideras P, Bentley D. Nature (London) 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 5.Gibson S, August A, Kawakami Y, Kawakami T, Dupont B, Mills G B. J Immunol. 1996;156:2716–2722. [PubMed] [Google Scholar]
- 6.Siliciano J D, Morrow T A, Desiderio S V. Proc Natl Acad Sci USA. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mano H, Yamashita Y, Sato K, Yazaki Y, Hirai H. Blood. 1995;85:343–350. [PubMed] [Google Scholar]
- 8.Rawlings D J, Witte O N. Semin Immunol. 1995;7:237–246. doi: 10.1006/smim.1995.0028. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J S, Teutsch M, Dong Z, Wortis H H. Proc Natl Acad Sci USA. 1996;93:10966–10971. doi: 10.1073/pnas.93.20.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uckun F M, Waddick K G, Mahajan S, Jun X, Takata M, Bolen J, Kurosaki T. Science. 1996;273:1096–1100. doi: 10.1126/science.273.5278.1096. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami Y, Miura T, Bissonnette R, Hata D, Khan W N, Kitamura T, Maeda-Yamamoto M, Hartman S E, Yao L, Alt F W, Kawakami T. Proc Natl Acad Sci USA. 1997;94:3938–3942. doi: 10.1073/pnas.94.8.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreotti A H, Bunnell S C, Feng S, Berg L J, Schreiber S L. Nature (London) 1997;385:93–107. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 13.Gibson T J, Hyvonen M, Musacchio A, Saraste M, Birney E, Kurosaki T. Trends Biochem Sci. 1994;19:349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings D J, Scharenberg A M, Park H, Wahl M I, Lin S, Kato R M, Fluckiger A C, Witte O N, Kinet J P. Science. 1996;271:822–825. doi: 10.1126/science.271.5250.822. [DOI] [PubMed] [Google Scholar]
- 15.Mano H, Yamashita Y, Miyazato A, Miura Y, Ozawa K. FASEB J. 1996;10:637–642. doi: 10.1096/fasebj.10.5.8621063. [DOI] [PubMed] [Google Scholar]
- 16.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 17.Datta K, Franke T F, Chan T O, Makris A, Yang S I, Kaplan D R, Morrison D K, Golemis E A, Tsichlis P N. Mol Cell Biol. 1995;15:2304–2310. doi: 10.1128/mcb.15.4.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stokoe D, Stephens L, Copeland T, Hawkins P. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 19.Salim K, Bottomley M J, Querfurth E, Zvelebil M J, Gout I, Scaife R, Margolis R L, Gigg R, Smith C I, Driscoll P C, et al. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 20.Siegall C B, Schwab G, Nordan R P, FitzGerald D J, Pastan I. Cancer Res. 1990;50:7786–7788. [PubMed] [Google Scholar]
- 21.Okamoto M, Lee C, Oyasu R. Cancer Res. 1997;57:141–146. [PubMed] [Google Scholar]
- 22.Lutticken C, Wegenka U M, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur A G, Wilks A F, Yasukawa K, Taga T, et al. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 23.Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 24.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen-Kiang S. Curr Top Microbiol Immunol. 1995;194:189–198. doi: 10.1007/978-3-642-79275-5_23. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi T. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 27.Boulton T G, Stahl N, Yancopoulos G D. J Biol Chem. 1994;269:11648–11655. [PubMed] [Google Scholar]
- 28.Grasso, A., Wen, D., Pretlow, T. & and Kung, H. Oncogene 15,, 2705–2716. [DOI] [PubMed]
- 29.Hunter T. J Biol Chem. 1982;257:4843–4848. [PubMed] [Google Scholar]
- 30.Ohara O, Dorit R, Gilbert W. Proc Natl Acad Sci USA. 1989;86:5673–5677. doi: 10.1073/pnas.86.15.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frohmann M. PCR Methods Appl. 1994;4:540–558. [Google Scholar]
- 32.Powis G, Bonjouklian R, Berggern M, Vlahos C. Cancer Res. 1994;54:2419–2423. [PubMed] [Google Scholar]
- 33.Klippel A, Reinhard C, Kavanaugh W M, Apell G, Escobedo M A, Williams L T. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, et al. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattsson P T, Vihinen M, Smith C I. BioEssays. 1996;18:825–834. doi: 10.1002/bies.950181009. [DOI] [PubMed] [Google Scholar]
- 36.Hyvonen M, Saraste M. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Tsukada S, Satterthwaite A, Havlik M H, Park H, Takatsu K, Witte O N. Immunity. 1995;2:451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 38.Bang Y J, Pirnia F, Fang W G, Kang W K, Sartor O, Whitesell L, Ha M J, Tsokos M, Sheahan M D, Nguyen P, et al. Proc Natl Acad Sci USA. 1994;91:5330–5334. doi: 10.1073/pnas.91.12.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kojima T, Fukuda M, Watanabe Y, Hamazato F, Mikoshiba K. Biochem Biophys Res Commun. 1997;236:333–339. doi: 10.1006/bbrc.1997.6947. [DOI] [PubMed] [Google Scholar]
- 40.Cohen R J, Glezerson G, Taylor L F, Grundle H A, Naude J H. J Urol. 1993;150:365–3668. doi: 10.1016/s0022-5347(17)35484-8. [DOI] [PubMed] [Google Scholar]
- 41.Aprikian A, Cordon-Cardo C, Fair W, Reuter V E. Cancer. 1993;71:3952–3965. doi: 10.1002/1097-0142(19930615)71:12<3952::aid-cncr2820711226>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 42.Abrahamsson, P. A. (1996) Prostate 6 (Suppl.), 3–8. [DOI] [PubMed]
- 43.Huttner A, Lei T, Fahlbusch R, Schrell W, Adams E F. Life Sciences. 1996;58:1323–1329. doi: 10.1016/0024-3205(96)00098-7. [DOI] [PubMed] [Google Scholar]