Abstract
The oncogenic fusion tyrosine kinase nucleophosmin/anaplastic lymphoma kinase (NPM/ALK) induces cellular transformation in anaplastic large-cell lymphomas (ALCLs) carrying the t(2;5) chromosomal translocation. Protein-protein interactions involving NPM/ALK are important for the activation of downstream signaling pathways. This study was aimed at identifying novel NPM/ALK-binding proteins that might contribute to its oncogenic transformation. Using a proteomic approach, several RNA/DNA-binding proteins were found to coimmunoprecipitate with NPM/ALK, including the multifunctional polypyrimidine tract binding proteinassociated splicing factor (PSF). The interaction between NPM/ALK and PSF was dependent on an active ALK kinase domain and PSF was found to be tyrosine-phosphorylated in NPM/ALK-expressing cell lines and in primary ALK+ ALCL samples. Furthermore, PSF was shown to be a direct substrate of purified ALK kinase domain in vitro, and PSF Tyr293 was identified as the site of phosphorylation. Y293F PSF was not phosphorylated by NPM/ALK and was not delocalized in NPM/ALK+ cells. The expression of ALK fusion proteins induced delocalization of PSF from the nucleus to the cytoplasm and forced overexpression of PSF-inhibited proliferation and induced apoptosis in cells expressing NPM/ALK. PSF phosphorylation also increased its binding to RNA and decreased the PSF-mediated suppression of GAGE6 expression. These results identify PSF as a novel NPM/ALK-binding protein and substrate, and suggest that PSF function may be perturbed in NPM/ALK-transformed cells.
Introduction
Anaplastic large-cell lymphoma (ALCL) comprises a group of CD30/Ki-1+ T-cell or null-cell lymphoid neoplasms.1 A subset of ALCL can be characterized by the expression of fusion proteins involving the anaplastic lymphoma kinase (ALK).2 ALK is a receptor tyrosine kinase normally expressed in specific tissues of the central nervous system during embryogenesis.3,4 Due to chromosomal translocations involving the ALK gene at 2p23, ALK is also aberrantly expressed in lymphoid tissues. To date, 11 ALK fusion proteins have been detected in ALCL, the most common of which is nucleophosmin (NPM)/ALK, occurring in 70% of patients with ALK+ ALCL.5 NPM/ALK, the product of the t(2;5)(p23;q35) translocation, encodes a chimeric 80-kDa protein consisting of the N-terminal portion (amino acids 1-117) of NPM fused to the cytoplasmic portion of ALK (amino acids 1058-1620).6,7 NPM is a ubiquitously expressed protein normally localized in the nucleus that has been implicated in nuclear/cytoplasmic trafficking, the cell cycle, centrosome duplication, and maintenance of genomic stability.8–13 The N-terminal portion of NPM contains a homodimerization domain, which is responsible for the formation of NPM/ALK oligomers capable of transphosphorylation and activation of the ALK kinase domain.14 NPM/ALK has a nuclear and cytoplasmic localization, whereas other ALK chimeric proteins (eg, ATIC/ALK and CLTC/ALK) possess diffuse or granular cytoplasmic immunostaining patterns.15,16 The constitutive activation of ALK is sufficient to induce cellular transformation in vitro14,17,18 and lymphoid/myeloid neoplasms in xenograft or transgenic mice models.19–22
NPM/ALK exerts its transforming potential via its ability to interact and activate several antiapoptotic and mitogenic signaling transducers (ie, PI-3K, JAK, STAT, and PLC-γ).17,23–26 The interaction of NPM/ALK with these signaling molecules is either direct or mediated by adapter proteins containing SH2- or phosphotyrosine-binding domains.17,23 NPM/ALK has been shown to interact with the adapter proteins IRS1, SHC, GRB2, and CRKL,14,17,18,23 and to coimmunoprecipitate with PI-3K, STAT3, STAT5, JAK3, JAK2, NIPA (nuclear interacting partner of ALK), and p130CAS.27–29 The significance of these interactions in NPM/ALK-mediated oncogenesis has been partially investigated. For example, activation of the PI-3K pathway is required for the growth of BaF3-NPM/ALK–transformed cells in mice, while STAT3 is essential for NPM/ALK-induced lymphomagenesis in a transgenic mouse model.30,31 In contrast, mutation of docking sites within NPM/ALK for SHC and IRS1 did not affect the transforming ability of NPM/ALK in vitro, while mutation of the PLC-γ docking site only impaired mitogenic, but not antiapoptotic, signaling.17 Since multiple pathways are targeted by NPM/ALK, it is possible that functional redundancy exists between pathways. Furthermore, the oncogenicity of NPM/ALK is likely to be the result of a complex interplay between these signaling pathways and possibly other as yet unidentified, downstream effectors. We report the identification of novel ligands of NPM/ALK, including several multifunctional RNA/DNA-binding proteins such as the polypyrimidine tract-binding protein-associated splicing factor (PSF), the nuclear RNA-binding protein 54 kDa (p54nrb), translocated in liposarcomas (FUS/TLS), and EWS (expressed in Ewing sarcoma).
Materials and methods
Antibodies
The polyclonal anti-PSF and the monoclonal anti-ALK1 antibodies have been previously described.32,33 The anti-ALK11 polyclonal antibody was kindly supplied by Dr S. W. Morris (St Jude Research Hospital, Memphis, TN). The polyclonal anti–β-actin (Cell Signaling Technology, Danvers, MA), the polyclonal anti–lamin B1 (Abcam, Cambrige, United Kingdom), the polyclonal anti-SHC (Upsate Biotech Lake Placid, NY), the monoclonal antihemagglutinin (HA; clone HA-11; Covance, Berkeley Antibody Company, Berkeley, CA), the monoclonal anti–histone H1 (Upstate Biotechnology, Lake Placid, NY), the monoclonal F2 anti-PARP (Santa Cruz Biotechnology, Santa Cruz, CA), the monoclonal anti-PSF (Sigma-Aldrich, St Louis, MO), and the antiphosphotyrosine (4G10; Upstate Biotechnology) antibodies were used as suggested by the manufacturers. Normal rabbit IgG1 was obtained from Amersham (Arlington Heights, IL). Horseradish peroxidase (HRP)–conjugated anti-mouse or anti-rabbit secondary antibodies were obtained from Bio-Rad (Hercules, CA). The Alexa Fluor 488 goat anti–mouse IgG was obtained from Molecular Probes (Eugene, OR), and the Cy5 conjugated affinity goat anti–rabbit IgG was obtained from Rockland (Gilbertsville, PA).
Plasmids
Full-length PSF cDNA cloned into the bacterial expression vector pET 15b (Novagen, Madison, WI) has been previously described.34 To clone pCR3.1 containing full length HA-tagged PSF (pCR3.1-HA-PSF), the open reading frame (ORF) of PSF was cloned into pCR3.1 (Invitrogen, San Diego, CA) with BamHI and XhoI, then an HA tag was created at the 3′ end of the ORF of PSF. Site-directed mutagenesis of Tyr293 to Phe (Y293F) in HA-PSF was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). pcDNA3.0 (Invitrogen) containing wild-type NPM/ALK (pcDNA3-NA) or a kinase-dead NPM/ALK possessing the K210R mutation (pcDNA3-NA-K210R) were kindly provided by S. W. Morris (St Jude Research Hospital, Memphis, TN). pSG5 (Stratagene) containing GCN4/ALK was created from a pSG5 vector already containing GCN4/RAR. RAR was removed by digestion with EcoRV (blunt) and BglII and replaced by ALK cytoplasmic domain ORF. To clone GCN4/ALK into the MSCV-IRES-GFP retroviral expression system (BD Biosciences, San Jose, CA), GCN4/ALK was excised from pSG5 with BglII and MSCV-IRES-GFP was digested with Xho1. GCN4/ALK and MSCV-IRES-GFP were blunt-ended with EcoR1 and ligated. For the retroviral construct MigR1-HA-PSF, HA-tagged PSF was excised from pCR3.1-HA-PSF with BamHI and XhoI, and subcloned into the BglII/XhoI sites of the bicistronic green fluorescent protein (GFP)–containing MigR1 vector.
Cell lines
The murine IL-3–dependent pro-B BaF3 cell line, the SUDHL-1 (t(2:5)+) human ALCL-derived cell line, the K562 (Ph+ BCR/ABL+) human chronic myeloid leukemia (CML)–derived cell line, human embryonic kidney (HEK) 293T cells, and the Jurkat T-cell line were obtained from DSMZ (Braunschweig, Germany). The human ALCL (t(2:5)+) JB6 cell line was kindly provided by Dr F. Turturro (Louisiana State University Health Science Center/Feist-Weiller Cancer Center, Shreveport, LA). BaF3 cells were stably transfected with 10 μg pcDNA3-NA (BaF3-NA), pcDNA3-NA-K210R (BaF3-KD), or pcDNA3-Bcr/Abl (BaF3-BA) by electroporation. Clonal cell lines were obtained by limiting dilution. A stable BaF3 cell line expressing GCN4/ALK was generated using MSCV-GCN4/ALK. BaF3 cells (106) were infected with supernatant from Phoenix packaging cells (kind gift of G.P. Nolan, Stanford University School of Medicine, Stanford, CA) transfected with retroviral construct MSCV-GCN4/ALK. GCN4/ALK-expressing cells were selected by culturing in the absence of IL-3. All cells were maintained in a humidified atmosphere at 37°C and 5% CO2.
Expression and purification of recombinant proteins
N-terminally 6xHis-tagged PSF protein was produced using BL21 competent cells (Stratagene) and pET15b-PSF. Cells were harvested and lysed by sonication in 50 mM NaH2PO4 and 300 mM NaCl (pH 8.0). Lysates were clarified by centrifugation at 15 000g for 30 minutes at 4°C. PSF was purified using NI-NTA agarose resin (QIAGEN, Hilden, Germany) in batch mode following instructions. 6xHis-tagged ALK protein containing residues Leu1073-Ala1459 (ALK_HUMAN, GenBank accession no. Q9UM73), which includes the predicted kinase domain, was produced using a Baculovirus expression system and purified as described elsewhere.35 GST-tagged ALK (Leu1073-Ala1459) was expressed using the Bac-to-Bac Baculorvirus expression system (Invitrogen) and purified using GSTrap columns (GE Healthcare, Chalfont St Giles, United Kingdom).
Immunoprecipitation and Western blotting
Cells (20 × 106/sample) were washed in ice-cold PBS and lysed on ice in lysis buffer (50 mM Tris HCl [pH 7.4], 1% Triton X-100, 5 mM EDTA, 150 mM NaCl, 1 mM Na3VO4, 1 mM NaF, 1 mM PMSF, and protease inhibitor cocktail). Cell lysates were clarified by centrifugation at 12 000g for 20 minutes at 4°C, and protein was quantified using the Bradford assay (BioRad Laboratories, Hercules, CA). Frozen lymph nodes were obtained from 2 patients with ALCL and normal spleen tissue was obtained from a healthy donor, after informed consent (kindly provided by Dr P. Collini, Pathology Department, National Cancer Institute, Milan, Italy). Tissues were homogenized and lysed as described previously.36 Proteins were immunoprecipitated by incubating 100 μg (cell lines), or 1.5 mg (patient tissue) of total protein overnight with anti-PSF and anti-ALK1 at 4°C, followed by a 1-hour incubation with 30 μL protein A–Sepharose (GE Healthcare) at 4°C. Immunocomplexes were washed 5 times with ice-cold lysis buffer, denatured, and subjected to SDS-PAGE. Western blots were performed with anti-PSF, anti-ALK1, and anti-pTyr primary antibodies.
Tryptic digestion and mass spectrometric peptide sequencing
Anti-ALK1 immunocomplexes obtained by immunoprecipitation of 400 × 106 cells were resolved by SDS-PAGE and detected by silver staining. Bands of interest were excised from silver stained gels, reduced, alkylated, and digested overnight with bovine trypsin as described.37 A total of 1 μL of supernatant containing the generated tryptic peptides was loaded onto the matrix-assisted laser desorption/ionization (MALDI) target using the dried droplet technique and α-cyano-4-hydroxycinnamic acid (HCCA) acid as matrix. MALDI–time-of-flight (TOF) mass measurements were performed on a Voyager-DE STR TOF mass spectrometer (Applied Biosystems, Framingham, MA) operated in the delayed extraction and reflector mode. Spectra, internally calibrated, were processed via the Data Explorer software (Applied Biosystems). Proteins were unambiguously identified by searching a comprehensive nonredundant protein database using the program ProFound.38
Kinase assays
Full-length purified 6xHis-tagged PSF was phosphorylated in 50 μL of reaction buffer containing 25 mM HEPES (pH 7.5), 10 mM MgCl2, 10 mM MnCl2, 0.037 MBq (1 μCi; 4500 Ci/mmol) [γ32P]ATP, 20 μM ATP, 2 mM DTT, purified 6xHIS-ALK (0.4 μg), and purified 6xHis-PSF (0.25 μg). After incubation at 30°C for 30 minutes, reactions were terminated by the addition of 15 μL of 5 × Laemmli buffer. Samples were denatured, subjected to SDS-PAGE, and visualized by autoradiography. Synthetic PSF peptides were synthesized as previously described39 (Tyr293, Tyr251, Tyr470, Tyr488-490, Tyr597-602, Tyr698) or purchased from Sigma. Genosys, Hoverhill, United Kingdom (Tyr527 and Tyr624). PSF peptides (400 μM) were phosphorylated in the presence of 10 U of GST-ALK as described elsewhere.39 One unit was defined as the amount of GST-ALK transferring 1 pmol phosphate per minute to the random polymer polyGlu4Tyr (0.1 mg/mL) under standard conditions.
Nuclear and cytoplasmic extracts
Cells (3 × 107) were washed in ice-cold PBS, resuspended in a buffer containing 20 mM HEPES (pH 7), 2 mM MgCl2, 10 mM KCl, 0.5% NP40, and protease inhibitors, and homogenized using a Wheaton A Dounce homogenizer (20 strokes; Wheaton Science Products, Millville, NJ). Lysates were clarified by centrifugation at 1500g for 5 minutes. The resulting supernatant was subsequently centrifuged at 15 000g for 10 minutes. The soluble fraction, representing the cytoplasmic extract, was immediately transferred to a prechilled tube and stored on ice. The insoluble fraction, which contains nuclei, was washed twice with ice-cold buffer and then resuspended in buffer supplemented with 0.5 M NaCl and rocked gently for 30 minutes at 4°C to extract nuclear proteins. The extracted material was then centrifuged at 15 000g for 10 minutes, and the soluble nuclear extract fraction was transferred to a prechilled tube and placed on ice. Extracts were stored at −80°C until use.40
Immunofluorescence
293T cells were seeded on glass coverslips and transiently transfected with 5 μg pCR3.1-HA-PSF alone or together with 5 μg of pcDNA3-NA, pcDNA3-NA-K210R, or pG5-GCN4/ALK, using the calcium phosphate precipitation method. At 24 hours after transfection, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 and 2% BSA in PBS at room temperature (RT) for 10 minutes. Cells were incubated overnight at 4°C with the primary antibody, washed 3 times with PBS 0.2% Triton X-100, and incubated for 1 hour at room temperature with the relevant secondary antibodies. The nuclear staining was performed by incubating with 50 μg/mL propidium iodide for 30 minutes at 37°C. The coverslips were then washed and mounted. Fluorescence was detected using the Nikon Eclipse 600 confocal microscope (Nikon, Melville, NY).
Proliferation and apoptosis assays
Parental BaF3, BaF3-NA, BaF3-BA, and BaF3-KD cells (5 × 106) were transiently transfected by electroporation (260 V and 1050 μF) with 10 μg pCR3.1 or pCR3.1-HA-PSF. Cells were then resuspended in medium with (BaF3-Par, BaF3-KD) or without IL-3 (BaF3-NA, BaF3-BA), and cell proliferation was measured at 24, 48, and 72 hours after transfection by 3H-thymidine incorporation as described previously.25 Apoptosis was assessed by detecting PARP cleavage by immunoblotting with the anti-PARP monoclonal antibody or by annexin V–fluorescein isothiocyanate (FITC) binding using the Apoptosis Detection Kit (Bender MedSystems Diagnostic, Vienna, Austria). Samples were analyzed using a FACScalibur and CellQuest software (BD Bioscience, San Jose, CA).
Colony formation assay
The amphotropic-packaging cell line Phoenix A (G. P. Nolan, Stanford University School of Medicine, Stanford, CA) was transiently transfected41 with MigR1-HA-PSF, and the infectious supernatant was used to infect JB6, SUDHL-1, and Jurkat cell lines. After infection, cells were sorted for GFP expression using a FACSCalibur. GFP+ cells (103) were plated in 0.9% Metho-Cult M3234 semisolid medium (Stem Cell Technologies, Vancouver, BC, Canada), and colonies were scored 9 to 12 days later.42
Results
Identification of proteins coimmunoprecipitating with NPM/ALK
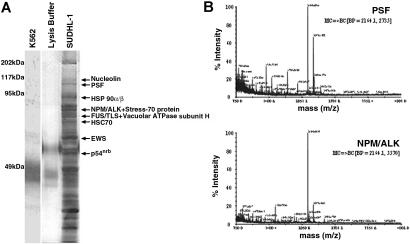
NPM/ALK was immunoprecipitated from SUDHL-1 cells, and associating proteins were visualized by SDS-PAGE and silver staining. Several proteins with diverse molecular weights (MWs) coimmunoprecipitated with NPM/ALK (Figure 1A), but were not present in anti-ALK1 immunoprecipitates from NPM/ALK− K562 cells or lysis buffer alone. Coimmunoprecipitating proteins with a MW greater than 50 kDa were analyzed by MALDI-TOF, and 10 were identified. These included the heat-shock proteins HSP 90α/β,43,44 previously identified as NPM/ALK interacting proteins,45 and novel ligands such as HSC 70,46 V-ATPase subunit-H,47 Stress-70 protein,48 nucleolin,49 PSF,50 FUS/TLS,51 nonO/p54nrb,52 and EWS53 (Table 1). NPM/ALK itself was also positively identified demonstrating the reliability of the mass spectrometry sequencing. Tandem mass spectrometry (MS/MS) profiles identifying the PSF and NPM/ALK proteins are shown in Figure 1B.
Figure 1.
Identification of proteins coimmunoprecipitating with NPM/ALK by mass spectrometry. (A) Silver-stained SDS-PAGE gel of anti-ALK1 immunoprecipitates from NPM/ALK− K562 cells (lane 1), lysis buffer alone (lane 2), and NPM/ALK+ SUDHL-1 cells (lane 3). Bands were excised from the gel, trypsin-digested, and analyzed by MALDI-TOF. Identified proteins are indicated by arrows. (B) MS/MS sequencing of peptides identifying PSF (GenBank accession no. P23246) and NPM/ALK (GenBank accession no. AAA58698) proteins.
Table 1.
Summary of proteins coimmunoprecipitating with NPM/ALK from SUDHL-1 cells, identified by MALDI-TOF
| NCBI | Protein | MW, kDa | Description |
|---|---|---|---|
| P19338 | Nucleolin | 77 (theoretical), 110 (apparent) | Abundant RNA/DNA-binding nucleolar protein. Plays a major role in ribosome biogenesis and implicated in other processes (eg, pre-mRNA metabolism, transcriptional repression, DNA repair, replication, and recombination).49 |
| P23246 | PSF | 74 (theoretical), 100 (apparent) | RNA/DNA-binding nuclear protein involved in pre-mRNA processing, transcriptional repression, DNA repair replication, and recombination.50 |
| P07900 | HSP 90-α | 90 | Cytoplasmic protein belonging to the heat-shock 90 family of molecular chaperones.43 |
| P08238 | HSP 90-β | 90 | Cytoplasmic protein belonging to the heat-shock 90 family of molecular chaperones.44 |
| AAA58698 | NPM/ALK | 80 | Oncogenic fusion tyrosine kinase formed by the t(2;5) chromosomal rearrangement in ALCL.7 |
| P38646 | Stress-70 protein | 75 | Mitochondrial protein belonging to the heat-shock 70 family. Implicated in the control of cell proliferation and cellular aging, and may act as a molecular chaperone.48 |
| P35637 | FUS/TLS | 54 (theoretical), 68-75 (apparent) | Oncogenic nuclear RNA/DNA-binding protein involved in pre-mRNA processing, transcriptional activation, and DNA recombination.51 |
| Q01844 | EWS | 68 (theoretical) | RNA binding protein involved in oncogenic fusion proteins with various transcription factors in sarcoma. EWS fusion proteins are potent transcriptional activators.53 |
| P11142 | HSC70 | 70 | Heat-shock cognate 70-kDa protein is a member of the heat-shock 70 family of molecular chaperones.46 |
| Q9UI12 | Vacuolar-ATP synthase subunit-H | 56 | Regulatory subunit of the universal proton pump required for the acidification of endosomes and lysosomes.47 |
| Q15233 | p54nrb | 54 | RNA/DNA-binding nuclear protein involved in pre-mRNA processing, transcriptional activation, and DNA repair replication.52 |
Proteins were identified in 6 independent experiments.
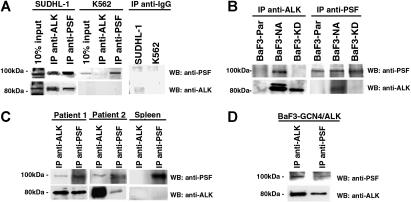
The binding of NPM/ALK to PSF was further investigated by reciprocal immunoprecipitation and Western blotting using NPM/ALK+ cells (SUDHL-1– and NPM/ALK-transfected BaF3 cells [BaF3-NA]) and NPM/ALK− cells (K562 and parental BaF3 cells [BaF3-Par]). PSF was detected in anti-ALK1 immunoprecipitates derived from t(2;5) SUDHL-1 and BaF3-NA cells (Figure 2A-B). Accordingly, NPM/ALK was detected in anti-PSF immunoprecipitates from the same cells. Coimmunoprecipitation of PSF and NPM/ALK was also observed in primary lymph node tissue obtained from 2 patients with NPM/ALK+ ALCL, but not in spleen tissue from a healthy donor (Figure 2C). Furthermore, PSF was unable to interact with the kinase-dead NPM/ALK mutant expressed in BaF3 cells (BaF3-KD; Figure 2B). Thus, PSF is a bona fide NPM/ALK ligand, and this interaction is strictly dependent on NPM/ALK kinase activity.
Figure 2.
PSF coimmunoprecipitates with ALK fusion proteins expressed in cell lines and ALCL tumors. Reciprocal immunoprecipitation (IP) and Western blotting (WB) experiments were performed using anti-ALK1 and anti-PSF antibodies on lysates derived from (A) NPM/ALK+ SUDHL-1 cells and NPM/ALK− K562 cells; (B) BaF3-NPM/ALK (BaF3-N/A), BaF3-Parental (BaF3-PAR), and BaF3 cells expressing kinase-dead NPM/ALK (BaF3-KD); (C) lymph node tissue taken from 2 patients with NPM/ALK+ ALCL and NPM/ALK− spleen tissue; and (D) BaF3 cells expressing GCN4/ALK (BaF3-GCN4/ALK). As an internal control, SUDHL-1 and K562 cells were subjected to IP with an anti-IgG antibody (A).
To determine whether PSF can interact with ALK fusions other than NPM/ALK, we assessed its ability to coimmunoprecipitate with the GCN4/ALK fusion protein. GCN4/ALK contains the coiled-coil homodimerization domain of the yeast GCN4 protein fused to the complete cytoplasmic domain of ALK.54 PSF was able to interact with GCN4/ALK (Figure 2D), indicating that the NPM moiety is dispensable for the PSF-ALK interaction, and suggests that the PSF-ALK interaction is mediated through the ALK kinase domain.
PSF is a direct target of NPM/ALK tyrosine kinase activity
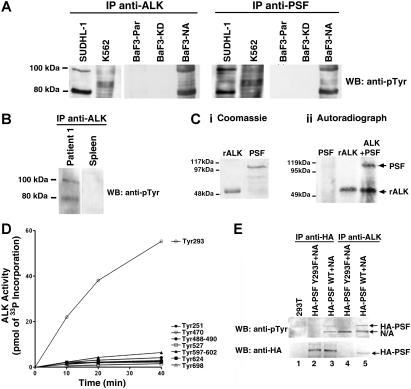
Since NPM/ALK has a constitutive tyrosine kinase activity, and PSF binds to NPM/ALK in cells, we examined whether PSF could be a substrate of NPM/ALK. Initially, the tyrosine phosphorylation status of PSF in cell lines expressing or not expressing NPM/ALK was examined. A 100-kDa tyrosine-phosphorylated protein corresponding to PSF was detected in anti-ALK and anti-PSF immunoprecipitates from SUDHL-1 and BaF3-NA cells (Figure 3A). In contrast, no such tyrosine-phosphorylated band was found in anti-ALK and anti-PSF immunoprecipitates from K562, BaF3-KD, and BaF3-Par cells. The fact that PSF was not phosphorylated in K562 cells, which express the constitutively active tyrosine kinase Bcr/Abl, suggests that PSF tyrosine phosphorylation is specifically dependent on ALK activity. PSF was also found to be tyrosine-phosphorylated in anti-ALK1 immunoprecipitates from lysates of tissue from 1 lymph node from a patient with ALCL, but not from normal spleen (Figure 3B). Due to insufficient material, it was not possible to assess PSF tyrosine phosphorylation in ALCL lymph nodes from patient 2.
Figure 3.
PSF is tyrosine-phosphorylated in NPM/ALK-expressing cells and is a substrate of ALK tyrosine kinase activity. (A) Anti-ALK1 and anti-PSF immunoprecipitates (IPs) from SUDHL-1, K562, BaF3-Par, BaF3-KD, and BaF3-NA cells were Western blotted with an antiphosphotyrosine (anti-pTyr) antibody. (B) Anti-ALK1 immunoprecipitates from lymph nodes derived from patient 1 with NPM/ALK+ ALCL and NPM/ALK− spleen tissue were Western blotted using the anti-pTyr antibody. (C) A radioactive in vitro kinase assay was performed with purified recombinant 6xHisALK kinase domain (rALK) and purified full-length 6xHis-PSF. Protein purity was checked by coomassie staining (Ci). Kinase assay was performed in the presence of rALK kinase domain alone (0.4 μg), PSF alone (0.25 μg), or rALK and PSF together. Samples were analyzed by SDS-PAGE and visualized by autoradiography (Cii). Bands corresponding to PSF and ALK are indicated by arrows. (D) Time courses of the phosphorylation of PSF peptides (400 μM) by GST-ALK. The peptide sequences are as follows: Tyr251, RGGRQHHPPYHQQHHQGP; Tyr293, PGEKTYTQRCRLFVGNLPADIT; Tyr470, EKLAQKNPMYQKERETPTR; Tyr488-490, TFEYEYSQRWKSLDEMEKQQR; Tyr527, EMEDAYHEHQANLLRQDLMRRQ; Tyr597-602, REESYSRMGYMDPRERDMR; Tyr624, MNMGDPYGSGGQKFPPLGGG; and Tyr698, GRGREEYEGPNKKPRF (target tyrosine in bold). (E) Immunoprecipitation with anti-HA (lanes 2,3) and anti-ALK1 (lanes 4,5) monoclonal antibodies of lysate derived from 293T cells transiently transfected with wild-type or mutant Y293F HA-PSF together with NPM/ALK. Immunoprecipitates were resolved by SDS-PAGE, and Western blotting with anti-pTyr and anti-HA antibodies was performed. Lane 1 shows lysate from nontransfected 293T cells.
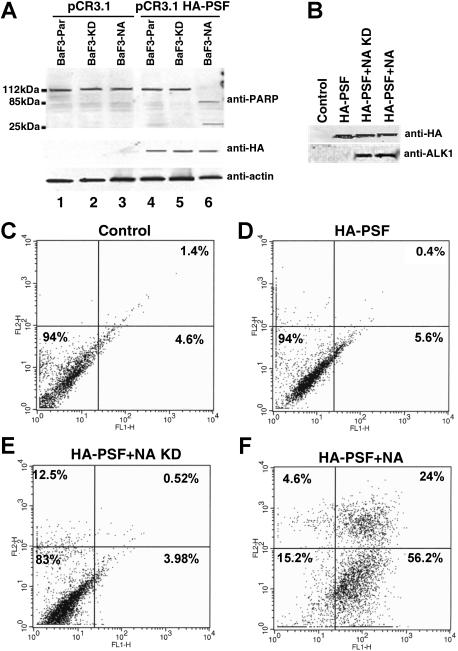
To determine if PSF could be directly phosphorylated by ALK, an in vitro kinase assay was performed using purified recombinant 6xHis-tagged ALK kinase domain and full-length 6xHis-tagged PSF. Purified ALK kinase domain possesses autophosphorylation activity and is able to phosphorylate PSF (Figure 3C), but not the unrelated GST and BSA proteins (data not shown). In an attempt to map the PSF tyrosine residue/s that is/are phosphorylated by ALK, we synthesised 8 synthetic PSF peptides containing the potential ALK tyrosine phosphorylation site. Peptides were chosen on the basis of the ALK substrate consensus sequence39,55 and phosphorylation site prediction programs (eg, NetPhos 2.0 Server).56 The ability of purified ALK kinase domain to phosphorylate these peptides was determined by an in vitro kinase assay. Only the peptide containing Tyr293 of PSF (target tyrosine in bold: PGEKTYTQRCRLFVGNLPADIT) was phosphorylated by ALK (Figure 3D). To confirm that PSF Tyr293 is indeed a bona fide ALK phosphorylation site, we mutagenized Tyr293 to Phe and transiently coexpressed the PSF Y293F mutant and NPM/ALK in 293T cells. We observed that mutant PSF was no longer tyrosine phosphorylated (Figure 3E top panel, lanes 2 and 4), unlike control wild-type PSF (Figure 3E top panel, lanes 3 and 5). Mutant PSF did not coimmunoprecipitate with NPM/ALK (Figure 3E; lower panel, lane 4). These data suggest that PSF is a substrate of NPM/ALK, and that tyrosine phosphorylation occurs at Tyr293 of PSF and is critical for the physical association of NPM/ALK and PSF.
ALK fusion proteins alter PSF subcellular localization
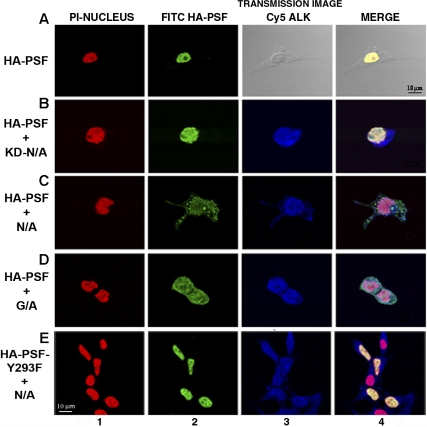
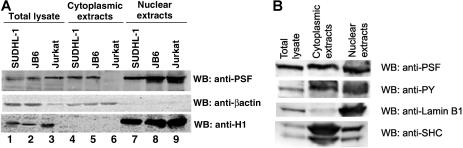
PSF normally displays a nuclear localization pattern, whereas NPM/ALK is localized in both the cytoplasm and nucleus.14,57,58 Since NPM/ALK binds PSF, we investigated whether this association affects the subcellular localization of PSF. Immunofluorescence and confocal microscopy experiments were performed in 293T cells transiently transfected with HA-PSF alone or cotransfected with HA-PSF and NPM/ALK, NPM/ALK KD, or GCN4/ALK (Figure 4). Since no anti-PSF antibody suitable for immunofluorescence was available, ectopically expressed HA-PSF was evaluated. In 293T cells, HA-PSF showed a characteristic nuclear staining pattern when expressed alone (Figure 4A). However, expression of HA-PSF together with active ALK fusion proteins, NPM/ALK or GCN4/ALK, resulted in HA-PSF localization in both the nucleus and cytoplasm (Figure 4C,D). In contrast, kinase-dead NPM/ALK did not affect the nuclear localization of HA-PSF (Figure 4B), consistent with its inability to coimmunoprecipitate with NPM/ALK (Figure 2B). To confirm the delocalization of PSF in NPM/ALK-expressing cells, we prepared nuclear and cytoplasmic protein extracts from JB6, SUDHL-1, and Jurkat cells, and detected PSF in these extracts by Western blotting. In the negative-control Jurkat cells, PSF was only detected in the nuclear extract (Figure 5A lanes 6 and 9), whereas in SUDHL-1 and JB6 cell lines, PSF was detected in both the nuclear and cytoplasmic extracts (Figure 5A lanes 4, 5, 7, and 8). Both nuclear and cytoplasmic fractions appear to contain phosphorylated PSF (Figure 5B). Interestingly, coexpression of the mutant Y293F PSF with active NPM/ALK resulted in the localization of mutant PSF only in the nucleus (Figure 4E). Therefore, these results demonstrate that ALK fusion proteins can induce the relocalization of PSF from the nucleus to the cytoplasm and indicate that Y293 phosphorylation is needed for PSF delocalization.
Figure 4.
PSF is delocalized to the cytoplasm in cells expressing ALK fusion proteins. Indirect immunofluorescence was performed on 293T cells transiently transfected with HA-PSF alone, or together with kinase-dead NPM/ALK (KD-N/A), NPM/ALK (N/A), GCN4/ALK (G/A), or with mutant HA-PSF-Y293F together with NPM/ALK (N/A). Fluorescence was detected by confocal microscopy. Nuclei were detected using propidium iodide (PI) shown in red (column 1). HA-PSF was detected by FITC staining shown in green (column 2). NPM/ALK was detected by Cy5 staining shown in blue (column 3). A transmission image in place of Cy5 staining is shown for cells transfected with HA-PSF alone. Images were acquired using a Nikon 60×/1.4 NA oil-immersion objective lens, a medium solution of 95% glycerol in PBS, and a Biorad Laser Sharp 2000 camera; image processing was done with Adobe Photoshop CS2 version 9.0.
Figure 5.
PSF content in cytoplasmic and nuclear extracts derived from human cell lines. Nuclear and cytoplasmic extracts were prepared from the NPM/ALK− cell line, Jurkat, and the NPM/ALK+ cell lines, SUDHL-1 and JB6. (A) Total cell lysates (lanes 1-3), cytoplasmic extracts (lanes 4-6), and nuclear extracts (lanes 7-9) were resolved by SDS-PAGE, and PSF was detected by Western blotting (top panel). The purity of the extracts were controlled by Western blotting with anti-β-actin for the cytoplasm (middle panel) and anti-histone H1 for the nucleus (bottom panel). (B) Nuclear and cytoplasmic extracts from SUDHL-1 cells were probed with antiphosphotyrosine antibody.
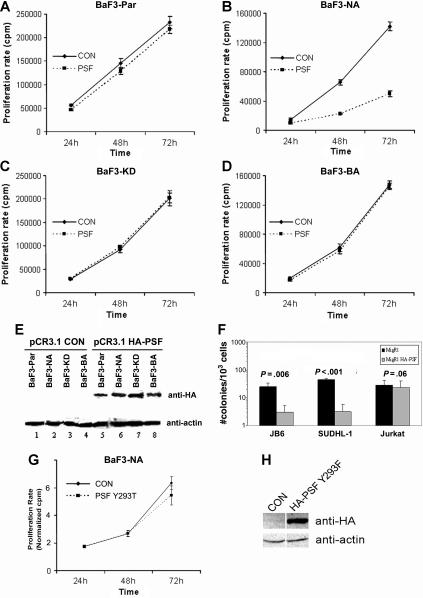
Forced PSF expression induces growth arrest and apoptosis, and reduces the clonogenic potential of NPM/ALK+ cells
We investigated the effect of overexpression of PSF on cells expressing and not expressing NPM/ALK. Since NPM/ALK expression is associated with phosphorylation and delocalization of PSF into the cytoplasm, it is plausible that PSF might contribute to the oncogenic activity of NPM/ALK. We transiently overexpressed HA-tagged PSF in BaF3-NA cells and observed a marked inhibition of proliferation in these cells compared with cells transduced with empty vector alone (Figure 6B). In contrast, PSF overexpression did not affect the growth of BaF3-Par cells, BaF3-KD cells, or BaF3 cells stably transformed with BCR/ABL (BaF3-BA; Figure 6A,C,D). The overexpression of HA-PSF was confirmed by Western blotting (Figure 6E). Unlike wild-type PSF, the mutant Y293F PSF had no effect on the proliferation of BaF3-NA cells (Figure 6G,H). We subsequently evaluated the effect of ectopic PSF expression on the clonogenicity of NPM/ALK+ cell lines. A retroviral vector carrying HA-tagged PSF (MigR1-HA-PSF) was used to transduce the NPM/ALK+ human cell lines, JB6 and SUDHL-1, and the NPM-ALK− Jurkat cell line. The effect of PSF overexpression on clonogenic potential was assessed in transduced cells using a methylcellulose colony formation assay. Overexpression of HA-PSF induced a marked decrease in the number of colonies derived from the NPM/ALK+ human cell lines, while having no effect on the NPM/ALK− Jurkat cells (Figure 6F). Transduction with MigR1 alone in JB6, SUDHL-1, and Jurkat cells had no effect on colony formation (Figure 6F). These results indicate that PSF overexpression exerts growth arrest specifically in NPM/ALK-expressing cells.
Figure 6.
PSF overexpression reduces the growth rate and clonogenic potential of NPM/ALK-expressing cells. BaF3-Par (A), BaF3-NA (B), BaF3-KD (C), and BaF3-BA (D) cells were transiently transfected with pCR3.1 encoding HA-PSF (PSF) or with empty vector (CON). Cell proliferation was measured by 3H-thymidine incorporation at 24, 48, and 72 hours after transfection. Results are shown as the mean (± SD) and are representative of 3 independent experiments. (E) Expression of HA-PSF was verified by Western blotting using anti-HA antibody at 72 hours, and protein loading was controlled by antiactin Western blotting. (F) The NPM/ALK+ cell lines, JB6 and SUDHL-1, and the NPM/ALK− cell line, Jurkat, were transduced with the MigR1-HA-PSF construct (▩), or the MigR1 retrovirus alone (■). Cells were selected for GFP positivity, and clonogenic potential was measured by colony formation in methylcellulose. Results are expressed as the mean (± SEM) of 3 independent clonogenic assays. Significance was determined by comparing MigR1 alone versus MigR1-HA-PSF in a 2-tailed paired samples t test. (G) BaF3-NA cells were transiently transfected with pCR3.1 encoding HA-Y293T PSF or with empty vector (CON). Error bars are standard deviation. (H) Transfection efficiency was controlled by anti-HA immunoblotting at 48 hours in control (CON) and HA-PSF Y293F samples loaded on the same gel. Total protein loading was controlled by antiactin immunoblotting.
The decrease of BaF3-NA cell proliferation was also accompanied by the induction of apoptosis, demonstrated by the cleavage of the caspase substrate, PARP59 (approximately 112 kDa), to approximately 85- and 25-kDa proteolytic products (Figure 7A). The induction of apoptosis by PSF overexpression in the presence of NPM/ALK was also evident when using the Annexin V binding assay (Figure 7F). Apoptosis was not observed in nontransfected controls or in cells transfected with HA-PSF alone or together with kinase-dead NPM/ALK (Figure 7C-E). The level of HA-PSF and NPM/ALK expression in transfected cells was controlled by Western blotting (Figure 7B). Therefore, forced overexpression of PSF induces apoptosis in NPM/ALK-expressing cells.
Figure 7.
PSF overexpression induces apoptosis in NPM/ALK-expressing cell lines. (A) BaF3-Par, BaF3-KD, and Baf3-NA cells were electroporated with pCR3.1 plasmid alone (lanes 1-3) and with pCR3.1 HA-PSF (lanes 4-6). After 30 hours of culture, the cell extracts from 5 × 106 cells were lysed and subjected to denaturing SDS-PAGE and Western blotting with an anti-PARP antibody. PSF expression and protein loading were controlled by anti-HA and antiactin Western blotting. (B-F) 293T cells were transiently transfected with pCR3.1 empty vector (CON) or HA-PSF alone or together with kinase-dead or wild-type NPM/ALK. The efficiency of transfection was determined by anti-HA and anti-ALK1 Western blotting at 72 hours after transfection (B). Apoptosis was assessed at the same time point by Annexin V analysis in control cells (C), cells transfected with HA-PSF alone (D), or together with kinase-dead NPM/ALK (E) (HA-PSF + N/A-KD), or wild-type NPM/ALK (F) (HA-PSF + NA). C-F, live cell percentages are shown in the bottom left panels; apoptotic cell percentages in the bottom right panels, and dead cell percentages in the top right panels.
PSF phosphorylation alters its RNA binding and transcriptional repression activities.
To further verify if NPM/ALK-mediated phosphorylation of PSF alters its biological functions, we studied 2 known activities of PSF: its RNA-binding ability60,61 and its transcriptional repressor activity of GAGE6.62 To study the first activity, the binding of purified PSF to a labeled, specific oligo RNA60 was evaluated. As shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), the RNA probe specifically recognized PSF as evidenced by the formation of a band; this binding was increased by the NMP/ALK-mediated phosphorylation of PSF (lane 2 versus 4). To evaluate the transcriptional activity of PSF on GAGE6, 293T cells were transfected with PSF and/or NPM/ALK, and the expression of GAGE6 assessed by real-time polymerase chain reaction (PCR). The results (Figure S2) show that NPM/ALK induced a substantial increase in GAGE6 expression levels in PSF-transfected cells that were not observed or greatly reduced when PSFY293F and NPM/ALK were transfected.
Discussion
NPM/ALK induces cellular transformation by constitutive activation of multiple antiapoptotic and proliferative signaling pathways through interactions with components of signaling cascades.5,29 NPM/ALK also associates with the chaperones HSP90 and HSP70 that regulate its degradation,45 and with NPM that localizes NPM/ALK in the nucleus.14 Hence, NPM/ALK ligands play an important role in mediating not only downstream signaling, but also impact on its subcellular localization. In this study, we identified novel NPM/ALK ligands, including 5 nuclear RNA/DNA-binding proteins: PSF, p54nrb, FUS/TLS, EWS, and nucleolin. A study by Crockett et al employing a similar proteomics approach to identify NPM/ALK-interacting partners identified a number of cytoplasmic signaling proteins, including adaptor molecules, kinases, and phosphatases.63 The RNA/DNA-binding proteins reported here were not identified, most likely due to differences in experimental conditions. However, a more recent publication identified an RNA-binding protein associated to NPM/ALK.64 The association of PSF with NPM/ALK was confirmed in ALCL-derived cell lines and patient samples, and required an active ALK kinase domain. PSF also bound to GCN4/ALK, suggesting that the interaction may be mediated via the ALK portion of the fusion protein. Therefore, PSF binding may be a common feature of variant ALK fusion proteins.
PSF is a multifunctional nuclear factor that has been implicated in diverse reactions in the nucleus.65 It is normally associated with subnuclear structures known as speckles, and with the nuclear membrane and matrix.57,58,66 PSF consists of an N-terminal proline/glutamine-rich domain that mediates interactions with proteins and DNA, 2 RNA-recognition motifs (RRMs) involved in interactions with proteins and RNA, and a C-terminal region containing 2 nuclear localization signals (NLSs).50 PSF forms multiprotein complexes that are involved in pre-mRNA splicing,50,60,67 gene transcription,32,34,68,69 DNA repair,70 DNA recombination,71,72 and cytoplasmic mRNA stability.73 PSF is most commonly associated with the closely related p54nrb nuclear factor,52 and has also been reported to interact with EWS and TLS/FUS.74 Since these proteins were also identified as novel NPM/ALK ligands, it is possible that NPM/ALK interacts with multiprotein complexes containing PSF and these other RNA/DNA-binding proteins.
We also showed that PSF is a substrate of ALK in vitro and is tyrosine-phosphorylated in NPM/ALK-expressing cell lines and in primary ALCL lymph node tissue. Mapping of the phosphorylation sites in PSF identified Tyr293 as the phosphorylated residue. Interestingly, the Y293F PSF mutant did not coimmunoprecipitate with NPM/ALK, which suggests that PSF phosphorylation may stabilize the interaction with NPM/ALK, or with another adaptor protein present in the NPM/ALK complex. The effect of tyrosine phosphorylation on PSF function has not been described previously, although both PSF and p54nrb have been identified as tyrosine-phosphorylated proteins associating with the nuclear envelope in neuroblastoma cells.75 By contrast, serine/threonine phosphorylation of PSF, p54nrb, TLS/FUS, and the serine-arginine (SR) family of splicing factors represents a mechanism through which the multiple function(s) of these RNA/DNA-binding proteins are regulated.71,74,76–80 For example, PKC-mediated phosphorylation of PSF, p54nrb, and TLS/FUS regulates RNA binding/processing and DNA recombination reactions.71,74,76–78 In light of the regulation of PSF activity by SR phosphorylation, it is possible that NPM/ALK may alter PSF function by tyrosine phosphorylation. The experiments performed here suggest that tyrosine phosphorylation of PSF can increase its RNA binding and inhibit the transcriptional repressor function of PSF.
PSF function may also be affected by its delocalisation to the cytoplasm in cells expressing active ALK fusion proteins, including NPM/ALK+ ALCL cell lines. Since kinase-dead NPM/ALK did not cause delocalization of PSF, it is conceivable that delocalization is dependent on tyrosine phosphorylation, possibly by stabilizing the PSF-NPM/ALK interaction. This hypothesis is supported by the finding that the mutant Y293F PSF was not phosphorylated, did not associate with NPM/ALK, and was not delocalized in NPM/ALK-expressing cells. The finding that both nuclear and cytoplasmic PSF fractions are tyrosine-phosphorylated in NPM/ALK+ cells is compatible with the subcellular localization of NPM/ALK inside cells.
The delocalization of PSF to the cytoplasm may alter its nuclear-associated functions. The fact that NPM/ALK-mediated phosphorylation of PSF increases its RNA binding activity (Figure S1), and that the combined transfection of NPM/ALK and PSF leads to an increase in GAGE6 transcription (Figure S2), are compatible with this hypothesis, which however will require further confirmatory experiments.
The modulation of other nuclear proteins via relocalization to the cytoplasm has been reported. For example, the oncogenic fusion protein PSF-TFE3, identified in papillary renal cell carcinoma,81 causes endogenous TFE3 and p53 to relocalize into the cytoplasm, thereby stimulating their degradation and inactivation.82 Another example is offered by the cytoplasmic relocation of NPM caused by C-terminal mutations, which are currently considered the primary leukemogenic event that disrupts normal NPM function in a cohort of patients with acute myeloid leukemia.83
From these data, it could be hypothesized that NPM/ALK-mediated phosphorylation of PSF may impact on the expression of factors regulating cell growth and apoptosis. Indeed, other ALK-associated factors such as NIPA protect NPM/ALK-expressing BaF3 cells from apoptosis through a mechanism requiring NPM/ALK activity and NIPA phosphorylation.27 Interestingly, overexpression of PSF, but not of Y293F PSF, specifically inhibited proliferation, clonogenic potential, and induced apoptosis in NPM/ALK+ cells. The interpretation of these results requires caution. The data showing that Y293F PSF did not inhibit growth could be interpreted in 2 ways: that phosphorylation of overexpressed PSF by NPM/ALK is necessary for such effects, or that the mutation of residue 293, which is located very close to the RRM1 domain (residues 297-369), affects other functions of PSF. The mechanisms by which overexpression of PSF leads to growth inhibition remain to be established and will be the subject of further studies.
In conclusion, we report here that NPM/ALK associates with and phosphorylates several RNA/DNA-binding proteins, in particular the multifunctional nuclear factor PSF. The data also show that PSF delocalizes to the cytoplasm in cells containing constitutively active forms of ALK through a mechanism that involves NPM/ALK-dependent phosphorylation of Tyr293 of PSF. Data were also provided showing that forced PSF expression impairs cell growth, induces apoptosis, and decreases clonogenic potential of NPM/ALK-expressing cells. Therefore, PSF might play a role in NPM/ALK-mediated lymphomagenesis through mechanisms that require further investigation. Whether PSF could be a target for other oncogenes also remains to be determined. Such studies should lead to a greater understanding of the mechanisms of ALK-mediated transformation.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr S. W. Morris (St Jude Research Hospital, Memphis, TN) for providing the ALK11 polyclonal antibody, pcDNA3-NA, and pcDNA3-NA-K210R plasmids; Dr P. Collini (Pathology Department, NCI Milan, Italy) for providing ALCL patient samples; and Dr Turturro (LSUHSC/Feist-Weiller Cancer Center, Shreveport, LA) for providing JB6 cell line. The authors are especially grateful to Lorenzo Bertola, Edoardo Marchesi, and Elvezia Bonacina (NCI Milan, Italy) for technical assistance, Barbara Cimbro for the preparation GST-ALK, and Valentina Belloni (University of Milano-Bicocca, Italy).
This work was supported in part by the Italian Association for Cancer Research (AIRC); Min. San. Ricerca Finalizzata (2003); Centro Nazionale Ricerche (CNR), Ministero Universita'E Ricerca-CO Fińanziamento (MIUR-COFIN), and Progetti di Ricerca di Interesse Nazionale (PRIN) programs (2004); EU (Prokinase network, no. 503467); Canadian Institute of Health Research (CIHR); Canadian Fund for Innovation (CFI); National Cancer Institute-Canada (NCI-C); National Institutes of Health (NIH) grants CA92318 (P.W.T) and CA095512 (D.P.); and by the Leukemia Research Fund.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.G. designed research, performed research, and wrote the paper; R.H.G. wrote the paper; S.R. performed research; P.S. performed research; C.C. performed research; A.B. performed research; P.W.T. contributed vital material and analyzed data; C.J.T. performed research; C.-J.H. contributed vital material; E.C. contributed vital material; K.P. contributed vital material; M.P. performed research; R.G.P. analyzed data; H.R. designed research; A.V. contributed vital material; A.D.-D. performed research; O.M. performed research; D.P. designed research, analyzed data, and wrote the paper; and C.G.-P. designed research, supervised work, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Annamaria Galietta, Department of Clinical Medicine, University of Milano-Bicocca, Via Cadore 48, Monza 20052, Italy; e-mail: galietta_am@yahoo.it.
References
- 1.Stein H, Foss HD, Durkop H, et al. CD30(+) anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood. 2000;96:3681–3695. [PubMed] [Google Scholar]
- 2.Morris SW, Xue L, Ma Z, Kinney MC. Alk+ CD30+ lymphomas: a distinct molecular genetic subtype of non-Hodgkin's lymphoma. Br J Haematol. 2001;113:275–295. doi: 10.1046/j.1365-2141.2001.02574.x. [DOI] [PubMed] [Google Scholar]
- 3.Morris SW, Naeve C, Mathew P, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 4.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 5.Pulford K, Morris SW, Turturro F. Anaplastic lymphoma kinase proteins in growth control and cancer. J Cell Physiol. 2004;199:330–358. doi: 10.1002/jcp.10472. [DOI] [PubMed] [Google Scholar]
- 6.Shiota M, Fujimoto J, Semba T, Satoh H, Yamamoto T, Mori S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- 7.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 8.Zatsepina OV, Rousselet A, Chan PK, Olson MO, Jordan EG, Bornens M. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112:455–466. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]
- 9.Okuda M, Horn HF, Tarapore P, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 10.Dumbar TS, Gentry GA, Olson MO. Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry. 1989;28:9495–9501. doi: 10.1021/bi00450a037. [DOI] [PubMed] [Google Scholar]
- 11.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 12.Shu X, Fry AM, Tulloch B, et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet. 2005;14:1183–1197. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- 13.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 14.Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Z, Cools J, Marynen P, et al. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- 16.Touriol C, Greenland C, Lamant L, et al. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like). Blood. 2000;95:3204–3207. [PubMed] [Google Scholar]
- 17.Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol Cell Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto J, Shiota M, Iwahara T, et al. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc Natl Acad Sci U S A. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiarle R, Gong JZ, Guasparri I, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 20.Miething C, Grundler R, Fend F, et al. The oncogenic fusion protein nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) induces two distinct malignant phenotypes in a murine retroviral transplantation model. Oncogene. 2003;22:4642–4647. doi: 10.1038/sj.onc.1206575. [DOI] [PubMed] [Google Scholar]
- 21.Kuefer MU, Look AT, Pulford K, et al. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 22.Jager R, Hahne J, Jacob A, et al. Mice transgenic for NPM-ALK develop non-Hodgkin lymphomas. Anticancer Res. 2005;25:3191–3196. [PubMed] [Google Scholar]
- 23.Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319–4327. [PubMed] [Google Scholar]
- 24.Zamo A, Chiarle R, Piva R, et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21:1038–1047. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- 25.Ruchatz H, Coluccia AM, Stano P, Marchesi E, Gambacorti-Passerini C. Constitutive activation of Jak2 contributes to proliferation and resistance to apoptosis in NPM/ALK-transformed cells. Exp Hematol. 2003;31:309–315. doi: 10.1016/s0301-472x(03)00007-9. [DOI] [PubMed] [Google Scholar]
- 26.Nieborowska-Skorska M, Slupianek A, Xue L, et al. Role of signal transducer and activator of transcription 5 in nucleophosmin/ anaplastic lymphoma kinase-mediated malignant transformation of lymphoid cells. Cancer Res. 2001;61:6517–6523. [PubMed] [Google Scholar]
- 27.Ouyang T, Bai RY, Bassermann F, et al. Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA). J Biol Chem. 2003;278:30028–30036. doi: 10.1074/jbc.M300883200. [DOI] [PubMed] [Google Scholar]
- 28.Ambrogio C, Voena C, Manazza AD, et al. p130Cas mediates the transforming properties of the anaplastic lymphoma kinase. Blood. 2005;106:3907–3916. doi: 10.1182/blood-2005-03-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coluccia AM, Gunby RH, Tartari CJ, Scapozza L, Gambacorti-Passerini C, Passoni L. Anaplastic lymphoma kinase and its signalling molecules as novel targets in lymphoma therapy. Expert Opin Ther Targets. 2005;9:515–532. doi: 10.1517/14728222.9.3.515. [DOI] [PubMed] [Google Scholar]
- 30.Slupianek A, Nieborowska-Skorska M, Hoser G, et al. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 2001;61:2194–2199. [PubMed] [Google Scholar]
- 31.Chiarle R, Simmons WJ, Cai H, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 32.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulford K, Lamant L, Morris SW, et al. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404. [PubMed] [Google Scholar]
- 34.Emili A, Shales M, McCracken S, et al. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunby RH, Tartari CJ, Porchia F, Donella-Deana A, Scapozza L, Gambacorti-Passerini C. An enzyme-linked immunosorbent assay to screen for inhibitors of the oncogenic anaplastic lymphoma kinase. Haematologica. 2005;90:988–990. [PubMed] [Google Scholar]
- 36.le Coutre P, Mologni L, Cleris L, et al. In vivo eradication of human BCR/ABL-positive leukemia cells with an ABL kinase inhibitor. J Natl Cancer Inst. 1999;91:163–168. doi: 10.1093/jnci/91.2.163. [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 39.Donella-Deana A, Marin O, Cesaro L, et al. Unique substrate specificity of anaplastic lymphoma kinase (ALK): development of phosphoacceptor peptides for the assay of ALK activity. Biochemistry. 2005;44:8533–8542. doi: 10.1021/bi0472954. [DOI] [PubMed] [Google Scholar]
- 40.Settleman J, Narasimhan V, Foster LC, Weinberg RA. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell. 1992;69:539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- 41.Neviani P, Santhanam R, Trotta R, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8:355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU. 1 correlates with negative regulation of the c-myb promoter. Blood. 1997;90:1828–1839. [PubMed] [Google Scholar]
- 43.Hickey E, Brandon SE, Smale G, Lloyd D, Weber LA. Sequence and regulation of a gene encoding a human 89-kilodalton heat shock protein. Mol Cell Biol. 1989;9:2615–2626. doi: 10.1128/mcb.9.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rebbe NF, Hickman WS, Ley TJ, Stafford DW, Hickman S. Nucleotide sequence and regulation of a human 90-kDa heat shock protein gene. J Biol Chem. 1989;264:15006–15011. [PubMed] [Google Scholar]
- 45.Bonvini P, Gastaldi T, Falini B, Rosolen A. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK(+) CD30(+) lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 2002;62:1559–1566. [PubMed] [Google Scholar]
- 46.Yamagishi N, Ishihara K, Hatayama T. Hsp105alpha suppresses Hsc70 chaperone activity by inhibiting Hsc70 ATPase activity. J Biol Chem. 2004;279:41727–41733. doi: 10.1074/jbc.M407947200. [DOI] [PubMed] [Google Scholar]
- 47.Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 48.Domanico SZ, DeNagel DC, Dahlseid JN, Green JM, Pierce SK. Cloning of the gene encoding peptide-binding protein 74 shows that it is a new member of the heat shock protein 70 family. Mol Cell Biol. 1993;13:3598–3610. doi: 10.1128/mcb.13.6.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 50.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 51.Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 52.Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohno T, Ouchida M, Lee L, Gatalica Z, Rao VN, Reddy ES. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 54.O'Donoghue SI, Junius FK, King GF. Determination of the structure of symmetric coiled-coil proteins from NMR data: application of the leucine zipper proteins Jun and GCN4. Protein Eng. 1993;6:557–564. doi: 10.1093/protein/6.6.557. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 56.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 57.Meissner M, Dechat T, Gerner C, Grimm R, Foisner R, Sauermann G. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J Cell Biochem. 2000;76:559–566. [PubMed] [Google Scholar]
- 58.Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 59.Gu Y, Sarnecki C, Aldape RA, Livingston DJ, Su MS. Cleavage of poly(ADP-ribose) polymerase by interleukin-1 beta converting enzyme and its homologs TX and Nedd-2. J Biol Chem. 1995;270:18715–18718. doi: 10.1074/jbc.270.32.18715. [DOI] [PubMed] [Google Scholar]
- 60.Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berglund JA, Fleming ML, Rosbash M. The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA. 1998;4:998–1006. doi: 10.1017/s1355838298980499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crockett DK, Lin Z, Elenitoba-Johnson KS, Lim MS. Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene. 2004;23:2617–2629. doi: 10.1038/sj.onc.1207398. [DOI] [PubMed] [Google Scholar]
- 64.Fawal M, Armstrong F, Ollier S, et al. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 65.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 66.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8:1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 69.Urban RJ, Bodenburg Y. PTB-associated splicing factor regulates growth factor-stimulated gene expression in mammalian cells. Am J Physiol Endocrinol Metab. 2002;283:E794–798. doi: 10.1152/ajpendo.00174.2002. [DOI] [PubMed] [Google Scholar]
- 70.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 71.Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Straub T, Grue P, Uhse A, et al. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- 73.Zolotukhin AS, Michalowski D, Bear J, et al. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deloulme JC, Prichard L, Delattre O, Storm DR. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J Biol Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- 75.Otto H, Dreger M, Bengtsson L, Hucho F. Identification of tyrosine-phosphorylated proteins associated with the nuclear envelope. Eur J Biochem. 2001;268:420–428. doi: 10.1046/j.1432-1033.2001.01901.x. [DOI] [PubMed] [Google Scholar]
- 76.Rosenberger U, Lehmann I, Weise C, Franke P, Hucho F, Buchner K. Identification of PSF as a protein kinase Calpha-binding protein in the cell nucleus. J Cell Biochem. 2002;86:394–402. doi: 10.1002/jcb.10233. [DOI] [PubMed] [Google Scholar]
- 77.Perrotti D, Bonatti S, Trotta R, et al. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perrotti D, Iervolino A, Cesi V, et al. BCR-ABL prevents c-jun-mediated and proteasome-dependent FUS (TLS) proteolysis through a protein kinase CbetaII-dependent pathway. Mol Cell Biol. 2000;20:6159–6169. doi: 10.1128/mcb.20.16.6159-6169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang HY, Fu XD. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–298. [PubMed] [Google Scholar]
- 81.Clark J, Lu YJ, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 82.Mathur M, Das S, Samuels HH. PSF-TFE3 oncoprotein in papillary renal cell carcinoma inactivates TFE3 and p53 through cytoplasmic sequestration. Oncogene. 2003;22:5031–5044. doi: 10.1038/sj.onc.1206643. [DOI] [PubMed] [Google Scholar]
- 83.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.