Abstract
Human N-acetyltransferase 2 (NAT2) genetic polymorphisms may modify drug efficacy and toxicity and individual cancer susceptibility from carcinogen exposure. A411T (L137F) and G364A (D122N) are two single nucleotide polymorphisms (SNPs) that coexist with other SNPs in human NAT2 alleles NAT2*5I and NAT2*12D, respectively. Cloning and expression in COS-1 cells showed that both A411T and G364A reduced NAT2 immunoreactive protein to an undetectable level without causing changes in mRNA level. Missense mutants displayed different effects on sulfamethazine N-acetylation activity for both L137 (wild-type: 70.2±5.2; L137F: 1.34±0.03; L137W: non-detectable; L137I: 34.2±2.0; L137G: 0.52±0.04 nmol/min/mg) and D122 (wildtype: 70.2±5.2; D122R: non-detectable; D122Q: non-detectable; D122E: 1.72±0.24 nmol/min/mg). To further test our hypothesis that A411T (L137F) and G364A (D122N) accelerate protein degradation, various NAT2 alleles were cloned and expressed in E. coli, which does not possess the ubiquitin-mediated degradation pathway. In contrast to the expression in mammalian cells, recombinant NAT2 possessing either of these two SNPs showed no reduction in immunoreactive NAT2 level when expressed in E. coli. These findings suggest that both A411T (L137F) and G364A (D122N) enhance NAT2 degradation, resulting in reduced NAT2 protein and catalytic activity for NAT2 5I and NAT2 12D.
Keywords: N-acetyltransferase-2, Single nucleotide polymorphism, protein degradation, slow acetylator phenotype
INTRODUCTION
N-acetyltransferase 2 (NAT2) [EC.2.3.1.5] is an important phase II metabolic enzyme for many drugs and xenobiotics [1]. NAT2 detoxifies aromatic amines such as sulfamethazine (SMZ) and 4-aminobiphenyl by N-acetylation but also bioactivates aromatic and heterocyclic amines to DNA-reactive mutagenic compounds via O-acetylation of their N-hydroxylated metabolites [2].
The gene encoding this enzyme, NAT2, is highly polymorphic in humans [3]. To date sixteen single nucleotide polymorphisms (SNPs) have been identified in the human NAT2 protein coding region, forming thirty-six human NAT2 alleles by different combinations of these SNPs (http://www.louisville.edu/medschool/pharmacology/NAT.html). The reference NAT2 allele (named NAT2*4) encodes an NAT2 protein with high N-acetylation activity and confers rapid acetylator phenotype. Studies in bacteria [4], yeast [5], and COS-1 cells [6] have shown that some nonsynonymous SNPs such as G191A (R64Q), T341C (I114T) and G590A (R197Q) lead to decreased NAT2 catalytic activity. Therefore, genetic polymorphism of NAT2 accounts for the variation of NAT2 phenotype, and thus individual responses to drugs and carcinogens that are NAT2 substrates. NAT2 phenotype may modify individual cancer risks upon carcinogen exposure. It has been found that slow NAT2 acetylators have higher risk of cigarette-related urinary bladder cancer [7]. In some molecular epidemiological studies, NAT2 polymorphism has been reported to modify individual risk for various cancers such as colon [8], breast [9-11] and prostate [12, 13] upon carcinogen exposure. Functional characterization of the effects of each SNP on NAT2 expression will enhance understanding of the genotype-phenotype relationships necessary to provide a better foundation for molecular epidemiological studies.
Two human NAT2 alleles, NAT2*5I and NAT2*12D, were identified recently. In NAT2*5I (genbank#230252), a new SNP A411T (L137F) was found coexisting with SNPs T341C (I114T), C481T (silent) and A803G (K268R). The other allele NAT2*12D (genbank#230251) consists of a new SNP G364A (D122N) together with A803G (K268R). Thus, we undertook a study to characterize these new SNPs and NAT2 alleles to further our understanding of the genotype/phenotype relationship in human populations.
MATERIALS AND METHODS
Plasmid Construction
Human NAT2 mammalian expression plasmids were constructed as described previously [14]. In brief, NAT2*4, NAT2*5I and NAT2*12D protein coding regions were each amplified from genomic DNA by polymerase chain reaction (PCR) with sense primer: 5'-actgatgctagcatggacattgaagcatattttgaaag-3', and anti-sense primer: 5'-actgatctcgagctaaatagtaagggatccatcaccag-3'. The PCR products were digested with restriction enzymes Nhe I and Xho I at 37 °C for 2 h, and then ligated into like-treated mammalian expression vector pcDNA5/FRT (Invitrogen, San Diego, CA) between a cytomegalovirus (CMV) promoter and a bovine growth hormone (BGH) polyadenylation sequence. The ligation products were then transformed into One Shot TOP 10 chemically competent E. coli (Invitrogen). For each of the three NAT2 alleles, eight clones were randomly picked and correct sequences of the insert were further confirmed by automated DNA sequencing.
Individual SNPs (A411T, G364A, T341C, C481T, A803G) and site-directed mutations (L137W, L137I, L137G, D122E, D122Q) were introduced into the NAT2*4 plasmid by a PCR-based method previously described [21]. The sequences of the mutagenesis primers are listed in Table 1.
Table 1.
Primers used for site-directed mutagenesis (mismatches of the oligonucleotides used for mutagenesis are underlined).
| NAME | SEQUENCE | Amino Acid Change |
|---|---|---|
| 411T-for | 5'-GTGGCAGCCTCTAGAATTTATTTCTGGGAAGGATCAG-3' | L137F |
| 411T-rev | 5'-CTGATCCTTCCCAGAAATAAATTCTAGAGGCTGCCAC-3' | |
| 341C-for | 5'-CTCCTGCAGGTGACCACTGACGGCAGGAATTAC-3' | I114T |
| 341C-rev | 5'-GTAATTCCTGCCGTCAGTGGTCACCTGCAGGAG-3' | |
| 481T-for | 5'-GAGAGAGGAATCTGGTACTTGGACCAAATCAGGAGAGAG-3' | None |
| 481T-rev | 5'-CTCTCTCCTGATTTGGTCCAAGTACCAGATTCCTCTCTC-3' | |
| 803G-for | 5'-GAGGTTGAAGAAGTGCTGAGAAATATATTTAAGATTTCCTTGGGG-3' | K268R |
| 803G-rev | 5'-CCCCAAGGAAATCTTAAATATATTTCTCAGCACTTCTTCAACCTC-3' | |
| L137W-for | 5'-GTGGCAGCCTCTAGAATGGATTTCTGGGAAGGATCAGC-3' | L137W |
| L137W-rev | 5'-GCTGATCCTTCCCAGAAATCCATTCTAGAGGCTGCCAC-3' | |
| L137 I-for | 5'-GTGGCAGCCTCTAGAAATAATTTCTGGGAAGGATCA-3' | L137I |
| L137I-rev | 5'-TGATCCTTCCCAGAAATTATTTCTAGAGGCTGCCAC-3' | |
| L137G-for | 5'-TGTGGCAGCCTCTAGAAGGAATTTCTGGGAAGGATCAG-3' | L137G |
| L137G-rev | 5'-CTGATCCTTCCCAGAAATTCCTTCTAGAGGCTGCCACA-3' | |
| 364A-for | 5'-GCAGGAATTACATTGTCAATGCTGGGTCTGGAAGC-3' | D122N |
| 364A-rev | 5'-GCTTCCAGACCCAGCATTGACAATGTAATTCCTGC-3' | |
| D122E-for | 5'-GGAATTACATTGTCGAGGCTGGGTCTGGAAGC-3' | D122E |
| D122E-rev | 5'-GCTTCCAGACCCAGCCTCGACAATGTAATTCC- 3' | |
| D122Q-for | 5'-CAGGAATTACATTGTCCAGGCTGGGTCTGGAAGC-3' | D122Q |
| D122Q-rev | 5'-GCTTCCAGACCCAGCCTGGACAATGTAATTCCTG-3' |
All bacterial expression plasmids were made by site-directed mutagenesis based on a NAT2*4 containing PKK322 plasmid [15] using the corresponding primers listed in Table 1. The plasmids were amplified in NovaBlue competent cells (Novagen, Madison, WI) and confirmed by automated DNA sequencing.
Cell Culture, Transfection, and Lysate Preparation
COS-1 cells (#CRL-1650, ATCC) were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum and 4 mM L-glutamine. Cells (1.2 × 106) were plated in 100 mm culture dishes for 16 h prior to transfection. Cells were transfected with 3.6 μg NAT2 plasmid DNA together with 0.4 μg pCMV-sport-βgal plasmid (Invitrogen) using Lipofectamine and Plus reagent (Invitrogen). After 48 h, transfected cells were washed with ice-cold phosphate buffered saline (PBS), trypsinized and collected in a conical tube by centrifugation at 600 × g for 2 min at 4 °C. In proteasome inhibitor assays, 5 μM MG-132 (Sigma-Aldrich, Atlanta, GA) or lactacystin (Boston Biochem, Cambridge, MA) was added to the culture medium 24 h post-transfection and cells were harvested after 0, 1, 2, and 4 h of treatment. Cell pellets were either subjected to RNA isolation (described below) or suspended in homogenizing buffer (20 mM sodium phosphate buffer, pH 7.4; 1 mM EDTA, 1 mM freshly prepared dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 1 μg/ml aprotinin) and stored at −80 °C. Lysates were made by three cycles of freeze-thawing. Following centrifugation at 15,000 × g for 10 min at 4 °C, the supernatants were isolated for immunoblotting and catalytic activity measurements.
Measurement of NAT2 Catalytic Activity
Sulfamethazine (SMZ) N-acetylation activity was measured to determine the catalytic activity, kinetic parameters and thermostability of variant NAT2 proteins with a high performance liquid chromatography method described previously [16]. Briefly, cell lysate was incubated with 1 mM acetyl coenzyme A (Ac-CoA) and 500 μM SMZ at 37 °C for 10-20 min. To determine the apparent Km of SMZ for NAT2 4 and NAT2-411T, a fixed concentration of Ac-CoA (1 mM) and SMZ concentrations ranging from 50 μM to 5000 μM were used. The apparent Km of SMZ was calculated by linear regression of Eadie-Hofstee plots. Water was substituted for Ac-CoA as the negative control. SMZ and N-acetyl-SMZ were separated and detected by absorbance at 260 nm. Total protein in cell lysate was measured by the Bradford assay using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA). β-galactosidase activity was measured using the ortho-nitrophenyl- β-D-galactopyranoside method as previously described [14].
Measurement of NAT2 Protein
Cell lysates (20-50 μg of total protein in the supernatant) were subjected to SDS-PAGE on 12% Tris-Glycine gels (Cambrex, Walkersville, MD). Proteins were then transferred onto nitrocellulose membranes (Amersham, Arlington Heights, IL) at 100 V for 1 h. The blots were subsequently treated with blocking buffer (20 mM Tris-HCl, 500 mM NaCl, 0.5% Tween 20, 5% non fat dry milk) at room temperature for 1 h and then probed with primary antibody (1:2000 dilution) specific for human NAT2 [14]. The immunoblots were visualized with the Pierce Pico Western chemiluminescent system (Pierce Biotech, Rockford, IL). The blots were stripped and re-probed with anti-α-tubulin IgG (Sigma-Aldrich) as the loading control.
Measurement of NAT2 mRNA
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA) from transfected COS-1 cells. RNA samples were treated with DNA-free DNase Treatment and Removal Reagent (Ambion, Austin, TX). cDNA was synthesized from 1 μg total RNA with oligo (dT) primer using SuperScript III reverse transcriptase (Invitrogen). Reactions without reverse transcriptase were performed in parallel as a negative control for DNA contamination. Real time RT-PCR was performed as described previously [14]. Each sample was tested in duplicate with separate tube of African green monkey β-actin as the internal control. Samples obtained from three independent experiments were used to calculate the means and standard deviations.
NAT2 Protein Aggregation Assay
Transfected cells were suspended in homogenizing buffer and passed through a 30.5 gauge needle. The preparation of Triton X-100 soluble and insoluble fractions was performed according to the method described by Klucken et al. [17]. The suspension was supplemented with 1% (v/v) of Triton X-100 and incubated for 30 min on ice. After a 60 min centrifugation (15,000 × g at 4 °C), the supernatant was isolated from the pellet, which was subsequently redissolved in 2% SDS-containing lysis buffer and sonicated for 10 seconds. Both the supernatant and the redissolved pellet were subjected to NAT2 immunoassay as described above.
Recombinant NAT2 Expression in E. coli
Human NAT2 proteins were transformed and expressed in JM105 E. coli according to the method described previously [15] with minor modifications. In brief, competent JM105 cells were prepared by calcium chloride treatment followed by an overnight aging at 4°C. Ten ng plasmid DNA was used to transform 50 μl competent cells with 30 min incubation on ice followed by a 45 sec heat shock at 42 °C. The colonies were picked and grown in LB-Ampicillin media. When OD600 reached 0.6, one mM isopropyl-β-D-thiogalactopyranoside was added to induce protein expression. E. coli cells were incubated for an additional 3 h and then pelleted by centrifugation at 4 °C and lysed by sonication (30 sec × 7). The lysates were subjected to centrifugation at 15,000 × g for 20 min at 4 °C and the supernatants were isolated for NAT2 catalytic activity and immunoassays as described above.
Statistical Analysis
Differences in catalytic activities or mRNA levels among NAT2 allozymes were tested for significance by one way analysis of variance followed by Bonferroni tests for multiple comparisons.
RESULTS
Recombinant Expression of NAT2*4, NAT2*5I, NAT2*12D and NAT2 Alleles with Individual SNPs in COS-1 Cells
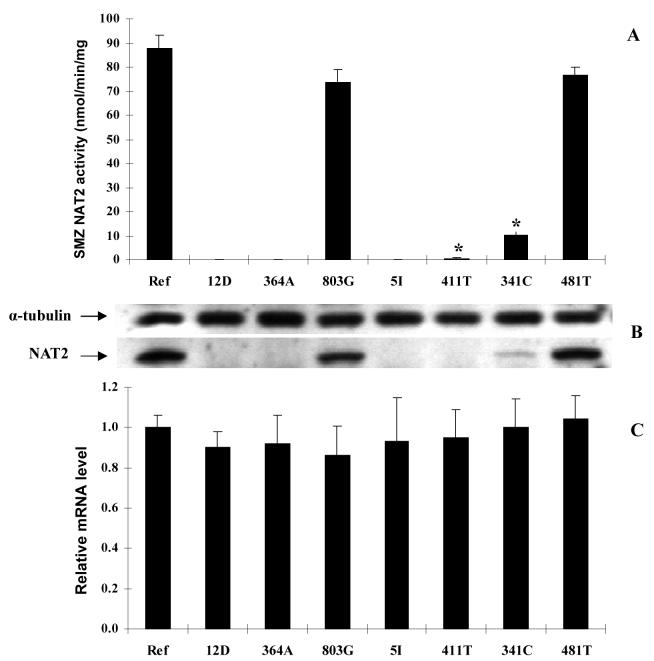
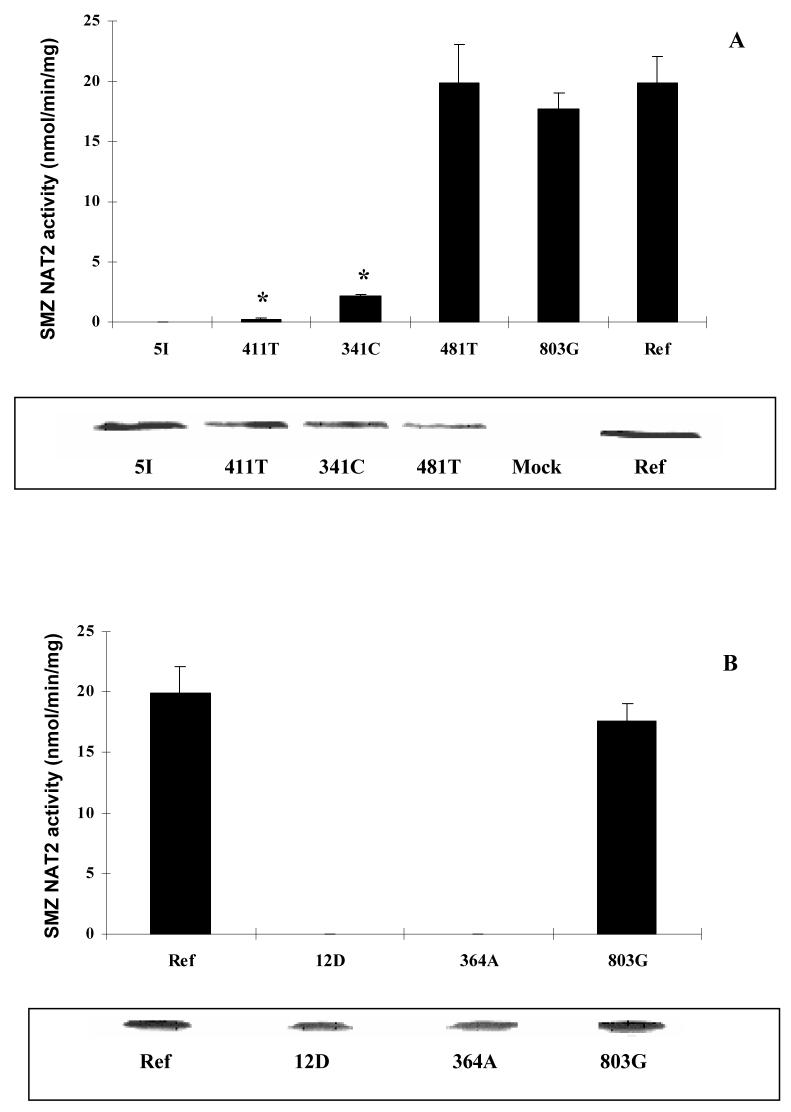
SMZ NAT2 activities in COS-1 cells expressing different NAT2 alleles were measured and normalized to β-galactosidase activities (Figure 1A). All transient transfections were performed in triplicate in three independent experiments. A pcDNA5/FRT plasmid without NAT2 insertion was used as the negative control and there was no SMZ NAT activity detected in cells transfected with this plasmid. Cell viability after transfection was above 95%, by Trypan Blue exclusion. Mammalian expression of NAT2*5I (possessing the combination of A411T, T341C, C481T and A803G) and NAT2*12D (with the combination of G364A and A803G) resulted in undetectable SMZ NAT2 activities in cell lysates. In order to differentiate the contribution of each individual SNP in these alleles, we constructed human NAT2 alleles possessing only one of the SNPs (NAT2*411T, NAT2*364A, NAT2*341C, NAT2*481T or NAT2*803G). We measured the NAT2 catalytic activity in COS-1 cells transfected with NAT2 alleles possessing each one of these SNPs and found that they had different effects on NAT2 activity. Compared to the NAT2 4 reference protein, C481T and A803G did not alter SMZ NAT2 catalytic activity, whereas all other SNPs reduced SMZ NAT2 activity to various degrees. NAT2 activity was reduced around 8-fold by T341C and 80-fold by A411T, suggesting that both T341C and A411T contribute to reduced catalytic activity of NAT2 5I. A411T did not significantly change the apparent Km of SMZ (NAT2-411T: 554 ± 134 μM, NAT2 4: 601 ± 141 μM, p>0.05). In NAT2*12D, the G364A SNP alone was sufficient to reduce SMZ NAT2 catalytic activity below the detection limit.
Figure 1.
Effect of individual SNPs and combinations (alleles) on catalytic activity, immunoreactive protein level, and the steady state mRNA level in COS-1 cells. (A): SMZ NAT2 activities in COS-1 cells expressing NAT2 alleles with different SNPs. Each bar represents mean ± SD of at least three independent transfections. NAT2 activities for NAT2 12D, NAT2 5I and NAT2-364A were below the limit of detection (0.3 nmol/min/mg). *Significantly lower than NAT2 4 (Ref), p<0.01. (B): Western blot of COS-1 cells expressing different NAT2 alleles as the order showed in (A). Cell lysates (20 μg total protein) were separated on 12% Tris-Glycine PAGE gel and then blotted on nitrocellulose membrane. The blots were probed with anti-NAT2 or anti-α-tubulin primary antibody. (C): Relative NAT2 mRNA levels in transfected COS-1 cells. The NAT2 mRNA was first normalized to internal β-actin and then expressed relative to the NAT2 mRNA level in NAT2*4 transfected COS-1 cells (defined as 1.0). Each bar shows the average and S.D. of the relative level based on three independent transfections. None of the mRNA levels differed significantly (p>0.05) from NAT2*4 (Ref).
NAT2 protein levels were determined by immunoblotting following a separation of total proteins in the COS-1 cell lysates and were normalized to α-tubulin level (Figure 1B). The human NAT2 protein (33 kD) was absent in cells transfected with the empty pcDNA5/FRT plasmid. The intensities of NAT2 immunoblots for each allozyme correlated with their catalytic activities. Neither C481T nor A803G reduced levels of immunoreactive NAT2 protein. T341C decreased NAT2 protein level in cell lysate. A411T and G364A, either alone or combined with other SNPs as in allele NAT2*5I and NAT2*12D, reduced the NAT2 protein below levels that could be detected by the western blot.
Steady state NAT2 mRNA levels were also measured in COS-1 cells expressing each of the human NAT2 alleles or alleles with individual SNPs. The CT (cycle threshold) values for NAT2 amplification in the reactions without reverse transcriptase (-RT control) were above 39, whereas those with reverse transcriptase had values around 21, indicating no significant plasmid DNA contamination. The result of real time RT-PCR showed that none of the SNPs, either alone or in combination, significantly decreased NAT2 mRNA level (p>0.05) (Figure 1C).
Effects of Leu137 on N-acetyltransferase 2 Expression in COS-1 Cells
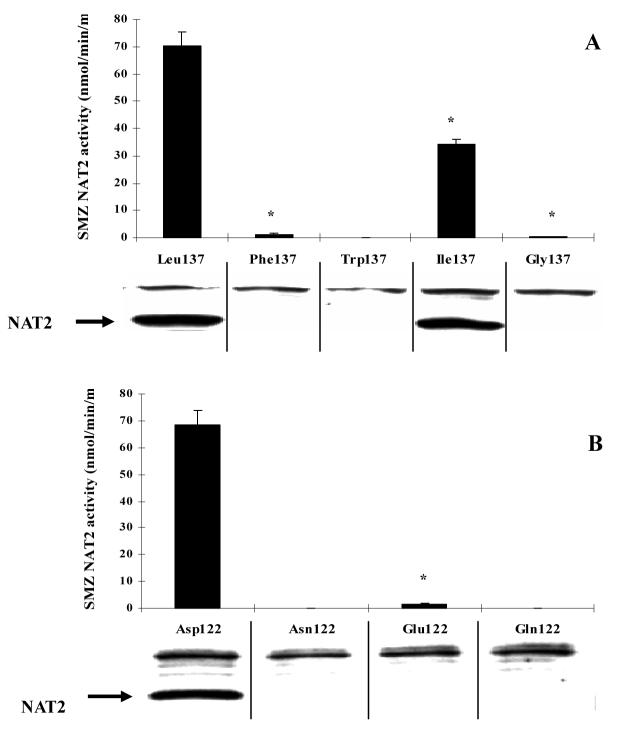
The A411T SNP caused the amino acid change L137F and resulted in a large decrease of enzyme activity. Site-directed mutagenesis was used to construct three NAT2 mutants by replacing Leu137 with tryptophan, isoleucine or glycine. SMZ NAT2 catalytic activity and protein of the mutant NAT2 alleles expressed in COS-1 cells are shown in Figure 2A. The L137I mutant reduced NAT2 immunoreactive protein and NAT2 catalytic activity to less than half of the reference NAT2 4. All other SNPs reduced immunoreactive NAT2 protein level below those detectable by western blot. While the L137G mutant still possessed a trace level of catalytic activity, the L137W mutant reduced catalytic activity below the detection level (0.3 nmol/min/mg).
Figure 2.
SMZ NAT2 catalytic activity and western blot of NAT2 allozymes differing only at the 137th amino acid (A) or the 122nd amino acid (B). Each bar represents mean ± SD of six independent transfections. NAT2 mutant with Phe137, Ile137, Gly137 or Glu122 had significantly (*: p<0.01) lower activity compared to NAT2 4. Catalytic activity of NAT2 with Trp137, Asn122 and with Gln122 were below the limit of detection (0.3 nmol/min/mg).
Effects of Asp122 on N-acetyltransferase 2 Expression in COS-1 Cells
Mutants replacing the Asp122 with glutamic acid or glutamine were constructed by site-directed mutagenesis. Like the D122N SNP, D122Q also reduced NAT2 catalytic activity and protein to undetectable levels. The D122E mutant reduced NAT2 catalytic activity about 40-fold, whereas the protein level was too low to be detected by western blot (Figure 2B).
Effects of SNPs on N-acetyltransferase 2 Aggregation and Thermostability in COS-1 Cells
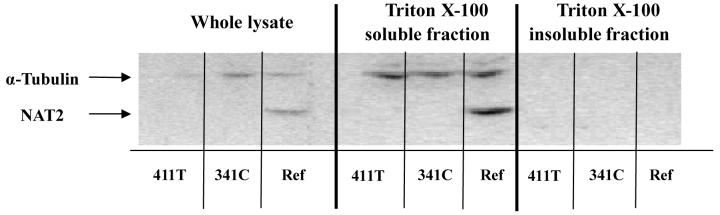
To determine whether the decreased NAT2 protein was related to protein aggregation caused by A411T (L137F), we measured the immunoreactive NAT2 protein in whole cell lysate, as well as in TritonX-100 soluble and insoluble fractions following recombinant expression of NAT2 4 and NAT2-411T (Figure 3). NAT2 4 had more immunoreactive protein than NAT2-411T in TritonX-100 soluble fractions, which is consistent with the result in Figure 1B. No NAT2 band was observed in the TritonX-100 insoluble fraction following recombinant expression of NAT2 4 or NAT2-411T, consistent with absence of protein aggregation caused by the A411T SNP in COS-1 cells.
Figure 3.
Western blot transfected cells were suspended in lysis buffer. Following one cycle of freeze-thawing, the suspension was supplemented with 1% (w/w) of Triton X-100 and incubated for 30 min on ice. After a 60 min centrifugation (15,000 × g at 4 °C), the supernatant was isolated from the pellet, which was subsequently redissolved in 2% SDS-containing lysis buffer and sonicated for 10 seconds. Both the supernatant and the redissolved pellet were subjected to western blot as described above. Ref: NAT2 4.
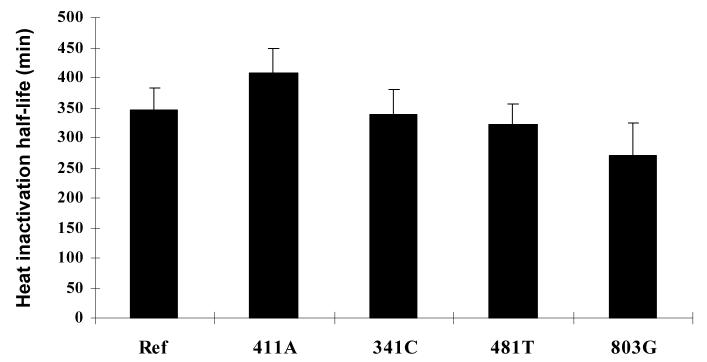
Thermostability of different NAT2 allozymes was measured in cell lysates of COS-1 cells transfected with human NAT2 alleles. As shown in Figure 4, the in vitro half-life of different NAT2 allozymes at 37 °C did not differ significantly (p>0.05). We were unable to measure the heat inactivation rate of NAT2-364A since the catalytic activity of this variant protein was below the detection level.
Figure 4.
Effect on NAT2 thermostability by different SNPs. Lysates were incubated at 37 °C for two hours. An aliquot was removed every 15 min from the incubation and assayed for SMZ NAT2 activity. The first order inactivation half-life was determined by linear regression. Three independent transfections were conducted to calculate the mean and standard deviation. None of the heat inactivation half-lives differed significantly (p>0.05) from NAT2 4 (Ref).
Recombinant Expression of NAT2*4, NAT2*5I, NAT2*12D and NAT2 Alleles with Individual SNPs in E. coli
Prokaryotic expression plasmids for human NAT2*4, NAT2*411T, NAT2*364A, NAT2*341C, NAT2*481T and NAT2*803G were constructed to study their expressions in E. coli. The bacterial lysates were subjected to NAT2 catalytic activity assay and western blot (Figure 5). No significant differences (p>0.05) in NAT2 protein level were observed for recombinant expression of any of the NAT2 allozymes, a finding strikingly different from what was observed in the mammalian cell expression system (Figure 1B). However, the catalytic activities of these NAT2 allozymes still varied similar to what we found in COS-1 cells. In bacterial lysates, NAT2-481T and NAT2-803G had catalytic activities comparable to the reference NAT2 4, whereas NAT2-411T and NAT2-341C had significantly lower (p<0.01) catalytic activities. The catalytic activity of NAT2-364A was below the limit of detection.
Figure 5.
Recombinant human NAT2 allozymes expressed in E. coli. NAT2 5I and individual SNPs are shown in top panel (A). NAT2 12D and individual SNPs are shown in bottom panel (B). NAT2-411T and NAT2-341C had catalytic activities significantly lower than NAT2 4 (Ref) (*: p<0.01). Catalytic activities of NAT2 5I, NAT2 12D and NAT2-364A were less than the limit of detection (0.3 nmol/min/mg). Western blots for each NAT2 allozyme are also shown underneath the respective bar graph. Ref: NAT2 4 expressed in E. coli; Mock: E. coli transformed with empty plasmid.
Effects of Proteosome Inhibitor Treatments on NAT2-411T Expression in COS-1 cells
Since the level of NAT2-411T protein was lower than our immunoblotting detection level, we were unable to measure the in vivo degradation rate of this protein by either cycloheximide treatment or [35S]-methionine incorporation. As an alternative, transfected cells were treated with MG-132 or lactacystin, two proteasome inhibitors, and harvested at different time points. As shown in Figure 6, treatment with either 5 μM of MG-132 or lactacystin increased NAT2-411T activity to levels about two-fold higher than in untreated cells. We also tried to measure the NAT2 protein level recovery by western blot but immunoreactive NAT2 levels remained below the western blot detection limit. Increasing MG-132 concentration or incubation time did not further enhance NAT2 catalytic activity or protein (data not shown).
Figure 6.
COS-1 cells were transfected with NAT2*411T (top) or NAT2*4 (bottom) 24 h before treatment with 5 μM MG-132 (open circles), 5 μM lactacystin (LAC, solid circles), or DMSO as a negative vehicle control (open squares). Cells were harvested at different time points after the treatment, and the cell lysates were tested for SMZ NAT2 catalytic activity. Each data point represents the mean ± standard error of three independent treatments.
DISCUSSION
We used COS-1 cells for functional characterization of the NAT2*5I and NAT2*12D alleles and to investigate the molecular mechanism of the A411T and G364A SNPs. COS-1 cells, derived from the kidney epithelial cells of Africa green monkey, are mammalian cells with undetectable basal SMZ NAT2 activity and have been used to express many human proteins including some NAT1 and NAT2 allozymes [18,19]. NAT2 expressed in COS-1 cells shows kinetic parameters similar to those of NAT2 in human liver [6, 20].
A411T (L137F)
NAT2-411T recombinantly expressed in E. coli had immunoreactive protein level similar to the reference NAT2 4 but had SMZ NAT2 catalytic activity about 1/8 of NAT2 4. However, when NAT2-411T was expressed in COS-1 cells, the activity was reduced to approximately 1/80 of NAT2 4, and the protein was undetectable by western blot. These results suggest that the decreased protein level caused by the A411T SNP was related to a mechanism existing in eukaryotic but not in prokaryotic cells, such as mRNA processing, post-translational modification and ubiquitin-mediated degradation. Our study found no change in mRNA level or protein thermostability, and no evidence of protein aggregation caused by the A411T SNP, suggesting that degradation is the mechanism for the reduction of NAT2 protein in mammalian cells. Although the NAT2-411T did not alter protein stability in vitro at 37°C, it is possible that NAT2 protein was misfolded and degraded inside the cell. Since NAT2-411T reduced NAT2 protein to levels lower than we could detect by immunoblotting, we were unable to measure the degradation rate of NAT2 protein. As an alternative, transfected cells were treated with proteasome inhibitors, and harvested at different time points. We found that both MG-132, a reversible proteasome inhibitor and lactacystin, an irreversible inhibitor were able to enhance NAT2 catalytic activity in NAT2-411T-expressing cells. Although we also tried to measure the NAT2 protein level by western blot, the NAT2 protein level after proteasome inhibitor treatment was still lower than the western blot detection limit.
Degradation has been reported as the mechanism of SNP-related phenotypic change in many proteins. Adding leupeptin did not increase NAT2-411T catalytic activity in transfected cells (data not shown), suggesting that the degradation is not lysosome-mediated. Enhanced proteasomal degradation has been reported as a principal mechanism for functional changes associated with certain polymorphisms in proteins such as thiopurine S methyltransferase [21], p53 [22], NAD(P)H: Quinone oxidoreductase 1 [23] and human N-acetyltransferase 1 [24]. Although this study provides no direct evidence for ubiquitination of the NAT2-411T variant protein, it is plausible that the ubiquitin/proteasome pathway may be a mechanism for the degradation of NAT2 since human N-acetyltransferase 1 is highly similar (>85% identity) to human NAT2. The ubiquitin-mediated proteasomal pathway is responsible for the clearance of cell cycle and transcription regulatory proteins as well as misfolded proteins [25]. In mammalian cells the specificity of degradation is determined mainly by different ubiquitin E3 ligases. However, in E. coli, the specificity of protein degradation depends on recognition by several types of proteases themselves, which means it is specific enough for some short-life transcriptional factors but not enough for misfolded enzymes. Characteristically the products of ubiquitin-mediated degradation are small pieces of peptides (around 3-5 amino acid long), and thus no degradation band should be seen in western blot, as we found with expression of NAT2-411T.
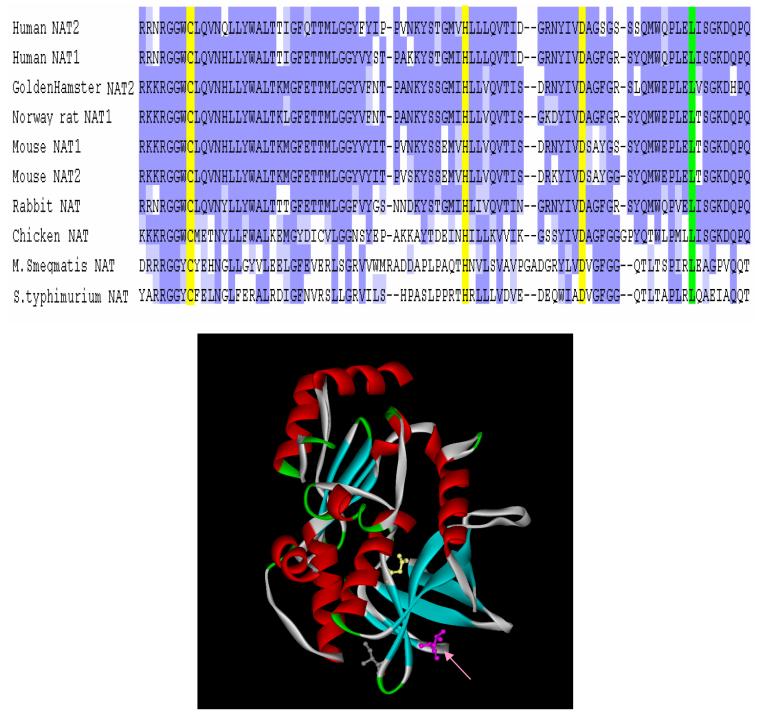
Multiple alignment by ClustalW [26] of N-acetyltransferases showed that L137 is conserved across species (Figure 7). Even though the similarity between prokaryotic NATs and eukaryotic NATs is not very high, especially at the C-terminal domain, several clusters of highly similar fragments, each 5-7 residues long, were found in the 1st and 2nd domain. Homology-based molecular modeling of human NAT2 (by MODELLER) showed that each of these short similar fragments corresponds to a β-sheet, suggesting conserved secondary structures in this region. Leu137 locates at the end of one conserved fragment, analogous to Thr114, whose replacement by isoleucine leads to enhanced protein degradation [14]. Site-directed mutagenesis showed that changing of Leu137 to Gly137, a β-sheet unfavorable residue according to the Chou-Fasman parameters [27], led to very low NAT2 protein level and catalytic activity. Replacement with a bulky/aromatic residue such as phenylalanine or tryptophan at this position also reduced catalytic activity by substantially reducing NAT2 protein level. However, substitution of Leu137 with isoleucine, a residue with similar structural and chemical characteristics to leucine, reduced NAT2 protein and catalytic activity only 2-fold. These results suggest that both proper size and chemical properties of the 137th amino acid are important for correct protein folding and protection from proteasomal degradation.
Figure 7.
Top: Multiple alignment of homologous N-acetyltransferases by ClustalW. This picture only shows a partial set of N-acetyltransferase sequences. Dark blue: amino acids identical among species; Light blue: amino acids somewhat similar among species; Yellow: putative catalytic triad: Cys68, His107 and Asn122 (numbered with human NAT2); Green: Leu137, which is conserved among all NATs listed. Bottom: Position of Leu137 (shown by arrow) on the human NAT2 model described in text. The picture is generated by Accelrys Discover Studio ViewPro.
G364A (D122N)
Structural studies on several prokaryotic N-acetyltransferase proteins have shown a common proposed catalytic triad composed of Cys69, His107 and Asp122, numbered according to the sequence of N-acetyltranferase from S. typhimurium [28-30]. These three amino acids are spatially superimposed in all the available crystal structures of prokaryotic N-acetyltransferases. In addition, homology-based molecular models of human N-acetyltransferase subunits also revealed a catalytic triad (Cys68-His107-Asp122 in human) with the same tertiary structure [31, 32].
Mutagenesis studies on prokaryotic NATs have shown that Asp122 in the catalytic triad is essential for N-acetyltransferase catalytic activity [33]. Specifically, the catalytic core aspartic acid of N-acetyltransferase from S. typhimurium and M. smegmatis, were mutated and the mutant proteins were expressed in E. coli. Replacement of the catalytic core aspartic acid with an asparagine caused undetectable N-acetyltransferase activity of both organisms. Similar to what we found with recombinant NAT2-364A, the D122N mutant of M. smegmatis N-acetyltransferase was expressed as a soluble protein in E. coli with N-acetyltransferase protein levels comparable to the wild-type protein. Asp122 is also structurally and catalytically essential for hamster NAT2. A recent study on the D122N mutant of hamster NAT2 showed that the mutant protein was catalytically inactive and was found only in the insoluble part of cell lysate when expressed in E. coli. The mutant protein expressed at similar levels to that of the wild type protein, following treatment to refold and dissolve the protein [34]. Our study in mammalian cells, however, found that the D122N substitution caused substantial reduction of NAT2 protein, analogous to L137F. These results suggest that even though Asp122 exists in the catalytic core while Leu137 does not, they both are critical to maintain the correct folding and thus structural stability of human NAT2.
In silico analysis of 59 N-acetyltransferase-like sequences from different species showed that the active site cysteine and histidine are fully conserved in these sequences, whereas the aspartic acid is not [29]. In this study, we showed that the D122N SNP was sufficient to reduce NAT2 catalytic activity and protein following mammalian expression of the human NAT2*12D allele. Mutation of Asp122 to a glutamine also caused undetectable NAT2 protein and catalytic activity, whereas mutation to glutamate resulted in low but detectable catalytic activity, suggesting the importance of the negative charge at this position. This finding is consistent with the observation that the Asp122 is highly but not fully conserved across species. In addition to the D122N SNP in the human NAT2*12D variant, Asp122 is replaced by Glu122 in N-acetyltransferases from four Bacillus species, but the catalytic activity of these enzymes are still unknown [33].
In summary, both NAT2*5I and NAT2*12D were characterized as slow acetylator alleles. NAT2*12D is the only allele in the human NAT2*12 cluster to be identified as a slow acetylator allele since it is unique among this cluster in possessing G364A (D122N). A411T (L137F) and G364A (D122N) reduced catalytic activity of human NAT2 in both mammalian cells and E. coli expression systems. However, loss of NAT2 protein was found for both variants in mammalian cells but not in E. coli. Degradation of misfolded NAT2 protein in mammalian cells is a likely mechanism for the functional effects of A411T (L137F) and G364A (D122N). Investigations on the structure and the catalytic mechanism of mammalian N-acetyltransferases are needed to better understand the NAT2 genotype-phenotype relationship.
Acknowledgments
Grant Support: This work was partially supported by USPHS grant CA-34627, a dissertation research award from the Susan G. Komen Breast Cancer Foundation (DISS0403147); and a grant from the Kentucky Lung Cancer Research Program.
REFERENCES
- 1.Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol Rev. 1985;37:25–79. [PubMed] [Google Scholar]
- 2.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 3.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 4.Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular genetics of human polymorphic N-acetyltransferase: enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric NAT2 allozymes. Hum Mol Genet. 1994;3:729–34. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- 5.Fretland AJ, Leff MA, Doll MA, Hein DW. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11:207–215. doi: 10.1097/00008571-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Blum M, Demierre A, Grant DM, Heim M, Meyer UA. Molecular mechanism of slow acetylation of drugs and carcinogens in humans. Proc Natl Acad Sci U S A. 1991;88:5237–5241. doi: 10.1073/pnas.88.12.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risch A, Wallace DM, Bathers S, Sim E. Slow N-acetylation genotype is a susceptibility factor in occupational and smoking related bladder cancer. Hum Mol Genet. 1995;4:231–236. doi: 10.1093/hmg/4.2.231. [DOI] [PubMed] [Google Scholar]
- 8.Le Marchand L, Hankin JH, Wilkens LR, Pierce LM, Franke A, Kolonel LN, Seifried A, Custer LJ, Chang W, Lum-Jones A, Donlon T. Combined effects of well-done red meat, smoking, and rapid N-acetyltransferase 2 and CYP1A2 phenotypes in increasing colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:1259–66. [PubMed] [Google Scholar]
- 9.Sillanpaa P, Hirvonen A, Kataja V, Eskelinen M, Kosma VM, Uusitupa M, Vainio H, Mitrunen K. NAT2 slow acetylator genotype as an important modifier of breast cancer risk. Int J Cancer. 2005;114:579–84. doi: 10.1002/ijc.20677. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–7. [PubMed] [Google Scholar]
- 11.van der Hel OL, Peeters PHM, Hein DW, Doll MA, Brobbee DE, Kromhout D, et al. NAT2 slow acetylation and GSTM1 null genotypes may increase postmenopausal breast cancer risk in long-term smoking women. Pharmacogenetics. 2003;13:399–407. doi: 10.1097/00008571-200307000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Hein DW, Leff MA, Ishibe N, Sinha R, Frazier HA, Doll MA, Xiao GH, Weinrich MC, Caporaso NE. Association of prostate cancer with rapid N-acetyltransferase 1 (NAT1*10) in combination with slow N-acetyltransferase 2 acetylator genotypes in a pilot case-control study. Environ Mol Mutagen. 2002;40:161–7. doi: 10.1002/em.10103. [DOI] [PubMed] [Google Scholar]
- 13.Costa S, Pinto D, Morais A, Vasconcelos A, Oliveira J, Lopes C, Medeiros R. Acetylation genotype and the genetic susceptibility to prostate cancer in a southern European population. Prostate. 2005;64:246–52. doi: 10.1002/pros.20241. [DOI] [PubMed] [Google Scholar]
- 14.Zang Y, Zhao S, Doll MA, States JC, Hein DW. The T341C (Ile114Thr) polymorphism of N-acetyltransferase 2 yields slow acetylator phenotype by enhanced protein degradation. Pharmacogenetics. 2004;14:717–23. doi: 10.1097/00008571-200411000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Hein DW, Doll MA, Rustan TD, Ferguson RJ. Metabolic activation of N-hydroxyarylamines and N-hydroxyarylamides by 16 recombinant human NAT2 allozymes: effects of 7 specific NAT2 nucleic acid substitutions. Cancer Res. 1995;55:3531–6. [PubMed] [Google Scholar]
- 16.Leff MA, Fretland AJ, Doll MA, Hein DW. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. J Biol Chem. 1999;274:34519–22. doi: 10.1074/jbc.274.49.34519. [DOI] [PubMed] [Google Scholar]
- 17.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces alpha-synuclein aggregation and toxicity. J Biol Chem. 2004;279:25497–502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 18.Butcher NJ, Ilett KF, Minchin RF. Substrate-dependent regulation of human arylamine N-acetyltransferase-1 in cultured cells. Mol Pharmacol. 2000;57(3):468–73. doi: 10.1124/mol.57.3.468. [DOI] [PubMed] [Google Scholar]
- 19.de Leon JH, Vatsis KP, Weber WW. Characterization of naturally occurring and recombinant human N-acetyltransferase variants encoded by NAT1. Mol Pharmacol. 2000;58(2):288–99. doi: 10.1124/mol.58.2.288. [DOI] [PubMed] [Google Scholar]
- 20.Grant DM, Blum M, Beer M, Meyer UA. Monomorphic and polymorphic human arylamine N-acetyltransferases: a comparison of liver isozymes and expressed products of two cloned genes. Mol Pharmacol. 1991;39:184–191. [PubMed] [Google Scholar]
- 21.Tai HL, Fessing MY, Bonten EJ, Yanishevsky Y, d'Azzo A, Krynetski EY, et al. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics. 1999;9:641–650. doi: 10.1097/01213011-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 23.Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, Ross D. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:Quinone oxidoreductase 1. Mol Pharmacol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 24.Butcher NJ, Arulpragasam A, Minchin RF. Proteasomal degradation of N-acetyltransferase 1 is prevented by acetylation of the active site cysteine. J Biol Chem. 2004;279:22131–22137. doi: 10.1074/jbc.M312858200. [DOI] [PubMed] [Google Scholar]
- 25.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevelige P, Jr., Fasman GD. In: Chou-Fasman Prediction of Secondary Structure, in Prediction of Protein Structure and the Principles of Protein Conformation. Fasman GB, editor. Plenum; New York: 1989. (ISBN 0-306-43131-9). [Google Scholar]
- 28.Sinclair JC, Sandy J, Delgoda R, Sim E, Noble ME. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat Struct Biol. 2000;7:560–4. doi: 10.1038/76783. [DOI] [PubMed] [Google Scholar]
- 29.Sandy J, Mushtaq A, Kawamura A, Sinclair J, Sim E, Noble M. The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis--an enzyme which inactivates the anti-tubercular drug, isoniazid. J Mol Biol. 2002;318:1071–83. doi: 10.1016/S0022-2836(02)00141-9. [DOI] [PubMed] [Google Scholar]
- 30.Westwood IM, Holton SJ, Rodrigues-Lima F, Dupret JM, Bhakta S, Noble ME, Sim E. Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase. Biochem J. 2005;385:605–12. doi: 10.1042/BJ20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues-Lima F, Delomenie C, Goodfellow GH, Grant DM, Dupret JM. Homology modelling and structural analysis of human arylamine N-acetyltransferase NAT1: evidence for the conservation of a cysteine protease catalytic domain and an active-site loop. Biochem J. 2001;356:327–34. doi: 10.1042/0264-6021:3560327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues-Lima F, Dupret JM. 3D model of human arylamine N-acetyltransferase 2: structural basis of the slow acetylator phenotype of the R64Q variant and analysis of the active-site loop. Biochem Biophys Res Commun. 2002;291:116–23. doi: 10.1006/bbrc.2002.6414. [DOI] [PubMed] [Google Scholar]
- 33.Sandy J, Mushtaq A, Holton SJ, Schartau P, Noble ME, Sim E. Investigation of the catalytic triad of arylamine N-acetyltransferases: essential residues required for acetyl transfer to arylamines. Biochem J. 2005;390:115–23. doi: 10.1042/BJ20050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Liu L, Hanna PE, Wagner CR. Catalytic mechanism of hamster arylamine N-acetyltransferase 2. Biochemistry. 2005;44:11295–306. doi: 10.1021/bi047564q. [DOI] [PubMed] [Google Scholar]