Abstract
T-bet plays a critical role in controlling IFNγ expression, Th1 polarization, and CD8 cytolytic development. Its regulation has been demonstrated to be mostly IFNγ/Stat1 dependent while IL-12/Stat4 independent. Here we show that IL-12/Stat4 binds to a distant highly conserved STAT-responsive T-bet enhancer, and induces IFNγ/Stat1-independent T-bet expression in CD8 T cells. Luciferase reporter assay showed that both Stat4 and Stat1 activate reporter gene expression from constructs containing a wild-type but not mutated T-bet enhancer. Studies in virus-infected mice demonstrated that the IL-12/Stat4/T-bet cascade operates in vivo and regulates IFNγ in CD8 T cells. Together, we provide a novel mechanism for T-bet regulation, and suggest that IL-12/Stat4/T-bet play an important role in CD8 effector responses.
Introduction
T-box expressed in T cells (T-bet), a member of the T-box family of transcription factor genes, was first identified from a Th1-cell cDNA library as a Th1-specific transcription factor,1 and was also cloned as Tbx21.2 Analysis of T-bet–deficient mice has shown that CD4 T cells lacking T-bet are severely impaired in their ability to produce IFNγ, susceptible to Leishmania major infection, and have a marked in vivo shift of the Th1/Th2 balance toward the Th2 pathway.3–5 These findings demonstrate a central role for T-bet in governing IFNγ expression and Th1 development in CD4 T cells. In addition to its critical role in CD4 T-cell differentiation, T-bet has also been demonstrated to play important roles in regulating IFNγ and cytotoxic T lymphocyte (CTL) activities in CD8 T cells.6–8 It binds directly to the promoters of perforin and granzyme B9, and dominant negative T-bet abolishes IFNγ production and CTL function in CD8 T cells.10
T-bet was initially suggested to be induced by IL-12/Stat4 signals,1,11 but later investigation demonstrated that it is regulated by IFNγ/Stat1.12–14 The molecular mechanism by which IFNγ/Stat1 directs T-bet expression remains unclear. Other signaling pathways have also been demonstrated to regulate T-bet. For example, IL-15 up-regulates T-bet expression in natural killer (NK) cells, and Stat4−/−-deficient NK cells have a significant defect in T-bet induction by IL-12 and IL-18.9 Very recently, it has been shown that retroviral-mediated Stat4 expression in CD4 T cells induces Stat1-independent T-bet expression.15 In addition to being positively regulated, T-bet has been shown to be negatively regulated by TGFβ, the Th2 transcription factor GATA3, and the Tec family tyrosine kinase Itk.15–19
Little is known how T-bet is regulated in CD8 T cells. By studying T-bet regulation in CD8 T cells during the early stage of activation, we found that IL-12/Stat4 efficiently induces IFNγ/Stat1-independent T-bet transcription. Chromatin immunoprecipitation assays uncovered a novel conserved T-bet enhancer, residing 13 kb upstream of the T-bet coding region, which responds to both IL-12/Stat4 and IFNγ/Stat1 signals. Our studies identified a novel T-bet enhancer and provide a new molecular basis for T-bet regulation by IL-12/Stat4 and IFNγ/Stat1.
Materials and methods
This study was approved by the institutional review board of Mount Sinai School of Medicine and informed consent was obtained in accordance with the Declaration of Helsinki. Animals involved in the research reported in this paper were done with approval from the IACUC.
Mice
BALB/c, C57BL/6, OT-1 transgenic mice specific for OVA257-264, Stat4 KO (BALB/c background), T-bet KO (BALB/c background), IFNγ KO (C57BL/6 background), and IFNγR KO (C57BL/6 background) mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). Stat1 KO (129S6 background) and 129S6 control mice were purchased from Taconic (Germantown, NY).
Reagents
Recombinant mouse cytokines IL-12 and IFNγ were purchased from BD Pharmingen (San Diego, CA); anti-CD3, FITC-labeled anti-CD4, FITC-labeled anti-B220, FITC-labeled anti-CD8, PE-labeled anti-CD8, PE-Cy5-anti-CD44, anti-CD28, and anti-IFNγ mAbs were purchased from BD Pharmingen. IFNα and IFNβ were purchased from PBL Biomedical Laboratories (Piscataway, NJ). CD4 and CD8 MicroBeads were purchased from Miltenyi Biotech (Auburn, CA). Ovalbumin peptides (OVA257–264) were purchased from American Peptide (Sunnyvale, CA). Antibodies to phosphorylated Stat1, phosphorylated Stat4, and Stat4 were from Zymed Lab (South San Francisco, CA); anti–T-bet Abs (H-210) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-Stat1 Abs and chromatin immunoprecipitation (ChIP) assay kit were purchased from Upstate Group (Lake Placid, NY). Mouse Stat4 was amplified by reverse-transcription–polymerase chain reaction (RT-PCR) and cloned into pcDNA3.1-expressing vector (Invitrogen, Carlsbad, CA), and human Stat1 or Stat4 expression plasmids were purchased from Panomics (Fremont, CA). An expressing vector for JAK220 was provided by Dr L Rui (University of Michigan, Ann Arbor, MI). AdV-GFP gene transfer vectors were amplified by our vector core facility and have been used previously.21 P32-dCTP was purchased from PerkinElmer (Boston, MA). EL-4 and NK92 cells were purchased from the ATCC (Manassas, VA), and U3A cells were maintained as previously described.22,23
Cell culture
Single cell suspensions were prepared from spleen or lymph node by filtering through nylon mesh (40-μm diameter). CD4 and CD8 T cells were positively selected by using CD4+ or CD8+ MicroBeads or sorted by Moflo (Cytomation, Denver, CO) after staining with FITC-anti-CD4, PE-anti-CD8, and PE-Cy5-anti-CD44, seeded into 96-well round-bottom plates coated with anti-CD3 mAb (5 μg/mL) at 4 × 104 cells/well, then further activated with soluble anti-CD28 mAb (1 μg/mL), with various cytokines as indicated. For antigen-presenting cell (APC)–dependent assays, B cells were purified by cell sorting after staining with FITC-anti-B220 (after sorting, cell purity is about 98%), and treated with mitomycin C (50 μg/mL; Sigma, St Louis, MO), for 50 minutes at 37°C. Purified OT1 CD8+ T cells (2 × 104) and mitomycin C–treated B cells (104) were incubated with OVA peptides (5 μg/mL) in round-bottom 96-well plates with cytokines for various times as indicated. Human CD8 T cells were purified with human CD8+ MicroBeads (Miltenyi Biotech) from healthy human donors.
Western blotting
Protein extracts were prepared and concentrations were determined as previously described.24 Proteins were then separated by 7.5% SDS-polyacrylamide; transferred to polyvinylidene difluoride membranes; probed with antiphosphorylated Stat4 (1:200 dilution), anti-Stat4 (1:500 dilution), antiphosphorylated Stat1 (1:250 dilution), anti-Stat1 (1:500 dilution), or anti-T-bet (1:200); subsequently incubated with HRP-conjugated anti–rabbit IgG; and detected by the enhanced chemiluminescence system (Amersham, Piscataway, NJ).
Quantitative RT-PCR
Total RNA was extracted from purified cells with TRIzol solution (Invitrogen). Reverse transcription was carried out using the Omniscript reverse-transcription system (Qiagen, Hilden, Germany) and random primers. Quantitative PCR was performed with the LightCycler system (Roche, Mannheim, Germany) and the SYBR Green PCR kit (Qiagen). All experiments were done at least 3 separate times, and expression of specific genes was normalized and expressed as percentage relative to housekeeping genes (cyclophilin A or GAPDH).
Chromatin immunoprecipitation (ChIP) and eletrophoretic mobility shift assay (EMSA)
ChIP was performed according to the manufacturer's protocol. To identify Stat4- or Stat1-binding sites, CD4 or CD8 T cells were purified and stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IL-12 or IFNγ for 24 hours. Cells were then cross-linked with formaldehyde, sonicated to obtain genomic DNA fragments between 500 to 1000 bp, immunoprecipitated with anti-Stat4 or anti-Stat1 Abs, cross-linking reversed by high-salt solution, and DNA purified, and real-time PCR was performed. Twenty pairs of primers were used. All experiments were done at least 3 separate times. EMSAs were performed as previously described25 with oligonucleotide probes based on 2 STAT-binding sites within the distal T-bet enhancer region (probe A: sense strand, CCC AGC TTC GAG GAA ACA, antisense strand, AGG TGT TTC CTC GAA GCT; probe B: sense strand, CTC CTT TTA GCG GAA AGC, antisense strand, CTC GCT TTC CGC TAA AAG; mutated A, sense strand, CCC AGC AAC GAG GAA ACA, antisense strand, AGG TGT TTC CTC G[b]TT GCT; mutated B, sense strand, CTC CTT AAA GCG GAA AGC, antisense strand, CTC GCT TTC CGC T[b]TT AAG), and a potential STAT-binding site located 940-bp upstream of the transcription initiation site (probe C, sense strand, CCG TAG TTA TTG GAA TAG, antisense strand, GTT CTA TTC CAA TAA CTA).
Luciferase reporter assay
Various T-bet enhancer regions were amplified from BALB/c mouse genomic DNA by PCR with primers carrying restriction sites for NheI and BglII, and cloned into a TransLucent reporter vector containing a minimal TA promoter and the TATA box from the herpes simplex virus thymidine kinase promoter (Panomics). Wild-type (sense, GCT AGC TTT TCC CAG CTT CGA GGA AAC ACC TCC TTT TAG CGG AAA GCG AGA TGG AGA TCT, antisense, AGA TCT CCA TCT CGC TTT CCG CTA AAA GGA GGT GTT TCC TCG AAG CTG GGA AAA GCT AGC) and mutated (A instead of T, mutant 1: sense, GCT AGC TTT TCC CAG C[b]AA CGA GGA AAC ACC TCC TTT TAG CGG AAA GCG AGA TGG AGA TCT, antisense, AGA TCT CCA TCT CGC TTT CCG CTA AAA GGA GGT GTT TCC TCG TTG CTG GGA AAA GCT AGC; mutant 2: sense, GCT AGC TTT TCC CAG CTT CGA GGA AAC ACC TCC TT[b]A AAG CGG AAA GCG AGA TGG AGA TCT, antisense, AGA TCT CCA TCT CGC TTT CCG CT[b]T TAA GGA GGT GTT TCC TCG TTG CTG GGA AAA GCT AGC; mutant 3: sense, GCT AGC TTT TCC CAG C[b]AA CGA GGA AAC ACC TCC TT[b]A AAG CGG AAA GCG AGA TGG AGA TCT, antisense, AGA TCT CCA TCT CGC TTT CCG CT[b]T TAA GGA GGT GTT TCC TCG TTG CTG GGA AAA GCT AGC) T-bet enhancer oligonucleotides were synthesized, annealed, and cloned into reporter vectors. HeLa cells were transfected with various constructs, treated with human IFNγ (100 ng/mL) for 24 hours, and luciferase activities were measured with Luciferase assay kit (Promega, Madison, WI) according to the manufacturer's protocol and normalized to background activity from TA-Luc control (Panomics). For cotransfection experiments, HeLa, U3A, or EL-4 cells were transfected with JAK2, with or without Stat1- or Stat4-expressing plasmids, in combination with various reporter constructs. Twenty-four hours after transfection, luciferase activities were measured and normalized to TA-Luc control reporter.
Results
IL-12 induces Stat1-independent T-bet expression in CD8 T cells
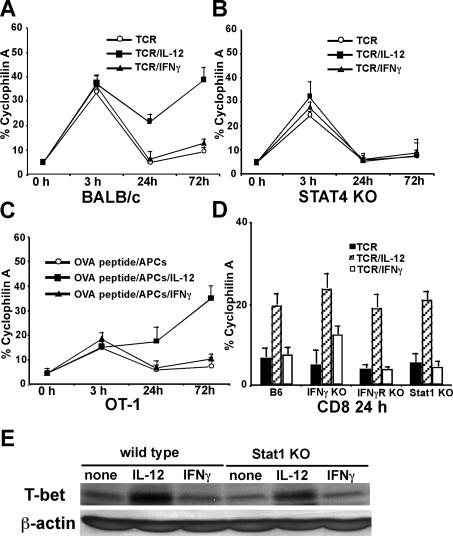
We first examined how IL-12 and IFNγ regulate T-bet in CD8 T cells during the early period of activation. Purified CD8 T cells were stimulated with plate-bound anti-CD3 mAbs (5 μg/mL) plus soluble anti-CD28 mAbs (1 μg/mL), with or without IL-12 or IFNγ for 3, 24, and 72 hours, and T-bet expression was measured by real-time RT-PCR. As shown in Figure 1A, anti-CD3 plus anti-CD28 mAbs induce rapid T-bet expression in CD8 T cells as early as 3 hours after stimulation, and neither IL-12 nor IFNγ affect TCR-induced immediate T-bet expression. The early T-bet expression induced by TCR stimulation declines 24 hours after stimulation, and exogenous IFNγ treatment does not increase or sustain this early T-bet expression. Interestingly, there are significantly higher levels of T-bet expressed in the IL-12 treatment group 24 and 72 hours after stimulation. Since Stat4 activation is crucial for IL-12 signal transduction and biologic functions,26 we performed similar experiments on Stat4-deficient CD8 T cells. We observed that Stat4 deficiency in CD8 T cells does not diminish TCR-mediated T-bet expression at 3 hours after stimulation, indicating that Stat4 does not play a role in TCR-induced immediate T-bet expression. However, at 24 or 72 hours after stimulation, IL-12 is no longer able to enhance T-bet expression in Stat4-deficient cells, indicating that up-regulation of T-bet by IL-12 requires Stat4 (Figure 1B). Similar results were obtained when OT1 transgenic CD8 T cells were stimulated with purified B cells as APCs loaded with OVA257-264 peptides (Figure 1C), indicating that IL-12 also enhances T-bet expression in CD8 T cells stimulated by antigenic peptides. To further test whether IL-12–mediated T-bet induction is dependent on IFNγ/IFNγR/Stat1 signals, IFNγ-deficient, IFNγR-deficient, and Stat1-deficient CD8 T cells were used. As shown in Figure 1D, IFNγ, IFNγR, or Stat1 deficiency do not abolish IL-12–mediated T-bet expression. Western blotting of T-bet expression on wild-type or Stat1-deficient CD8 T cells further demonstrated that IL-12 induces IFNγ/Stat1-independent T-bet expression in CD8 T cells (Figure 1E).
Figure 1.
IL-12 induces IFNγ-independent T-bet expression in CD8 T cells. Cells (5 × 105/mL) were stimulated, and real-time PCR for T-bet expression was normalized and expressed as percentage relative to cyclophilin A. (A) BALB/c CD8 T cells were stimulated by anti-CD3 plus anti-CD28 mAbs in combination with IL-12 or IFNγ for 3, 24, or 72 hours. (B) Stat4-deficient CD8 T cells were used. (C) OT1 CD8 T cells were stimulated with OVA peptide (5 μg/mL) and purified B cells as APCs in combination with IL-12 or IFNγ for 3, 24, or 72 hours. (D) C57BL/6 wild-type, IFNγ-deficient, IFNγR-deficient, or Stat1-deficient CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs in combination with IL-12 or IFNγ for 24 hours. (E) Western blotting of T-bet expression was performed on wild-type or Stat1-deficient CD8 T cells stimulated with anti-CD3 plus anti-CD28 mAbs in combination with IL-12 or IFNγ for 24 hours. Data shown are one representative of 3 independent experiments; standard errors of PCR triplicates are shown.
Sustained Stat4 activation is associated with T-bet induction
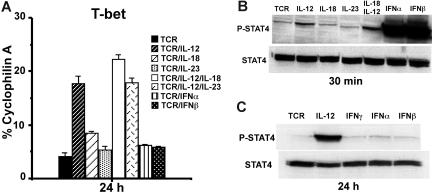
In addition to IL-12, IL-23 and type 1 IFNs (IFNα and IFNβ) also activate Stat4.27,28 To further investigate the role of Stat4 activation for CD8 T-bet regulation, we treated purified CD8 T cells with anti-CD3 plus anti-CD28 mAbs in combination with various cytokines (10 ng/mL). Real-time RT-PCR showed that only IL-12 but not IL-18, IL-23, IFNα, or IFNβ induces T-bet expression after 24 hours of stimulation (Figure 2A). Western blotting for Stat4 activation at 30 minutes after stimulation showed that IL-18 and IL-23 do not induce much Stat4 phosphorylation (Figure 2B), suggesting that their failure to induce T-bet is likely due to their inability to induce Stat4 activation in CD8 T cells. On the contrary, IFNα and IFNβ induce much greater Stat4 phosphorylation than IL-12 in CD8 T cells (Figure 2B), despite the fact that they fail to induce T-bet (Figure 2A). To determine if the failure to induce T-bet by IFNα and IFNβ may be due to their inability to sustain Stat4 activation compared with IL-12, Western blotting was performed 24 hours after stimulation. As shown in Figure 2C, IL-12, but not IFNα or IFNβ, maintains Stat4 phosphorylation at 24 hours after stimulation. These data suggest that sustained Stat4 activation is associated with T-bet induction in CD8 T cells.
Figure 2.
Sustained Stat4 activation is associated with T-bet induction. (A) BALB/c CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, in combination with indicated cytokines (10 ng/mL each) for 24 hours, and real-time RT-PCR was used to measure T-bet expression. Data shown are one representative of 3 independent experiments; standard errors of PCR triplicates are shown. (B) Western blotting for Stat4 activation. BALB/c CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs plus the indicated cytokines for 30 minutes. Antiphosphorylated Stat4 or anti-Stat4 Abs were used for Western blotting. (C) IL-12 but not IFNα or IFNβ induces sustained Stat4 activation at 24 hours after stimulation. CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IL-12, IFNα, or IFNβ. Data shown are one representative of 3 independent experiments; standard errors of PCR triplicates are shown.
IL-12/Stat4 signals are differentially regulated in CD8 and CD4 T cells
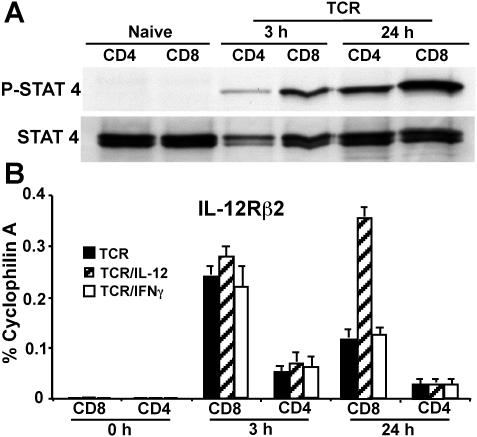
We next investigated if IL-12/Stat4-regulated early T-bet expression in CD8 but not CD4 T cells is due to differential IL-12/Stat4 activation. Purified CD8 and CD4 T cells were treated with IL-12 for 15 minutes, without anti-CD3 plus anti-CD28 mAb stimulation. As shown in Figure 3A, IL-12 fails to induce measurable Stat4 activation in either CD8 or CD4 T cells, likely due to the fact that naive T cells do not express much IL-12Rβ2, which is pivotal for IL-12/Stat4 signaling.29,30 When cells are stimulated with IL-12 in combination with TCR, IL-12 induces low and comparable Stat4 activation in both CD8 and CD4 T cells at 3 hours after TCR activation. However, at 24 hours after stimulation, IL-12 induces a much stronger Stat4 activation in CD8 than in CD4 T cells (Figure 3A), demonstrating there is differential IL-12/Stat4 signaling comparing CD8 with CD4 T cells. We next examined if differential Stat4 responses stimulated by IL-12 are due to differential IL-12Rβ2 regulation. Flow cytometric analysis failed to quantitate any meaningful IL-12Rβ2 expression in either naive or activated CD8 T cells within 24 hours of stimulation (not shown). We used real-time RT-PCR to measure IL-12Rβ2 expression. As shown in Figure 3B, both naive CD8 and CD4 T cells express very little IL-12Rβ2. After stimulation for 3 hours, IL-12Rβ2 is up-regulated by TCR in both CD8 and CD4 T cells, but it is more highly up-regulated in CD8 than CD4 T cells (9-fold vs 2-fold), and exogenous IL-12 or IFNγ has little effect on TCR-mediated IL-12Rβ2 up-regulation. At 24 hours after stimulation, IL-12Rβ2 expression declines somewhat, but is still expressed at higher levels in CD8 compared with CD4 T cells. Furthermore, IL-12 treatment enhances IL-12Rβ2 expression in CD8 but not CD4 T cells (Figure 3B). These data correspond with the Western blotting data for Stat4 activation (Figure 3A), suggesting that differential IL-12Rβ2 regulation between CD8 and CD4 T cells contributes to differential IL-12/Stat4 signaling.
Figure 3.
IL-12/Stat4 signals are differentially regulated in CD8 and CD4 T cells. (A) Western blotting for Stat4 activation in CD4 and CD8 T cells. BALB/c CD4 and CD8 T cells were stimulated with IL-12 for 15 minutes, without anti-CD3 plus anti-CD28 mAb stimulation, or stimulated with IL-12 in combination with anti-CD3 plus anti-CD28 mAbs for 3 hours or 24 hours. (B) Real-time RT-PCR for IL-12Rβ2 expression. CD4 or CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IL-12 or IFNγ for 3 or 24 hours. Data shown are one representative of 3 independent experiments; standard errors of PCR triplicates are shown.
Identification of a distant STAT-responsive T-bet regulatory region
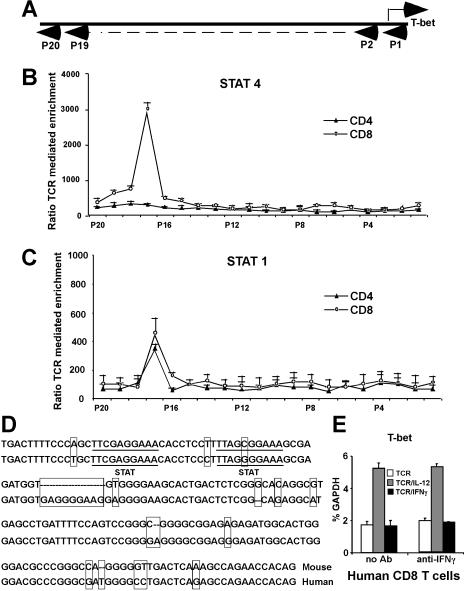
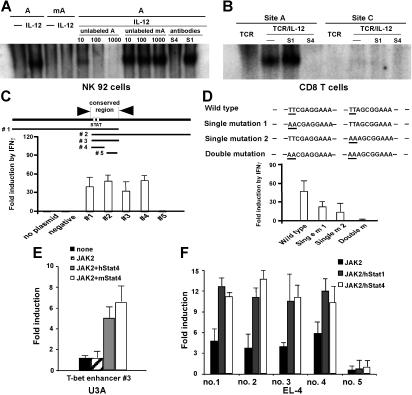
The chromatin immunoprecipitation (ChIP) assay–based genomic array has been used to identify potential targets for specific transcription factors.9,31 We next used it to identify T-bet promoters/enhancers with the potential to bind Stat4 and/or Stat1. The T-bet gene consists of 6 exons and is located on chromosome 11D within a genomic region spanning 43 823 bp from 8529064 to 8485242 (GenBank; NCBI). We focused on the upstream 5′-flanking region of exon 1 (beyond position 8515805, about 14 kb total). For every 500- to 600-bp interval, we constructed a pair of primers for real-time PCR to amplify 150-bp fragments. These real-time PCR primers and their amplified fragments served as tags to locate specific regions of the T-bet locus (Figure 4A). A total of 20 primer pairs were synthesized, and primer specificity was checked by conventional PCR on C57BL/6 genomic DNA (data not shown). Purified CD8 or CD4 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IL-12 for 24 hours. As shown in Figure 4B, there was more than a 30-fold enrichment by IL-12 for a Stat4-associated sequence located 13-kb upstream of the T-bet coding sequence in CD8 T cells (primer pair no. 17), while there is very little enrichment in CD4 T cells, indicating that IL-12–activated Stat4 binds to this region in CD8 T cells after 24 hours of stimulation. Since IFNγ-deficient T cells are very sensitive to exogenous IFNγ in T-bet regulation (Figure 1D), we used IFNγ-deficient CD8 or CD4 T cells to perform ChIP assay to identify potential binding sites for Stat1, and determine if Stat1 may bind to the same region as Stat4. As shown in Figure 4C, the ChIP assay showed there is about a 4-fold enrichment in both CD8 and CD4 T cells for the same sequence identified for Stat4 binding, indicating that this region interacts with both IL-12–stimulated Stat4 and IFNγ-stimulated Stat1. It should be noted that the lesser enrichment of this region by IFNγ/Stat1 compared with IL-12/Stat4 does not necessarily mean that Stat1 binds to the enhancer less efficiently or avidly than Stat4, since it is possible that anti-Stat1 antibodies are less efficient then anti-Stat4 antibodies for the ChIP assay.
Figure 4.
Identification of a conserved distant T-bet enhancer region. (A) ChIP assay–based genomic scanning. Twenty primer pairs were used for these experiments. (B) Sequences precipitated by anti-Stat4 mAbs. Purified CD8 and CD4 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IL-12 for 24 hours, and ChIP assays were performed with anti-Stat4. Sequences of T-bet regulatory regions precipitated by anti-Stat4 were quantitated by real-time PCR. Data are expressed as the ratio of TCR/IL-12–mediated to TCR alone–mediated enrichments. (C) Sequences precipitated by anti-Stat1 mAbs. Purified IFNγ-deficient CD4 and CD8 T cells were stimulated with anti-CD3 plus anti-CD28 mAbs, with or without IFNγ (10 ng/mL) for 24 hours, and ChIP assays were performed with anti-Stat1. Sequences of T-bet regulatory regions precipitated by anti-Stat1 were quantitated by real-time PCR. Data are expressed as the ratio of TCR/IFNγ-mediated to TCR alone–mediated enrichments. (B,C) Data shown are the means plus or minus SD from 3 independent experiments. (D) Alignment of mouse and human T-bet enhancer. Nucleotides differing between mouse and human are boxed. Potential STAT-binding motifs TTCGAGGAA and TTAGCGGAA are underlined. (E) IL-12 also induces IFNγ-independent T-bet in human CD8 T cells. Human CD8 T cells were stimulated with anti-CD3 plus anti-CD28 Abs, with or without anti-IFNγ Abs, in combination with human IL-12 or human IFNγ. T-bet expression was normalized and expressed as percentage relative to GAPDH. Data shown are one representative of 3 independent experiments; standard errors of PCR triplicates are shown.
Sequence analysis of the T-bet gene region between primer pairs no. 16 and no. 18 located 2 potential STAT-binding motifs, TTCGAGGAA and TTAGCGGAA, separated by 10 base pairs (Figure 4D), matching the characteristics of STAT-responsive elements.32–35 GenBank blasts using sequences near these 2 potential Stat4-binding sites (about 50 bp) resulted in identification of an almost identical 150-bp sequence (Figure 4C) in human chromosome 17 located upstream of TBLYM (human T-bet gene), further suggesting this region may play an important role in T-bet regulation. Using purified human CD8 T cells treated with human anti-CD3 plus anti-CD28 mAbs, in combination with human IL-12 or IFNγ, also demonstrated that IL-12 induces human T-bet expression (Figure 4E). Anti-IFNγ antibodies do not affect IL-12–mediated T-bet induction, suggesting that human T cells may use similar mechanisms for IL-12–dependent, IFNγ-independent T-bet induction.
Stat4 and Stat1 transactivate reporters through STAT-binding elements of T-bet enhancer
To determine if these 2 STAT-binding sites interact with IL-12–activated Stat4, EMSA experiments were performed. We first used the NK92 cell line that responds to IL-12 for Stat4 activation. As shown in Figure 5A, site A (A) binds to IL-12–activated Stat4, while the mutated site A (mA) does not. DNA-binding complex was competed by unlabeled probe but not mutated probe, and abolished by anti-Stat4 but not anti-Stat1 antibodies. Similar results were obtained for site B (not shown). EMSAs in CD8 T cells 24 hours after stimulation also show IL-12–activated Stat4 binds to site A, but not a potential STAT-binding site (probe C) located 940-bp upstream of the transcription initiation site (Figure 5B); similar results were obtained for site B (not shown). These findings further demonstrate that IL-12–activated Stat4 binds to the enhancer region in CD8 T cells. We next examined if the region we identified responds to Stat1 or Stat4 to transactivate reporter gene expression. As shown in Figure 5C, 5 separate constructs with or without the target STAT-binding elements were cloned into the TransLucent reporter vector containing a minimal TA promoter and the TATA box from the herpes simplex virus thymidine kinase promoter (Panomics), with or without the target STAT-binding elements. HeLa cells were transfected with the various reporter constructs, and then treated with human IFNγ for 24 hours. As shown in Figure 5C, all constructs containing STAT-binding elements respond to IFNγ by inducing reporter gene expression, while the constructs that do not contain STAT-binding sites fail to respond to IFNγ. To further determine if transactivation is mediated through STAT-binding elements, a series of mutants were generated. As shown in Figure 5D, mutating either binding site significantly reduced reporter gene expression, and mutating both STAT-binding elements abolished transactivation of the T-bet enhancer. These results demonstrate that the identified region interacts with IFNγ-stimulated Stat1 through STAT-binding elements to transactivate gene transcription. Since cotransfecting JAK2 with STAT molecules allows them to be phosphorylated and subsequently gain DNA-binding and transactivation activities,32 we performed cotransfection experiments to determine how the T-bet enhancer responds to Stat4. To exclude the potential for endogenous Stat1 to mask or interfere with Stat4 activity, the T-bet enhancer reporter construct containing the fully conserved region (construct no. 3) was cotransfected with a JAK2-expressing vector20 in combination with human or mouse Stat4-expressing plasmids into Stat1-deficient U3A cells.22,23 As shown in Figure 5E, expression of JAK2 alone does not transactivate reporter gene expression, while cotransfecting JAK2 with Stat4-induced reporter gene expression in U3A cells, demonstrating that Stat4 transactivates gene expression through the T-bet enhancer. The various reporter constructs were also used in EL-4 cells. Similar to IFNγ/Stat1-mediated reporter expression in HeLa cells, only constructs with STAT-binding motifs are significantly transactivated by JAK2 in combination with Stat1- or Stat4-encoding plasmids (Figure 5F), further demonstrating that the identified sequence specifically interacts with Stat1 and Stat4, and has enhancer activity.
Figure 5.
T-bet enhancer interacts with IL-12–activated Stat4 and transactivates reporter gene expression. (A). EMSA in NK92 cells. Nuclear extracts were prepared from NK92 cells treated with IL-12 for 15 minutes. A indicates site A within the identified distal T-bet enhancer; mA, mutated site A. Competition was performed with 10-, 100-, or 1000-fold unlabeled probe. Equal amount of specific anti-Stat1 (S1) and anti-Stat4 Abs (S4) was used for neutralization. (B) EMSA in CD8 T cells. Nuclear extracts were prepared from cells 24 hours after stimulation. Site A indicates the STAT-binding site within the identified distal T-bet enhancer; site C, a potential STAT-binding site located 940-bp upstream of the transcription initiation site. Equal amount of specific anti-Stat1 (S1) and anti-Stat4 (S4) Abs was used for neutralization. (C) T-bet enhancer constructs containing STAT-binding motifs respond to IFNγ/Stat1 by transactivating Luciferase reporter expression. Total of 5 constructs were made. No. 1 contains 5′ nearby region; no. 2 contains 3′ nearby region; no. 3 is the exact conserved region; no. 4 is the minimal conserved region containing 2 potential STAT-binding motifs; and no. 5 is the conserved region without STAT-binding motifs. HeLa cells were transfected with the indicated constructs and treated with human IFNγ for 24 hours, and luciferase activities were measured. (D) Point mutations were made, and reporter assays were performed. (E) JAK2-mediated Stat4 activation induces reporter gene expression. U3A cells were cotransfected with the conserved T-bet enhancer region (construct no. 3, 100 ng), along with JAK2 (100 ng), in combination with mouse or human Stat4 (500 ng) plasmids. (F) T-bet enhancer constructs containing STAT-binding motifs respond to both Stat1 and Stat4. The indicated T-bet enhancer reporter constructs were cotransfected with human Stat1 or Stat4, along with JAK2 into EL-4 cells. (C-F) Data shown are the means plus or minus SD from 3 independent experiments.
Adenovirus induces Stat4- but not Stat1-dependent CD8 T-bet expression in vivo
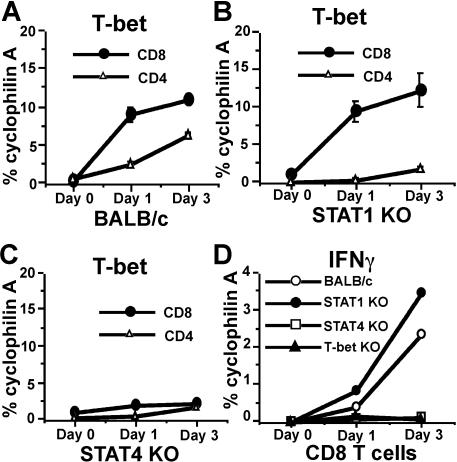
Adenovirus (AdV) elicits strong CD8, CD4, and NK-cell responses similar to many other viruses, and is widely used in gene therapy and vaccination.36 We used AdV gene transfer vectors as a model to investigate if the induction of CD8 T-bet by IL-12/Stat4 occurs in vivo with a microbial stimulus. AdV vector was injected into the hind footpad of BALB/c mice, which were killed after 1 or 3 days, draining lymph nodes (LNs) were harvested, CD8 and CD4 T cells were purified by flow cytometry, and real-time RT-PCR was performed to measure T-bet expression. As shown in Figure 6A, AdV administration induces T-bet expression in both CD8 and CD4 T cells. Stat1 deficiency abolishes AdV-induced T-bet expression in CD4 T cells (Figure 6B) but not in CD8 T cells. When Stat4-deficient mice were used, AdV failed to induce significant T-bet expression in CD8 T cells, indicating a critical role for Stat4 in regulating T-bet expression (Figure 6C). Stat4 deficiency also significantly limited AdV-mediated T-bet induction in CD4 T cells, which was anticipated, since IL-12/Stat4 signals play an important role for regulating IFNγ production in both APCs and CD4 T cells. Thus the low level of IFNγ production in Stat4-deficient mice resulted in less T-bet expression in CD4 T cells, in contrast to exogenous IFNγ in in vitro experiments that is fully capable of stimulating these CD4 T cells. IFNγ expression was also measured by real-time RT-PCR. As shown in Figure 6D, Stat4-deficient or T-bet–deficient CD8 T cells have highly impaired IFNγ expression, while Stat1-deficient and wild-type CD8 T cells have similar IFNγ responses, indicating that Stat4 and T-bet play important roles in CD8 IFNγ regulation in vivo. Together, these experiments demonstrate that the IL-12/Stat4/T-bet cascade in CD8 T cells has physiological importance in vivo.
Figure 6.
AdV induces Stat4- but not Stat1-dependent CD8 T-bet expression in vivo. (A) AdV-GFP gene transfer vectors were injected into footpads of BALB/c mice. On days 1 and 3, CD8 and CD4 T cells were purified from draining LNs, and real-time RT-PCR was used to measure T-bet expression. (B) Real-time RT-PCR for T-bet expression in CD8 and CD4 T cells from Stat1-deficient mice injected with AdV-GFP. (C) Real-time RT-PCR for T-bet expression in CD8 and CD4 T cells from Stat4-deficient mice injected with Adv-GFP. (D) Real-time RT-PCR for IFNγ expression in CD8 T cells from various strains injected with AdV-GFP.
Discussion
Diverse immune mechanisms have evolved to protect the host from a vast array of pathogens, and CD8 T cells are key components of the effector arm of the immune response. While the master role of T-bet in regulating IFNγ expression, type 1 immune responses, and its regulation in CD4 T cells has been extensively documented, little is known about how T-bet is regulated in CD8 T cells or its precise role in IFNγ production and CTL function. Here, we found that IL-12–mediated Stat4 induces IFNγ/Stat1-independent T-bet expression in CD8 T cells. We showed that differential T-bet regulation by IL-12 in CD8 and CD4 T cells is due to differential IL-12/Stat4 signal regulation. We identified a novel T-bet enhancer, and provided a molecular basis for T-bet regulation. In addition, we observed that the IL-12/Stat4/T-bet cascade is relevant in vivo and regulates IFNγ expression in CD8 T cells during an antiviral immune response.
Studies by Murphy and colleagues have been instrumental in elucidating IL-12Rβ2 regulation and its role in governing IL-12 responsiveness during CD4 T-cell polarization (Afkarian et al,14 Presky et al,29 and Szabo et al30). Similar to naive CD4 T cells, we found that naive CD8 T cells do not express IL-12Rβ2, and consequently do not respond to IL-12 for Stat4 phosphorylation. However, after TCR activation, IL-12Rβ2 is up-regulated to a much higher level in CD8 than in CD4 T cells, resulting in strong and sustained Stat4 activation (Figures 2,3). Sustained activation may enable Stat4 to bind to the distal T-bet enhancer and induce T-bet expression (Figure 4). Since the presence of even low levels of IL-4 inhibits IL-12Rβ2 expression,30 and our previous data showed that TCR activation leads to transient IL-4 production only in CD4 but not CD8 T cells (Y.Y., unpublished data, July 2006), it is possible that the initial presence of TCR-induced IL-4 in CD4 but not CD8 T cells may determine differences in early IL-12Rβ2 expression, and subsequently determine IL-12–mediated differences in Stat4 activation and T-bet regulation.
STATs bind to a palindromic core DNA motif TTCN2-4GAA present in cytokine-inducible genes,32–35 and the arrangement in pairs for the STAT-binding motif allows the cooperative binding of 2 STAT dimers,32,37 and the mismatch half site TTA instead of TTC is permissible.35 In this report, we identified a conserved T-bet enhancer, which consists of 2 potential STAT-binding motifs TTCGAGGAA and TTAGCGGAA that are 10 base pairs apart and match the characteristic STAT-responsive elements. In vivo ChIP assay showed that this T-bet enhancer binds to both IFNγ-activated Stat1 and IL-12–activated Stat4 (Figure 4). EMSA experiments showed that these STAT-binding motifs interact with IL-12–activated Stat4. In vitro reporter assays further confirmed that this site responds to Stat4 by transactivating a reporter gene (Figure 5). The results demonstrated that the T-bet enhancer also responds to Stat1 (Figure 5), suggesting that the enhancer itself may not selectively respond to Stat1 or Stat4, and upstream signaling cascades controlling Stat1 or Stat4 activation determine how and when T-bet is activated. Since NK and NKT cells are responsive to IL-12 but not IFNγ, our results may explain the previous finding that IL-12 is able to induce T-bet mRNA in NK and NKT cells.9 It is currently not known how the T-bet enhancer interacts with the proximal T-bet promoter as well as other signaling molecules in directing T-bet gene transcription. Although there is a potential STAT-responsive element TTATTGGAA located 940-bp upstream of the transcription initiation site, ChIP and EMSA assays failed to detect interactions between this site and Stat4 after stimulation (Figures 4 and 5B). Enhancer knockout mice will be needed to define the precise role of the T-bet enhancer region, and how it may affect Th1 development in CD4 T cells and CTL activity in CD8 T cells.
The role of T-bet in controlling IFNγ expression of CD8 T cells is less dominant than in CD4 T cells. T-bet is required for optimal production of IFNγ in CD8 T cells stimulated by antigenic stimuli, but not for CD8 T cells stimulated with anti-CD3 plus anti-CD28 mAbs.7,38 In vivo studies showed that T-bet plays an important role for IFNγ production in CD8 T cells following lymphocytic choriomeningitis virus (LCMV) infection, but not for primary Listeria monocytogenes infection.6–8 Here, we found that T-bet deficiency results in decreased IFNγ expression in CD8 T cells following AdV vector injection (Figure 6). The molecular mechanisms that determine differential requirements for T-bet in IFNγ regulation in CD8 T cells activated by different types of antigenic stimulation are currently not known. In addition to T-bet, CD8 T cells express another member of the T-box transcription factor family, eomesodermin (Eomes, Tbr2), which plays complementary roles to T-bet in regulating IFNγ production and CD8 effector development. It is possible that alternative requirements for T-bet in IFNγ regulation may be due to differential Eomes regulation in CD8 T cells depending on the type of antigenic stimulation.
In agreement with our finding that AdV administration induces Stat4- but not Stat1-dependent CD8 T-bet expression in vivo, it has been recently showed that IL-12 positively regulates T-bet in effector CD8 T cells during infection with Listeria monocytogenes.39 CD8 T cells from T-bet–deficient OT1 TCR transgenic mice are markedly less efficient at killing peptide-loaded target than wild-type CTLs, both in vitro and in vivo,7 and CD8-mediated responses after LCMV infection or DNA vaccination are severely compromised in T-bet–deficient mice.6 These findings all indicate that the IL-12/Stat4/T-bet cascade is physiologically important. The identification of a novel enhancer may provide a new target to control CD8 effector responses.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R01 AI 62855 (Y.D.), AI 41428 (J.S.B), and AI 62765 (J.S.B).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.Y. performed research, analyzed data, and wrote the paper; J.C.O. performed research; Y.D. designed the research and contributed to interpretation and writing; and J.S.B. contributed to data interpretation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yaozhong Ding, Department of Gene and Cell Medicine, Box 1496, Mount Sinai School of Medicine, 1 Gustave L. Levy Place, New York, NY 10029; email: yaozhong.ding@mssm.edu.
References
- 1.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhang WX, Yang SY. Cloning and characterization of a new member of the T-box gene family. Genomics. 2000;70:41–48. doi: 10.1006/geno.2000.6361. [DOI] [PubMed] [Google Scholar]
- 3.Finotto S, Neurath MF, Glickman JN, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 4.Neurath MF, Weigmann B, Finotto S, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettelli E, Sullivan B, Szabo SJ, Sobel RA, Glimcher LH, Kuchroo VK. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juedes AE, Rodrigo E, Togher L, Glimcher LH, von Herrath MG. T-bet controls autoaggressive CD8 lymphocyte responses in type 1 diabetes. J Exp Med. 2004;199:1153–1162. doi: 10.1084/jem.20031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan BM, Juedes A, Szabo SJ, von Herrath MG, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Way SS, Wilson CB. Cutting edge: immunity and IFN-gamma production during Listeria monocytogenes infection in the absence of T-bet. J Immunol. 2004;173:5918–5922. doi: 10.4049/jimmunol.173.10.5918. [DOI] [PubMed] [Google Scholar]
- 9.Townsend MJ, Weinmann AS, Matsuda JL, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 10.Pearce EL, Mullen AC, Martins GA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 11.Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21:479–483. doi: 10.1016/s0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 12.Mullen AC, High FA, Hutchins AS, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 13.Lighvani AA, Frucht DM, Jankovic D, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afkarian M, Sedy JR, Yang J, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 15.Usui T, Preiss JC, Kanno Y, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNγ gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelik L, Constant S, Flavell RA. Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J Exp Med. 2002;195:1499–1505. doi: 10.1084/jem.20012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 18.Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. 2004;21:67–80. doi: 10.1016/j.immuni.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 20.Rui L, Carter-Su C. Identification of SH2-bbeta as a potent cytoplasmic activator of the tyrosine kinase Janus kinase 2. Proc Natl Acad Sci U S A. 1999;96:7172–7177. doi: 10.1073/pnas.96.13.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Ding Y, Schroppel B, et al. Differential chemokine and chemokine receptor gene induction by ischemia, alloantigen, and gene transfer in cardiac grafts. Am J Transplant. 2003;3:1216–1229. doi: 10.1046/j.1600-6143.2003.00207.x. [DOI] [PubMed] [Google Scholar]
- 22.McKendry R, John J, Flavell D, Muller M, Kerr IM, Stark GR. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci U S A. 1991;8:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Chen D, Tarcsafalvi A, Su R, Qin L, Bromberg JS. Suppressor of cytokine signaling 1 inhibits IL-10-mediated immune responses. J Immunol. 2003;170:1383–1391. doi: 10.4049/jimmunol.170.3.1383. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Ochando J, Yopp A, Bromberg JS, Ding Y. IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J Immunol. 2005;174:2720–2729. doi: 10.4049/jimmunol.174.5.2720. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y, Qin L, Zamarin D, et al. Differential IL-10R1 expression plays a critical role in IL-10 mediated immune regulation. J Immunol. 2001;167:6884–6892. doi: 10.4049/jimmunol.167.12.6884. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 27.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen KB, Watford WT, Salomon R, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 29.Presky DH, Yang H, Minetti LJ, et al. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci U S A. 1996;93:14002–14007. doi: 10.1073/pnas.93.24.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the interleukin (IL)-12Rβ2 subunit expression in developing T helper (TH1) and TH2 cells. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinmann AS. Novel ChIP-based strategies to uncover transcription factor target genes in the immune system. Nat Rev Immunol. 2004;4:381–386. doi: 10.1038/nri1353. [DOI] [PubMed] [Google Scholar]
- 32.Schindler U, Wu P, Rothe M, Brasseur M, McKnight SL. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 33.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 34.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 35.Ehret GB, Reichenbach P, Schindler U, et al. DNA binding specificity of different STAT proteins: comparison of in vitro specificity with natural target sites. J Bio Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q, Muruve DA. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 2003;10:935–940. doi: 10.1038/sj.gt.3302036. [DOI] [PubMed] [Google Scholar]
- 37.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 39.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]